Isolation and Characterization of Intestinal Escherichia coli Clones from Wild Boars in Germany (original) (raw)
Abstract
Our understanding of the composition of Escherichia coli populations in wild boars is very limited. In order to obtain insight into the E. coli microflora of wild boars, we studied E. coli isolates from the jejunums, ileums, and colons of 21 wild boars hunted in five geographic locations in Germany. Ten isolates per section were subjected to clonal determination using pulsed-field gel electrophoresis. One representative isolate per clone was further investigated for virulence traits, phylogenetic affiliation, and antimicrobial susceptibility. Macrorestriction analysis of 620 isolates revealed a range of clone diversity among the sections and animals, with up to 9 and 16 different clones per section and animal, respectively. Most of the clones for a given animal were shared between two adjacent intestinal sections. The overall highest clonal diversity was observed within the colon. While the astA gene was present in a large number of clones, other virulence genes and hemolytic ability were detected only sporadically. Clones of all four ECOR groups dominated the intestinal sections. Phylogenetic analysis and the occurrence of virulence genes correlated with the isolation frequencies for clones. All E. coli clones from wild boars were susceptible to all antimicrobial agents tested. In conclusion, though several parameters (including an animal-specific and highly diverse E. coli clone composition, the simultaneous occurrence of single clones in two adjacent intestinal sections of a given animal, and a higher E. coli diversity in the large intestine than in the small intestine) of E. coli populations of wild boars were similar to those of previously described E. coli populations of conventionally reared domestic pigs, our data also indicate possible differences, as seen for the E. coli diversity in the large intestine, the occurrence of certain virulence genes and phylogenetic groups, and antimicrobial susceptibilities.
Escherichia coli is a very versatile bacterium, and E. coli strains can be grouped into nonpathogenic (commensal) and pathogenic strains; the latter cause intestinal or extraintestinal diseases (19, 32). Pathogenic E. coli strains which express virulence genes can colonize the gut and cause gastrointestinal disorders as well as intoxications (19, 26). Several E. coli pathotypes, such as Shiga toxin-producing E. coli, are responsible for major economic losses in the pig industry (41). Commensal E. coli strains are by definition members of the gastrointestinal autochthonous flora of most mammalian hosts (14) and are considered to maintain the physiological milieu of the gut, to support digestion, and to provide defense mechanisms against enteric pathogens.
The characterization of intestinal E. coli strains with respect to diversity, virulence trait profiles, phylogenetic affiliations, and antibiotic resistance provides important information about the health status of the harboring host and the risks for development of diseases, transmission of pathogens, or resistance to antibiotic treatment. Data from domestic pigs (Sus scrofa domestica) showed that E. coli populations are highly dynamic and individual (20, 36). In contrast to pathogenic E. coli strains, which typically carry certain virulence gene patterns associated with specific pathotypes (13), commensal E. coli strains rarely contain virulence genes (6). However, in several cases commensal porcine E. coli strains have also been reported to harbor a broad range of virulence-associated genes (35, 37, 40), and it is not known why the strains from clinically healthy domestic pigs possess such virulence trait genes.
Based on the E. coli reference collection (ECOR), E. coli strains can be classified into four main phylogenetic groups, each correlating with certain pathotypes. In studies of intestinal E. coli strains, the majority of commensal E. coli strains were shown to be members of ECOR groups A and B1, whereas pathogenic E. coli strains belonged to ECOR groups A and D. ECOR group B2 strains were in general very rarely found in the porcine intestine (2, 7, 11, 44). In contrast, nonporcine extraintestinal pathogenic E. coli (ExPEC) strains have been shown to be predominantly ECOR group B2 (3, 12, 18). Recently, Dixit and colleagues (11) demonstrated that specific ECOR groups colonized specific intestinal sections of domestic pigs.
The occurrence of antibiotic resistance in E. coli strains in domestic pigs is clearly connected to the use of antibiotics in pig production (22). Due to this application, pathogenic as well as commensal E. coli strains developed resistance. Commensal strains might serve as donors of antibiotic resistance genes to pathogenic E. coli (4, 5). This process of occurrence and transmission of antimicrobial resistance is of importance for general animal health.
In contrast to data about the E. coli microflora from conventionally reared domestic pigs, data for the E. coli microflora from wild boars (Sus scrofa) are not available. The main objectives of this study were to analyze the E. coli microflora of wild boars with respect to their diversity and phylogenetic affiliations, possession of selected virulence genes, and antimicrobial resistance. The data generated will provide basic information about the diversity and composition of E. coli populations within the intestine of the ancestor of our domestic pigs. Additionally, such data will help in our understanding of the evolution of commensal E. coli strains in domestic pigs due to conventional production practices.
MATERIALS AND METHODS
Sampling of digesta specimens from wild boars.
In the fall of 2004, 2005, and 2006, five hunts for wild boars in different regions in Brandenburg and Saxony, Germany, were attended. These regions have not been reported to suffer from recent epidemics in wild boar populations. Additionally, epidemiological surveys of the hunters from that region did not reveal any reports of diseased wild boars. Immediately after killing of the animals, which did not show any suspicious appearance or behavior before being killed, the intestine sections (jejunum, ileum, and colon) were removed as described elsewhere (37), transported on ice, and further processed within 3 h. The ages of the animals were estimated according to their tooth development.
Isolation of E. coli strains from intestinal sections.
Serial dilutions of digesta specimens were plated on MacConkey agar plates (Oxoid, Hampshire, United Kingdom) and incubated for 18 h at 37°C. Ten pink colonies per specimen (each representing a single isolate) were randomly chosen and streaked onto Chromagar orientation plates (Chromagar, Paris, France) and Gassner agar plates (Sifin, Berlin, Germany). They were assumed to be E. coli isolates when the colonies showed a typical pink color on Chromagar orientation and a blue/green color on Gassner agar plates after incubation at 37°C for 24 h. After macrorestriction analysis of the E. coli isolates, each clone was verified as E. coli using standard methods (42). The ability to hemolyse blood was tested after inoculation of the strains on Columbia agar plates (BD Difco, Heidelberg, Germany) containing 5% sheep erythrocytes after culture at 37°C for 24 h.
Ten pink colonies isolated from serial dilutions of individual digesta specimens (except for one animal, where only four E. coli colonies were isolated from the jejunum) were verified as E. coli. According to recent studies only 10 bacterial isolates are required to determine the most common clones in fecal samples, with a 90% chance of detection of a clone which has a frequency of at least 20% in a sample (38). E. coli isolates were assigned to clones based on macrorestriction analysis and preserved for further analysis. Four E. coli isolates initially derived from one wild boar each could not be recovered after storage and had to be removed from our study.
Assignment of individual E. coli clones and their phylogenetic affiliation.
To assign individual E. coli isolates to clones, we used pulsed-field gel electrophoresis (PFGE) as previously described (24). Bacteria were grown in LB medium overnight and adjusted to an optical density at 600 nm of 1.4. Samples (1.5 ml) were centrifuged and washed by resuspension twice in phosphate-buffered saline. The bacterial solutions were embedded in 1.2% pulsed-field-certified agarose (Bio-Rad, Munich, Germany) in TE buffer (10 mM Tris-HCl, 10 mM EDTA, pH 7.5). The solidified agar blocks were incubated for 24 h with proteinase K (Roth, Karlsruhe, Germany) in ESP buffer (500 mM EDTA, 1% sarcosyl, pH 9.5) and washed three times for 1.5 h each with TE buffer. Bacterial DNA was digested with 20 U XbaI at 37°C overnight. The digested blocks were embedded in a 1.2% pulsed-field-certified agarose gel, and DNA fragments were separated for 22 h at 6 V and 50 Hz and examined by ethidium bromide staining. If the XbaI digest failed, NotI was used instead. If both digests failed, we applied randomly amplified polymorphic DNA PCR (RAPD-PCR) for determination of clonal diversity. The primers used for RAPD-PCR were RAPD1 (5′-GGTGCGGGAA-3′), RAPD2 (5′-GTTTCGCTCC-3′), and RAPD4 (5′-AAGAGCCCGT-3′). PCR conditions consisted of denaturation for 5 min at 95°C followed by 45 cycles of 95°C for 1 min, 36°C for 1 min, and 72°C for 2 min and a final extension at 72°C for 10 min. The phylogenetic affiliations of the E. coli clones with respect to the ECOR groups (15) were determined by using a PCR technique described elsewhere (9).
Virulence gene determinations using PCR.
One representative isolate of each E. coli clone was tested for the occurrence of selected virulence genes. The selected virulence genes typical for porcine pathogenic E. coli strains were _stx_2e (coding for Shiga toxin 2e); faeG, fanA, fasA, fedA, and fimF41a (coding for fimbriae F4, F5, F6, F18, and F41, respectively); _est_-Ia and _est_-II (coding for heat-stable enterotoxins I and II, respectively); _eltB_-Ip (coding for heat-labile enterotoxin I), paa (coding for porcine adherence factor); _aida_-I (coding for adhesin involved in diffuse adhesion I), astA (coding for the heat-stable cytotoxin associated with enteroaggregative E. coli); and sepA (gene A coding for secretion of E. coli proteins). PCR was performed as previously described (6; B. T. Bosworth and T. A. Casey, presented at the 97th General Meeting of the American Society for Microbiology, Miami Beach, FL, 4 to 8 May 1997).
Antimicrobial susceptibility testing.
Forty-two E. coli clones from wild boars were tested for susceptibility to the following antimicrobial agents by the microdilution broth method as recommended by the Clinical and Laboratory Standards Institute (10, 27) (breakpoints for resistance are indicated in parentheses): ampicillin (≥32 μg/ml), amoxicillin-clavulanic acid (≥32/16 μg/ml), cefquinome (no breakpoint available), ceftiofur (no breakpoint available), cefalotin (≥32 μg/ml), cefazolin (≥32 μg/ml), chloramphenicol (≥32 μg/ml), enrofloxacin (no breakpoint available), gentamicin (≥16 μg/ml), neomycin (≥32 μg/ml) (25), spectinomycin (no breakpoint available), streptomycin (no breakpoint available), tetracycline (≥16 μg/ml), and trimethoprim-sulfamethoxazole (≥4/76 μg/ml). Although there were no breakpoints available for spectinomycin and streptomycin, for further analysis we defined E. coli as resistant to these antimicrobial agents if the MIC of spectinomycin for the E. coli isolate was ≥128 μg/ml and if that of streptomycin was ≥64 μg/ml. We assumed that these serum concentrations cannot be clinically achieved in pigs. Despite the fact that E. coli is naturally resistant to penicillin and spiramycin, we also included these antibiotics in our study to detect possible differences in the MICs between E. coli isolates from domestic and feral pigs. To perform the tests, commercially acquired microtiter plates (Sensititre; MCS Diagnostics, United Kingdom) were used. The susceptible E. coli population included all E. coli clones which had a MIC lower than the breakpoint or were defined as nonresistant in this study (see above for the description for spectinomycin and streptomycin). Additionally, as the tested E. coli clones from wild boars were susceptible to the tested antimicrobial substances (except for the natural resistance to penicillin and spiramycin), we compared MICs for these E. coli clones to MICs for susceptible E. coli clones isolated from intestinal sections of healthy domestic piglets (36, 37). These domestic piglets and their dams were not treated with antibiotics during and at least 3 months prior to the sampling period. E. coli clones from domestic piglets were therefore defined to be commensal E. coli clones in our study; they were isolated from weaning piglets (average age, 51 days) and comprised clones similar to the dominant E. coli clones from wild boars as well as minor clones. As the prevailing commensal E. coli strains of domestic pigs do carry antimicrobial resistance (up to 97% of all strains of one pig population) (references 1, 16, and 39 and our unpublished data) and thus it was impossible in practice to isolate comparable numbers of E. coli clones susceptible to all antimicrobial substances from a domestic pig population, we decided to use the following procedure. One isolate of each of 49 intestinal E. coli clones from domestic piglets was initially tested for antimicrobial susceptibilities, which was done in parallel to the testing of wild boar E. coli clones. If one E. coli clone was resistant to one antimicrobial substance, then the MIC from this clone for this substance was excluded but MICs from this clone for all other substances were included for further statistical analysis.
Evaluation of PFGE profiles, statistical analysis, and definitions.
Interpretation of PFGE profiles regarding similarity scores was performed using Bionumerics software (Applied Maths, Belgium) with the unweighted-pair group method using average linkages method, and Dice similarity indices (complete linkage; optimization, 1%; position tolerance, 1.3%) (Applied Maths, Belgium) were calculated. For statistical analysis of numerical parameters, nonparametric statistical tests were applied using Statgraphics Centurion XV (Statpoint Inc., VA). The Kruskal-Wallis test was applied for comparison of three or more groups and the Mann-Whitney U test for comparison of two groups. Associations between categorical variables were tested by the application of the chi-square test implemented in Microsoft Office Excel 2003. For all analysis, the significance level of α = 0.05 was applied.
Each E. coli colony as detected after initial screening on MacConkey agar plates was regarded as an individual isolate. A clone was defined as an E. coli group of isolates with a specific macrorestriction enzyme/RAPD-PCR pattern, whereas two clones differed by more than one band. The diversity of the coliform bacteria was measured as explained in detail by Katouli et al. (21) with Simpson's index of diversity (Di) (17, 21). This calculation enables comparison of diversities of populations with different numbers of isolates. A dominant clone was defined as a clone which represented ≥50% of typed isolates in one sample, and a minor clone was defined as a clone which represented ≤10% of typed isolates in one sample (38).
RESULTS
Wild boar sampling.
Twenty-one wild boars, which were killed between 2004 and 2006 in the states of Brandenburg and Saxony, Germany, were the subjects of this study. All boars were clinically healthy according to the postmortem observations, and the contents and the mucosa of their gastrointestinal tracts did not indicate any abnormalities. The ages of the wild boars were estimated to be 7 to 21 months.
Determination of clonal diversity.
Thirty E. coli isolates from each wild boar (10 isolates per intestinal section) were analyzed by macrorestriction and PFGE or RAPD-PCR for determination of clonal diversity. The total number of clones identified from all 21 wild boars was 108. On average, 5.1 ± 3.5 different E. coli clones per wild boar were detected in the digesta specimens, with 2.2 ± 1.5, 2.2 ± 1.4, and 3.0 ± 2.0 clones on average in the jejunum, ileum, and colon, respectively. However, the differences in average clone numbers observed between the different intestinal sections were not significant. The number of total clones per animal varied from 1 (boars 9 and 10) to 16 (boar 7) (Table 1).
TABLE 1.
Numbers of E. coli clones in different intestinal sections from wild boars
| Intestinal section | No. of clones from wild boar no.: | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
| Jejunum | 2 | 1 | 1 | 2 | 2 | 1 | 3 | 2 | 1 | 1 | 6 | 5 | 1 | 4 | 2 | 1 | 3 | 4 | 1 | 1 | 2 |
| Also in ileum only | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| Also in colon only | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Also in ileum and colon | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 |
| Ileum | 1 | 2 | 1 | 3 | 2 | 1 | 4 | 2 | 1 | 1 | 2 | 5 | 4 | 2 | 2 | 2 | 1 | 6 | 2 | 2 | 1 |
| Also in colon | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 3 | 0 | 1 | 2 | 1 | 3 | 1 | 1 | 1 |
| Colon | 2 | 2 | 4 | 4 | 5 | 4 | 9 | 5 | 1 | 1 | 1 | 2 | 6 | 3 | 2 | 3 | 1 | 3 | 1 | 1 | 3 |
| Total | 3 | 2 | 4 | 7 | 8 | 5 | 16 | 6 | 1 | 1 | 7 | 7 | 7 | 9 | 3 | 3 | 4 | 8 | 2 | 3 | 3 |
The diversity index describes the uniformity of distribution of clones in one sample; e.g., if only one clone appears in an intestinal section, the diversity index is zero. Diversity in the jejunum (Di = 0.32 ± 0.32) and in the ileum (Di = 0.34 ± 0.31) was not significantly different from that in the colon (Di = 0.44 ± 0.36). Totals of 51.1% and 36.1% of the clones isolated from the jejunum were also found in the ileum and colon, respectively, while 52% of clones isolated from the ileum were also found in the colon section. Together, 37% of clones isolated from the small intestine were also found in the large intestine (Table 1).
Virulence gene profiles and phylogenetic affiliation.
Since the ability to hemolyse erythrocytes is often linked with virulence in E. coli, the hemolytic ability of the clones was tested on Columbia agar containing sheep erythrocytes. Only one clone isolated from the jejunum of boar 11 showed hemolytic activity. We screened the clones for virulence genes typical for porcine pathogenic E. coli using PCR techniques. Clones were positive for the following genes: _est_-II (1.0% from all clones; one clone in one animal), _eltB_-Ip (2.9%; three clones in two animals), paa (2.9%; three clones in three animals), _aida_-I (4.8%; five clones in five animals), and astA (45.2%; 47 clones in 19 animals) (Table 2; see Fig. S1 in the supplemental material). Clones carrying either _paa, aida_-I, or astA could be found in all three intestinal sections. While clones carrying astA or _aida_-I were able to dominate an intestinal section, clones harboring the _est_-II and _eltB_-Ip genes were always minor clones. Up to seven different clones per animal were virulence gene positive. Different combinations of the virulence genes, i.e., astA plus _eltB_-Ip, astA plus _aida_-I, astA plus _est_-II plus _eltB_-Ip, and _eltB_-Ip plus paa, were observed.
TABLE 2.
Diversity and distribution of virulence genes of E. coli colonies from wild boarsa
| Wild boar no. | Intestinal section | Clone/virulence factor gene profile | No. of isolates |
|---|---|---|---|
| 6b | Jejunum | astA | 10 |
| Ileum | astA | 10 | |
| Colonc | astA | 5 | |
| astA | 2 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 7d | Jejunum | astA | 7 |
| _eltB_-Ip, paa | 1 | ||
| 0 | 1 | ||
| astA | 1e | ||
| Ileum | astA | 1e | |
| 0 | 7 | ||
| astA, est_-II, eltB-Ip_ | 1 | ||
| astA | 1 | ||
| Colon | 0 | 2 | |
| 0 | 1 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 0 | 1 | ||
| 9f | Jejunum | astA | 10e |
| Ileum | astA | 10e | |
| Colon | astA | 10e | |
| 14g | Jejunum | astA | 6 |
| 0 | 1 | ||
| astA | 1 | ||
| astA | 2 | ||
| Ileum | astA | 2 | |
| astA | 8 | ||
| Colon | 0 | 5 | |
| astA | 4 | ||
| astA | 1 | ||
| 17h | Jejunum | astA | 3 |
| _aida_-I | 5 | ||
| 0 | 2 | ||
| Ileum | 0 | 10 | |
| Colon | 0 | 10 | |
| 18i | Jejunum | astA | 2e |
| 0 | 3e | ||
| astA | 5 | ||
| Ileum | astA | 2e | |
| 0 | 2e | ||
| paa | 2 | ||
| 0 | 2 | ||
| astA | 1 | ||
| astA | 1 | ||
| Colon | astA | 1e | |
| paa | 4 | ||
| 0 | 5 |
All E. coli clones were assigned to one of the four ECOR groups using PCR, but two clones were excluded from these studies because their PCR results gave conflicting information. Possible correlations between ECOR groups, intestinal sections, virulence genes, and isolation frequencies were also analyzed. Clones of each of the four ECOR groups were found throughout the intestinal sections. In the jejunum 38.6% of the clones belonged to ECOR group B2, followed by ECOR groups A (31.8%), D (18.2%), and B1 (11.4%). This distribution was similar in the ileum (B2, 35.4%; A, 27.1%; D, 27.1%; and B1, 10.4%). In contrast, the colon section showed a more equal distribution of the ECOR groups (B2, 25.9%; A, 22.4%; D, 25.9%; and B1, 25.9%). Members of a single ECOR group were able to dominate an intestinal section (see Fig. S2 in the supplemental material). In two animals only ECOR group A members and in two animals only ECOR group B2 members were detected. ECOR group B2 members carried fewer virulence genes (35.7% of all clones carried at least one virulence gene) than group B1 (52.2%), group D (54.5%), and group A (75%) members, but such differences were again not significant. Statistically significant differences (P < 0.05) were observed for astA, for which 17.9% of ECOR group B2 members were positive compared to 39.1% of group B1, 54.5% of group D, and 75% of group A members. Other correlations were not considered due to the low abundance of the investigated virulence genes.
The isolation frequency was highest for ECOR group A clones (an average of 7.6 isolates per clone were isolated from one animal), followed by ECOR group B2 clones (6.3 isolates per clone), ECOR group D clones (5.4 isolates per clone), and ECOR group B1 clones (2.9 isolates per clone). Differences in the isolation frequencies between ECOR groups A and B1 were significant (P < 0.05). Details for each ECOR group and for the virulence gene astA are shown in Table 3. Irrespective of the phylogenetic affiliation, the average number of isolates per animal presenting a virulence gene-harboring clone was significantly higher (mean of 7.1 ± 8.0 isolates from one clone per animal; median, 5; minimum, 1; maximum, 30) than the number of isolates per clone without virulence genes (mean, 4.8 ± 6.2; median, 2; minimum, 1; maximum, 23) (P < 0.05).
TABLE 3.
Correlations between ECOR groups and virulence genes and frequency of isolation of clones
| ECOR group and virulence gene status (no. of clones)a | Isolation frequencyb | ||||
|---|---|---|---|---|---|
| Mean | SD | Median | Minimum | Maximum | |
| A | |||||
| + (21) | 9.1 | 10.0 | 5 | 1 | 30 |
| − (7) | 3.0 | 3.0 | 2 | 1 | 9 |
| astA (21)c | 9.1 | 10.0 | 5 | 1 | 30 |
| B1 | |||||
| + (12) | 4.2 | 3.4 | 2.5 | 1 | 10 |
| − (11) | 1.5 | 1.0 | 1 | 1 | 4 |
| astA (9) | 4.2 | 3.9 | 2 | 1 | 10 |
| B2 | |||||
| + (10) | 8.3 | 8.1 | 5.5 | 1 | 28 |
| − (19) | 6.6 | 7.7 | 2 | 1 | 21 |
| astA (5) | 12.2 | 10.1 | 12 | 2 | 28 |
| D | |||||
| + (12) | 5.0 | 6.3 | 2 | 1 | 23 |
| − (10) | 6.4 | 6.9 | 2 | 1 | 23 |
| astA (12)c | 4.7 | 6.1 | 2 | 1 | 23 |
| Total | |||||
| + (55) | 7.0 | 7.9 | 5 | 1 | 30 |
| − (47) | 4.7 | 6.1 | 2 | 1 | 23 |
| astA (47) | 7.4 | 8.5 | 4 | 1 | 30 |
Antimicrobial susceptibilities of E. coli clones from wild boars and comparison to susceptible E. coli clones from clinically healthy domestic piglets.
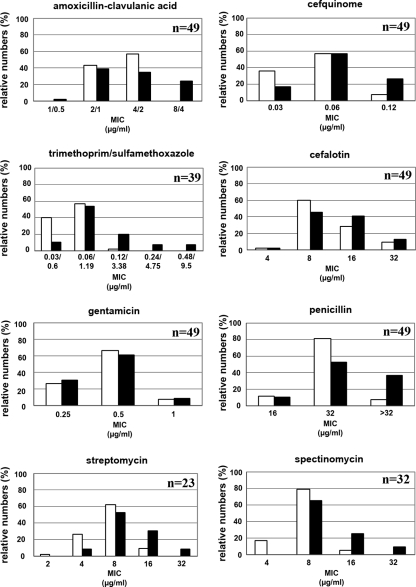
All E. coli clones from wild boars were susceptible to the tested antimicrobials. We further compared the susceptibilities of representative wild boar E. coli clones (n = 42) and susceptible E. coli clones from healthy domestic piglets (n = 49). The isolation strategies used for E. coli from both groups were similar (37), and susceptibility was tested in parallel. E. coli clones from wild boars were more sensitive than the susceptible E. coli population from domestic piglets for amoxicillin-clavulanic acid (P < 0.005), cefquinome (P < 0.05), penicillin (P < 0.005), spectinomycin (P < 0.0005), streptomycin (P < 0.05), and trimethoprim-sulfamethoxazole (P < 0.001), and susceptibilities tended to be higher for spiramycin (P = 0.061) and enrofloxacin (P = 0.077). Susceptibilities were not different for ampicillin (P = 0.518), cefalothin (P = 0.559), cefazolin (P = 0.152), ceftiofur (P = 0.255), chloramphenicol (P = 0.932), neomycin (P = 0.944), tetracycline (P = 0.693), and gentamicin (P = 0.865) (Fig. 1).
FIG. 1.
Antimicrobial susceptibilities of E. coli clones from wild boars and from clinically healthy piglets. Shown are the antimicrobial susceptibilities of E. coli clones from the susceptible E. coli population. Note that relative numbers (percentage of the susceptible population) are shown. This was chosen to avoid false representation using absolute numbers. Absolute numbers of E. coli clones from wild boars (always n = 42) would be in general higher, since data from resistant E. coli clones from domestic pigs were excluded. Means of susceptibility of E. coli clones from wild boars were always higher than means of susceptibility of susceptible E. coli clones from clinically healthy domestic piglets, with the exception of chloramphenicol, gentamicin, and tetracycline. Representative data for eight antibiotics are shown. Open bars, E. coli clones from wild boars; black bars, E. coli clones from domestic piglets; n, absolute numbers of susceptible E. coli clones from domestic pigs.
PFGE patterns of wild-boar-specific E. coli clones from this study and clones isolated from clinically healthy domestic pigs (100 clones from other studies [36, 37]) were compared. However, this analysis did not yield separate clusters for each animal group (see Fig. S3 in the supplemental material).
DISCUSSION
The first domestication of pigs started in the Near East about 9,000 years ago, while recent data suggest several centers of domestication of pigs later on. Thus, the ancestor of the European domestic pig population is the European wild boar, and interestingly, central Europe (especially Germany) was a center of pig domestication (23). Therefore, knowledge about the commensal intestinal bacterial flora of wild boars might be important for comparison with data generated from domestic pigs for two reasons: to monitor the processes of adaptation of intestinal bacteria to the host and its environment and to monitor the flow of bacterial strains between livestock production systems and the environment.
Parameters such as diversity, virulence gene profiles, phylogenetic analysis, and antimicrobial resistance provide important information about individual intestinal E. coli populations and possible correlations to health, diseases, and disease treatment strategies. In contrast to data from the intestinal E. coli microflora of conventionally reared domestic pigs, data from E. coli strains of wild boars are currently not available. In this study, we therefore characterized the intestinal E. coli microflora of three different intestinal sections from 21 wild boars. Although these data reflect only a limited number of wild boars, we compared these data with published data from conventionally reared, clinically healthy domestic pigs.
The overall E. coli microflora from wild boars was clearly individual and diverse. A broad variety of E. coli clones were able to simultaneously colonize a single animal, while many of these clones could be found in at least two different intestinal sections. The E. coli diversity in the large intestine was on average higher than that in the small intestine, as also seen in domestic pigs (37). However, the diversity of E. coli clones from colonic samples was lower in samples from wild boars than in colonic or fecal samples from domestic pigs. This observation was even more prominent when the wild boars of this study which were estimated to be older than 7 months were compared to adult pigs (Table 4), since E. coli diversity in adult swine seems to be higher than diversity in young piglets (20, 36). In this context it should be mentioned that two wild boars harbored only one dominant clone in all three intestinal sections investigated, a phenomenon which so far has not been reported for domestic pigs. A reason for the higher E. coli diversity in conventionally reared domestic pigs could be the fact that in conventional pig production large numbers of animals are housed together at high density, which favors the constant exchange of intestinal bacteria within individuals of the population. This process seems to be more likely than a possible reduction of the diversity as a result of the extremely controlled environment of modern pig production, with shielded long-range animal housing and clearly defined food and drug management leading to a tremendous restriction of the E. coli exchange from outside and permanent selection pressure due to constant feeding and medication.
TABLE 4.
Comparison of the diversity of E. coli populations between wild boars and conventionally reared domestic pigs
| Referencea | Pigs | Samples | Mean Di ± SD | |||
|---|---|---|---|---|---|---|
| Type | Age | n | Site | n | ||
| This study | Wild | 7-21 mo_b_ | 21 | Colon | 21 | 0.44 ± 0.35 |
| 20 | Domestic | 0.5-21 wk | 16 | Rectum | 180 | 0.86 ± 0.05 |
| Domestic | Sowsb | 4 | Rectum | 16 | 0.83 ± 0.08 | |
| 21 | Domestic | 1-9 wk | 10 | Rectum | 100 | 0.84 ± 0.14 |
| 37 | Domestic | 8 wk | 15 | Colon | 15 | 0.61 ± 0.25 |
| 36 | Domestic | 1-8 wk | 5 | Rectum | 40 | 0.44 ± 0.15 |
| Domestic | Sowb | 1 | Rectum | 4 | 0.92 ± 0.02 |
By screening E. coli clones for virulence genes, we found clones positive for _est_-II, _eltB_-Ip, _paa, aida_-I, and astA, which was the most common virulence trait. Of these clones, several E. coli clones harbored different virulence gene patterns, but none resembled a typical porcine pathogenic E. coli (13). Thus, even considering our small number of animals, virulence gene-positive E. coli clones seem to be common in the wild boar population. If one clone, independent of the phylogenetic origin, was virulence gene positive, then this clone was detected in more isolates than virulence gene-negative clones, indicating the possible role of virulence genes in colonization of the intestine or selection. Such a possible role of virulence genes in the intestinal colonization would also explain the occurrence of high numbers of virulence gene-positive E. coli clones in the intestines of clinically healthy, conventionally reared pigs and was previously also shown for ExPEC-typical virulence-associated genes of porcine E. coli clones (35-37, 40).
The occurrence of the astA gene in 45.2% of the clones from wild boars is substantially higher than that reported in different studies for E. coli isolates from diarrheic as well as clinical healthy domestic pigs (2, 6, 7, 8, 44). Enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1), which is encoded by the gene astA, was first described for enteroaggregative E. coli, an organism associated with persistent diarrhea in children. This gene was later found to be present in enterohemorrhagic E. coli, enterotoxigenic E. coli, enteropathogenic E. coli, and commensal E. coli (33, 34). However, the importance of the broad distribution of astA in E. coli clones from wild boars requires further clarification.
It has been proposed that the phylogenetic affiliation indicates a specific type/pathotype of E. coli (3, 12, 18, 43). In wild boars, isolates of each of the four ECOR groups were found in all intestinal sections. ECOR group B2 clones were most frequently isolated, followed by ECOR group A, D, and B1 clones. This is in contrast to other observations which indicated that ECOR group B2 members are very rare in the intestines of domestic pigs and predominantly comprise ExPEC (2, 7, 11, 43). Members of each ECOR group were able to dominate an intestinal section, and members of ECOR groups B2 and A were able to colonize as exclusive ECOR groups in single wild boars. Thus, we could not observe a significant correlation between single ECOR groups and specific intestinal sections as described for domestic pigs (11). We also calculated correlations between ECOR groups and the occurrence of virulence genes. The prevalence of the gene astA was lower in ECOR group B2 members than in other ECOR groups. Comparable data of ECOR group B2 members from domestic pigs are not available due to the low abundance of members of this group in other studies. However, recent studies of ExPEC showed as well that the prevalence of the gene astA was lower in ECOR group B2 members than in other ECOR groups (12). Isolates belonging to ECOR group A clones were significantly more frequent than isolates belonging to ECOR group B1 clones, indicating that the phylogenetic origin might play a role in the colonization of the intestine.
As expected, all clones from wild boars were susceptible to many antimicrobial substances. However, the comparison of the MICs for susceptible E. coli clones from both wild boars and domestic pigs revealed that wild boars carried E. coli clones which had significant lower MICs than susceptible E. coli clones from domestic pigs. The higher MICs of a variety of structurally unrelated antibiotics for E. coli clones from domestic pigs might suggest that in addition to the common occurrence of resistance genes in porcine E. coli strains (6, 16, 39), other general protective mechanisms have been selected under conditions related to the conventional pig production that seem to help E. coli resist antimicrobials. Such mechanisms include changes in the permeability of the outer membrane to antibiotics, specific drug transporters, and multidrug transporters (29-31). Some of the efflux pumps exhibit an extremely wide specificity covering practically all antibiotics, chemotherapeutic agents, detergents, dyes, and other inhibitors (28).
In conclusion, the E. coli microflora of wild boars is individual and diverse, and virulence genes are present. E. coli clones from wild boars obviously differ in several parameters (diversity, virulence genes, phylogenetic affiliation, and susceptibility to antimicrobial agents) from E. coli clones from clinically healthy conventional reared pigs, indicating that conventional animal production seems to affect porcine intestinal E. coli populations. However, these initial data from a limited number of wild boars warrant a larger study and in-depth analysis to further understand the nature of E. coli in domestic and wild pigs.
Supplementary Material
[Supplemental material]
Acknowledgments
This work was supported by grant FOR 438/1-1 from the Deutsche Forschungsgemeinschaft.
We thank Eberhard Kliemank (Wittichenau, Germany) and Stefan Rescher (Oberförsterei Drebkau, Revier Casel, Germany) for helping with sampling digesta specimens from wild boars. We thank John Turner (Berlin, Germany) for carefully reading the manuscript.
Footnotes
▿
Published ahead of print on 5 December 2008.
REFERENCES
- 1.Alali, W. Q., H. M. Scott, R. B. Harvey, B. Norby, D. B. Lawhorn, and S. D. Pillai. 2008. Longitudinal study of antimicrobial resistance among Escherichia coli isolates from integrated multisite cohorts of humans and swine. Appl. Environ. Microbiol. 74**:**3672-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, S., A. Tscholshiew, C. Menge, R. Weiss, G. Baljer, and R. Bauerfeind. 2007. Virulence and fitness gene patterns of Shiga toxin-encoding Escherichia coli isolated from pigs with edema disease or diarrhea in Germany. Berl. Munch. Tierarztl. Wochenschr. 120**:**307-316. [PubMed] [Google Scholar]
- 3.Bingen-Bidois, M., O. Clermont, S. Bonacorsi, M. Terki, N. Brahimi, C. Loukil, D. Barraud, and E. Bingen. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70**:**3216-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake, D. P., K. Hillman, D. R. Fenlon, and J. C. Low. 2003. Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. J. Appl. Microbiol. 95**:**428-436. [DOI] [PubMed] [Google Scholar]
- 5.Blake, D. P., R. W. Humphry, K. P. Scott, K. Hillman, D. R. Fenlon, and J. C. Low. 2003. Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J. Appl. Microbiol. 94**:**1087-1097. [DOI] [PubMed] [Google Scholar]
- 6.Boerlin, P., R. Travis, C. L. Gyles, R. Reid-Smith, N. Janecko, H. Lim, V. Nicholson, S. A. McEwen, R. Friendship, and M. Archambault. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71**:**6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, T. A., X. Y. Wu, I. Barchia, K. A. Bettelheim, S. Driesen, D. Trott, M. Wilson, and J. J. Chin. 2006. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72**:**4782-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, C., W. Cho, H. Chung, T. Jung, J. Kim, and C. Chae. 2001. Prevalence of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene in isolates in weaned pigs with diarrhea and/or edema disease. Vet. Microbiol. 81**:**65-71. [DOI] [PubMed] [Google Scholar]
- 9.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66**:**4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 3rd ed. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Dixit, S. M., D. M. Gordon, X. Y. Wu, T. Chapman, K. Kailasapathy, and J. J. Chin. 2004. Diversity analysis of commensal porcine _Escherichia coli_—associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology 150**:**1735-1740. [DOI] [PubMed] [Google Scholar]
- 12.Ewers, C., G. Li, H. Wilking, S. Kiessling, K. Alt, E. M. Antao, C. Laturnus, I. Diehl, S. Glodde, T. Homeier, U. Bohnke, H. Steinruck, H. C. Philipp, and L. H. Wieler. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297**:**163-176. [DOI] [PubMed] [Google Scholar]
- 13.Frydendahl, K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85**:**169-182. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, D. M., and J. Lee. 1999. The genetic structure of enteric bacteria from Australian mammals. Microbiology 145**:**2673-2682. [DOI] [PubMed] [Google Scholar]
- 15.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172**:**6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinton, M., D. J. Hampson, E. Hampson, and A. H. Linton. 1985. A comparison of the ecology of Escherichia coli in the intestine of healthy unweaned pigs and pigs after weaning. J. Appl. Bacteriol. 58**:**471-477. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26**:**2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183**:**78-88. [DOI] [PubMed] [Google Scholar]
- 19.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2**:**123-140. [DOI] [PubMed] [Google Scholar]
- 20.Katouli, M., A. Lund, P. Wallgren, I. Kuhn, O. Soderlind, and R. Mollby. 1995. Phenotypic characterization of intestinal Escherichia coli of pigs during suckling, postweaning, and fattening periods. Appl. Environ. Microbiol. 61**:**778-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katouli, M., L. Melin, M. Jensen-Waern, P. Wallgren, and R. Mollby. 1999. The effect of zinc oxide supplementation on the stability of the intestinal flora with special reference to composition of coliforms in weaned pigs. J. Appl. Microbiol. 87**:**564-573. [DOI] [PubMed] [Google Scholar]
- 22.Kim, L. M., J. T. Gray, B. G. Harmon, R. D. Jones, and P. J. Fedorka-Cray. 2005. Susceptibility of Escherichia coli from growing piglets receiving antimicrobial feed additives. Foodborne Pathog. Dis. 2**:**304-316. [DOI] [PubMed] [Google Scholar]
- 23.Larson, G., K. Dobney, U. Albarella, M. Fang, E. Matisoo-Smith, J. Robins, S. Lowden, H. Finlayson, T. Brand, E. Willerslev, P. Rowley-Conwy, L. Andersson, and A. Cooper. 2005. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307**:**1618-1621. [DOI] [PubMed] [Google Scholar]
- 24.Laturnus, C., J. Jores, I. Moser, P. Schwerk, and L. H. Wieler. 2005. Long-term clonal lineages within Campylobacter jejuni O:2 strains from different geographical regions and hosts. Int. J. Med. Microbiol. 294**:**521-524. [DOI] [PubMed] [Google Scholar]
- 25.Luhofer, G., A. Bottner, H. M. Hafez, M. Kaske, C. Kehrenberg, M. Kietzmann, D. Klarmann, G. Klein, P. Krabisch, T. Kuhn, A. Richter, C. Sigge, W. Traeder, K. H. Waldmann, J. Wallmann, C. Werckenthin, and S. Schwarz. 2004. Proposals of the working group “Antibiotic resistance” for the configuration of microtitre plates to be used in routine antimicrobial susceptibility testing of bacterial pathogens from infections of large food-producing animals and mastitis cases. Berl. Munch. Tierarztl. Wochenschr. 117**:**245-251. [PubMed] [Google Scholar]
- 26.Nagy, B., and P. Z. Fekete. 2005. Enterotoxigenic Escherichia coli in veterinary medicine. Int. J. Med. Microbiol. 295**:**443-454. [DOI] [PubMed] [Google Scholar]
- 27.NCCLS. 2004. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard. Informational supplement (May 2004). NCCLS document M31-S1. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 28.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1**:**516-523. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido, H. 1989. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 33**:**1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishino, K., Y. Inazumi, and A. Yamaguchi. 2003. Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 185**:**2667-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64**:**672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181**:**1753-1754. [DOI] [PubMed] [Google Scholar]
- 33.Savarino, S. J., A. Fasano, J. Watson, B. M. Martin, M. M. Levine, S. Guandalini, and P. Guerry. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc. Natl. Acad. Sci. USA 90**:**3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savarino, S. J., A. McVeigh, J. Watson, A. Cravioto, J. Molina, P. Echeverria, M. K. Bhan, M. M. Levine, and A. Fasano. 1996. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 173**:**1019-1022. [DOI] [PubMed] [Google Scholar]
- 35.Schierack, P., H. Steinruck, S. Kleta, and W. Vahjen. 2006. Virulence factor gene profiles of Escherichia coli isolates from clinically healthy pigs. Appl. Environ. Microbiol. 72**:**6680-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schierack, P., N. Walk, C. Ewers, H. Wilking, H. Steinruck, M. Filter, and L. H. Wieler. 2008. ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ. Microbiol. 10**:**1742-1751. [DOI] [PubMed] [Google Scholar]
- 37.Schierack, P., N. Walk, K. Reiter, K. D. Weyrauch, and L. H. Wieler. 2007. Composition of intestinal Enterobacteriaceae populations of healthy domestic pigs. Microbiology 153**:**3830-3837. [DOI] [PubMed] [Google Scholar]
- 38.Schlager, T. A., J. O. Hendley, A. L. Bell, and T. S. Whittam. 2002. Clonal diversity of Escherichia coli colonizing stools and urinary tracts of young girls. Infect. Immun. 70**:**1225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunde, M., K. Fossum, A. Solberg, and H. Sorum. 1998. Antibiotic resistance in Escherichia coli of the normal intestinal flora of swine. Microb. Drug Resist. 4**:**289-299. [DOI] [PubMed] [Google Scholar]
- 40.Taras, D., W. Vahjen, M. Macha, and O. Simon. 2006. Performance, diarrhea incidence, and occurrence of Escherichia coli virulence genes during long-term administration of a probiotic Enterococcus faecium strain to sows and piglets. J. Anim. Sci. 84**:**608-617. [DOI] [PubMed] [Google Scholar]
- 41.Wieler, L. H., A. Ilieff, W. Herbst, C. Bauer, E. Vieler, R. Bauerfeind, K. Failing, H. Klos, D. Wengert, G. Baljer, and H. Zahner. 2001. Prevalence of enteropathogens in suckling and weaned piglets with diarrhoea in southern Germany. J. Vet. Med. B 48**:**151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkle, S. 1979. Mikrobiologische und serologische Diagnostik, 3rd ed. VEB Gustav Fischer Verlag, Jena, Germany.
- 43.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60**:**1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, X. Y., T. Chapman, D. J. Trott, K. Bettelheim, T. N. Do, S. Driesen, M. J. Walker, and J. Chin. 2007. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl. Environ. Microbiol. 73**:**83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]