Deficient mismatch repair system in patients with sporadic advanced colorectal cancer (original) (raw)
Abstract
A deficient mismatch repair system (dMMR) is present in 10–20% of patients with sporadic colorectal cancer (CRC) and is associated with a favourable prognosis in early stage disease. Data on patients with advanced disease are scarce. Our aim was to investigate the incidence and outcome of sporadic dMMR in advanced CRC. Data were collected from a phase III study in 820 advanced CRC patients. Expression of mismatch repair proteins was examined by immunohistochemistry. In addition microsatellite instability analysis was performed and the methylation status of the MLH1 promoter was assessed. We then correlated MMR status to clinical outcome. Deficient mismatch repair was found in only 18 (3.5%) out of 515 evaluable patients, of which 13 were caused by hypermethylation of the MLH1 promoter. The median overall survival in proficient MMR (pMMR), dMMR caused by hypermethylation of the MLH1 promoter and total dMMR was 17.9 months (95% confidence interval 16.2–18.8), 7.4 months (95% CI 3.7–16.9) and 10.2 months (95% CI 5.9–19.8), respectively. The disease control rate in pMMR and dMMR patients was 83% (95% CI 79–86%) and 56% (30–80%), respectively. We conclude that dMMR is rare in patients with sporadic advanced CRC. This supports the hypothesis that dMMR tumours have a reduced metastatic potential, as is observed in dMMR patients with early stage disease. The low incidence of dMMR does not allow drawing meaningful conclusions about the outcome of treatment in these patients.
Keywords: mismatch repair, advanced colorectal cancer, chemotherapy, incidence, prognosis
In recent years the median overall survival (OS) for patients with advanced colorectal cancer (CRC) has significantly improved, mainly because of the availability of cytotoxic agents such as oxaliplatin and irinotecan, and the targeted drug bevacizumab (Punt, 2004). Despite these improvements cure is only rarely achieved, and not all patients respond to chemotherapy. Therefore, there is a need for predictive and prognostic tests that identify patients who may or may not benefit from systemic treatment.
Deficient mismatch repair (dMMR) is one of the genetic pathways that is involved in the development of CRC (Aaltonen et al, 1993; Ionov et al, 1993). Microsatellites are repetitive units in DNA. In normal circumstances insertions or deletions in these regions are repaired by the MMR system, which consists of several cooperating MMR proteins. Dysfunction of this system is causing dMMR. Known MMR gene products are: MLH1, MSH2, MSH6 and PMS2. A germ line mutation in one of these corresponding genes, most often MLH1 or MSH2, is the cause of dMMR in patients with Lynch syndrome, formerly Hereditary Non Polyposis Colorectal Cancer (HNPCC), which comprises 0.8–5% of all CRCs (Mecklin, 1987; Rodriguez-Bigas et al, 1997; Cunningham et al, 2001). Deficient mismatch repair is also observed in 10–20% of patients with sporadic CRC, usually caused by MLH1 promoter hypermethylation (Lothe et al, 1993; Kane et al, 1997; Cunningham et al, 1998; Peltomaki, 2003). Microsatellite analysis is the gold standard for the detection of dMMR in patients with a suspicion of Lynch syndrome, as well as in tumours with indecisive results of IHC. Lindor et al (2002) showed that immunohistochemistry (IHC) in colorectal tumours for MLH1 and MSH2 provides a rapid, cost-effective, sensitive (92.3%) and highly specific (100%) method for screening for DNA MMR defects, which was recently confirmed by our group (Overbeek et al, 2008).
Colorectal carcinomas with dMMR show several distinct pathological features, such as a location in the proximal colon, a poor histological differentiation, high numbers of tumour infiltrating lymphocytes, and are more often of mucinous type. Moreover patients with dMMR tumours have a better prognosis compared to patients with proficient MMR (pMMR) tumours (Sankila et al, 1996; Gryfe et al, 2000; Samowitz et al, 2001; Ward et al, 2001; Popat et al, 2005). Several studies investigated if chemosensitivity is implied in the better prognosis of patients with a dMMR tumour. In vitro studies have shown dMMR cell lines to be resistant to 5-fluorouracil (5FU) (Carethers et al, 1999; Arnold et al, 2003), but not to oxaliplatin (Fink et al, 1996; Sergent et al, 2002) or irinotecan (Magrini et al, 2002). In patients receiving adjuvant chemotherapy for CRC, conflicting results have been reported on the correlation between dMMR and outcome in retrospective studies (Elsaleh and Iacopetta, 2001; Ribic et al, 2003; Carethers et al, 2004; Benatti et al, 2005). Therefore, the ASCO 2006 and European 2007 guidelines do not recommend the use of dMMR as a prognostic and/or predictive marker in this setting (Locker et al, 2006; Duffy et al, 2007). Only few data from small, non-randomised studies are available on the function of dMMR in patients with advanced CRC (Popat et al, 2005). In most of these studies sporadic and hereditary CRC were not clearly differentiated. Histopathological and clinical differences between tumours with a mutation of a MMR gene and tumours with hypermethylation of the MLH1 promoter as a cause of dMMR have been described (Benatti et al, 2005; Jass, 2007), and therefore it seems reasonable to distinguish between these two types of dMMR tumours regarding response to therapy and survival (Poynter et al, 2008).
This is the first randomised study with chemotherapy in patients with advanced CRC which evaluates the incidence of dMMR and its correlation with clinical outcome.
Materials and methods
Study population
Tumour tissue was obtained from patients enrolled in a randomised phase III study, the CAIRO study of the Dutch Colorectal Cancer Group (DCCG), registered with ClinicalTrials.gov with the number NCT00312000, of which the results on survival have been published (Koopman et al, 2007). In this study 820 patients were randomised between first-line capecitabine, second-line irinotecan and third-line capecitabine+oxaliplatin (sequential treatment arm) vs first-line capecitabine+irinotecan, and second-line capecitabine+oxaliplatin (combination treatment arm). The primary endpoint was OS, and secondary endpoints included response rate, and progression-free survival (PFS). Assessment of tumour response was scheduled every three cycles (9 weeks) according to RECIST criteria (Therasse et al, 2000). Follow-up after completion of treatment was carried out every 3 months until death. For this study formalin-fixed paraffin-embedded material of the primary tumour and normal tissue was obtained from 515 patients out of 803 eligible patients of the CAIRO study. No tissue material was obtained in 288 patients because a resection of the primary tumour was not performed, or there was insufficient or non-available material.
Histology and pathology
Histopathologic evaluation was carried out by two independent observers, and in case of discordant results, the opinion of a third observer (pathologist: JvK) was final. Histopathological subtype and grade of differentiation were determined according to WHO criteria (Jass et al, 1990). Tumours were classified as right-sided (proximal of the splenic flexure), left-sided (distal of the splenic flexure) or rectal.
Immunohistochemistry
IHC was performed on formalin-fixed paraffin-embedded tissue. Of each paraffin-embedded block a 2 mm punch for assembling tissue microarrays (TMA's) was accomplished as previously described (Hendriks et al, 2003). Four 4 μm slides were assessed of every TMA and mounted on glass. The TMA slides were deparaffined and afterwards the tissues were rehydrated using xylene and ethanol. Water and phosphate-buffered saline (PBS) were used for washing of the slides. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in PBS for 30 min and slides were washed with water, after which heat-induced epitope retrieval was performed. The slides were stained with antibodies against MLH1 (clone G168-15; BD Biosciences, San Jose, CA, USA), MSH2 (clone GB12; Calbiochem, Darmstadt, Germany), MSH6 (clone 44; BD Biosciences) and PMS2 (clone A16-4; BD Biosciences). The scoring was performed by two blinded observers, and if the slide scoring was not unambiguous, the opinion of a third observer (pathologist: JvK) was final. Staining pattern of the MMR proteins was evaluated by using the normal epithelial, stromal and inflammatory cells as internal control. Protein expression was scored positive if at least one cancer cell nucleus showed staining, negative if none of the tumour cells showed staining with positive internal control, and not applicable if neither tumour nor stromal cells showed protein expression. In case of absence of MMR protein or not interpretable results of IHC, the IHC stainings on TMA were repeated on whole tissue slides for final scoring.
Microsatellite instability analysis
Microsatellite instability (MSI) analysis was performed for tumours of which the final IHC staining was not interpretable or with a negative staining for at least one of four mismatch repair proteins. In addition a random sample of 54 tumours was taken from the pMMR tumours. DNA was extracted from formalin-fixed paraffin-embedded tissues of tumour mucosa and corresponding normal mucosa by a standard procedure. Areas containing >50% tumour cells were selected by microscopic evaluation on a reference slide stained with H&E. Slides (50 μm thick) were made, and if necessary tumour cells were prepared using a scalpel. MSI status was determined by PCR and GeneScan analysis using two microsatellite markers (BAT 25 and BAT 26). If only one of these markers showed instability, the analysis was extended with four additional markers (BAT 40, D2S123, D5S346, D17S250) (Boland et al, 1998; Samowitz et al, 1999). A tumour was defined dMMR if at least two of the six markers showed instability, or pMMR if none of the markers showed any shift in mobility. MSI-low tumours with only one of the markers showing instability were included in the pMMR category. For the distinction between dMMR tumours caused by hypermethylation of the MLH1 promoter and mutation of one of the mismatch repair genes, dMMR tumours were further analysed for hypermethylation of the MLH1 promoter (Bettstetter et al, 2007).
Hypermethylation of MLH1 promoter
The DNA methylation status of the MLH1 promoter region was determined after bisulphite treatment of the DNA using the EZ DNA methylation KIT, ZYMO Research (Orange, CA, USA), as described before (Overbeek et al, 2007).
Statistical analysis
Patients were divided into three groups: pMMR tumours, dMMR tumours caused by hypermethylation of the MLH1 promoter and dMMR tumours without hypermethylation of the MLH1 promoter. Survival analysis was performed for patients with pMMR tumours vs dMMR tumours caused by hypermethylation of the MLH1 promoter and vs the total group of patients with a dMMR tumour, respectively. The association between dMMR and patient or tumour characteristics was investigated with an univariate logistic regression model. Patients were considered evaluable for response if they had completed at least three cycles of chemotherapy. Disease control was defined by stable disease with a duration of ⩾4 months or partial response or complete response. Differences in response and disease control rates were analysed by a _χ_2 (univariate) model. The PFS was calculated for first-line treatment, from the date of randomization to the first observation of disease progression or death from any cause reported after first-line treatment. OS and PFS curves were estimated using the Kaplan–Meier method and compared using a Cox proportional hazard model. All tests were two-sided and _P_-values of less than 0.05 were considered significant. All data received before May 2008 are included in this report with a median follow-up of 43 months. All analyses were performed using SAS 9.1 and S-plus 6.2 software.
Results
IHC, MMR and hypermethylation of the MLH1 promoter
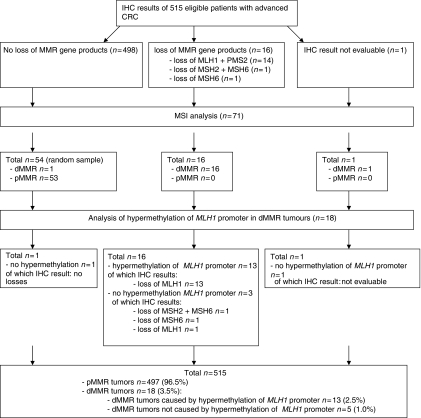
Figure 1 shows the results on IHC, MMR and hypermethylation of the MLH1 promoter. Samples of 515 eligible patients were available for IHC. In 498 tumours no loss for MMR gene products was observed, 14 tumours showed loss of MLH1 in combination with PMS2, 2 tumours showed loss of MSH6 of which one in combination with MSH2, and in one tumour the IHC staining was not evaluable. All these 17 tumours with loss/not evaluable IHC result of at least one MMR protein turned out to be dMMR by MSI analysis. In 54 tumours without loss of MMR gene products (random sample), dMMR was detected in 1 tumour by MSI analysis, resulting in a total of 18 dMMR tumours (3.5%). Hypermethylation of the MLH1 promoter was found in 13 out of these 18 tumours, all with protein loss of MLH1 by IHC. This resulted in 515 patients for the analysis: 18 with a dMMR tumour (3.5%), of which 13 patients with a dMMR tumour caused by hypermethylation of the MLH1 promoter, and 497 patients with a pMMR tumour.
Figure 1.
IHC results, MSI analysis and hypermethylation MLH1 promoter.
Patient characteristics and MMR status
Patient characteristics of the three groups of patients are presented in Table 1. The median age of the included patients was 63 years (range 31–81). Significant differences between the group of patients with dMMR caused by hypermethylation of the MLH1 promoter and the pMMR group were seen for the location (P<0.0001) and differentiation grade of the primary tumour (_P_=0.025). Patients with a dMMR tumour not caused by MLH1 promoter hypermethylation were younger than patients with a dMMR tumour caused by MLH1 promoter hypermethylation (_P_=0.0051). The univariate analysis showed an association of poorly differentiated features with an increased probability of exhibiting dMMR, patients with a poor/undifferentiated tumour have a 3.9 times increased risk of exhibiting dMMR compared to well/moderate differentiated tumours (_P_=0.025).
Table 1. Patient characteristics according to MMR status.
| Number of eligible patients | Group 1 pMMR (_n_=497) | Group 2 dMMR MLH1 hypermethylation (_n_=13) | Group 3 dMMR no MLH1 hypermethylation (_n_=5) | _P_-value# (1 vs 2) | _P_-value# (2 vs 3) |
|---|---|---|---|---|---|
| Age | |||||
| ⩽50 years | 51 (10%) | 0 (0%) | 2 (40%) | ||
| Median (range) | 63 (31–81) | 70 (54–78) | 57 (35–64) | 0.053 | 0.0051 |
| Gender | |||||
| Male | 315 (63%) | 7 (54%) | 4 (80%) | 0.49 | 0.29 |
| Location of the primary tumour | |||||
| Colon—left | 164 (33%) | — | — | <0.0001 | — |
| Colon—right | 122 (25%) | 13 (100%) | 5 (100%) | ||
| Rectosigmoid | 30 (6%) | — | — | ||
| Rectum | 161 (32%) | — | — | ||
| Unknown | 20 (4%) | — | — | ||
| Histology of primary tumour | |||||
| Adenocarcinoma | 438 (88%) | 10 (77%) | 2 (40%) | 0.10 | 0.14 |
| Mucinous adenocarcinoma | 38 (8%) | 3 (23%) | 3 (60%) | ||
| Adenosquamous carcinoma | 4 (<1%) | — | — | ||
| Undifferentiated carcinoma | 3 (<1%) | — | — | ||
| Unknown | 14 (3%) | — | — | ||
| Differentiation grade | |||||
| Well/moderate | 259 (52%) | 3 (23%) | 2 (40%) | 0.025 | 0.48 |
| Poor/undifferentiated | 222 (45%) | 10 (77%) | 3 (60%) | ||
| Unknown | 16 (3%) | — | — | ||
| Diagnosis of metastases | |||||
| <12 months before randomization | 292 (59%) | 8 (62%) | 1 (20%) | 0.84 | 0.11 |
| < 6 months before randomization | 244 (49%) | 7 (54%) | 1 (20%) | 0.77 | 0.18 |
| Number of sites involved | |||||
| 1 site of metastases | 241 (48%) | 7 (54%) | 1 (20%) | 0.73 | 0.18 |
| >1 site of metastases | 250 (50%) | 6 (46%) | 4 (80%) | ||
| Unknown | 6 (1%) | — | — | ||
| Primary tumour involved at start chemotherapy | 62 (12%) | 3 (15%) | 2 (40%) | 0.37 | 0.54 |
| Previous adjuvant therapy | 82 (17%) | 2 (15%) | 2 (40%) | 0.91 | 0.28 |
| Resection of primary tumour | 488 (98%) | 13 (100%) | 5 (100%) | 0.49 | — |
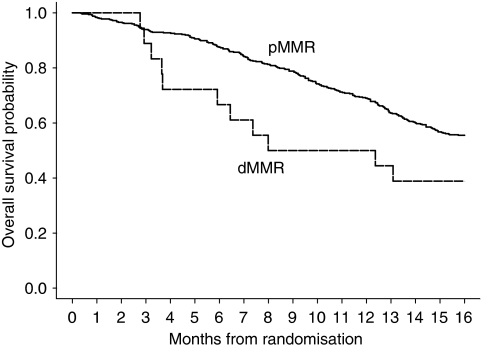
OS and PFS in relation to MMR status of the tumour
At the time of this analysis 447 patients have died, of which 15 patients exhibited dMMR. Table 2 presents the median OS and PFS for the pMMR and dMMR group of patients. The median OS was 17.9 months (95% CI 16.2–18.9), 7.4 months (95% CI 3.7–16.9) and 10.2 months (95% CI 5.9–19.8) for patients with a pMMR tumour, a dMMR tumour caused by hypermethylation of the MLH1 promoter, and the total group of patients with a dMMR tumour, respectively (Table 2; Figure 2). These differences were statistically not significant (Table 2). In the sequential treatment arm, the median OS for these groups of patients was 17.2 months (95% CI 14.7–18.8), 12.4 months (95% CI 3.2–->) and 12.7 months ((95% CI 7.4–22.2), respectively, and in the combination treatment arm 18.3 months (95% CI 16.2–20.6), 6.2 months (95% CI 3.6–31.3) and 6.2 months (95% CI 3.6–31.3), respectively. All these differences were statistically not significant (Table 2).
Table 2. OS and PFS according to MMR status with the number of events in italic.
| Number of patients | PMMR (_n_=497) | dMMR MLH1 hypermethylation (_n_=13) | dMMR total (_n_=18) | _P_-value# pMMR vs dMMR hypermethylation | _P_-value# pMMR vs dMMR total |
|---|---|---|---|---|---|
| Overall survival (months) | |||||
| Median (95% CI) | 17.9 (16.2–18.9) | 7.4 (3.7–16.9) | 10.2 (5.9–19.8) | 0.27 | 0.41 |
| n=440 | n=11 | n=15 | |||
| Sequential treatment | 17.2 (14.7–18.8) | 12.4 (3.2––>) | 12.7 (7.4–22.2) | 0.58 | 0.47 |
| Median (95% CI) | n=230 | n=4 | n=7 | ||
| Combination treatment | 18.3 (16.2–20.6) | 6.2 (3.6–31.3) | 6.2 (3.6–31.3) | 0.25 | 0.58 |
| Median (95% CI) | n=210 | n=7 | n=8 | ||
| PFS (months) | |||||
| Median (95% CI) | 6.9 (6.3–7.7) | 4.3 (2.4–6.6) | 4.0 (2.3–6.5) | 0.85 | 0.28 |
| n=490 | n=12 | n=17 | |||
| Sequential treatment | 5.8 (4.9–6.3) | 6.6 (2.2–->) | 4.2 (2.2–10.6) | 0.27 | 0.72 |
| Median (95% CI) | n=247 | n=4 | n=7 | ||
| Combination treatment | 8.3 (7.6–8.7) | 4.0 (2.3–6.5) | 4.0 (2.3–6.5) | 0.06 | 0.02 |
| Median (95% CI) | n=243 | n=8 | n=10 |
Figure 2.
OS by MMR status.
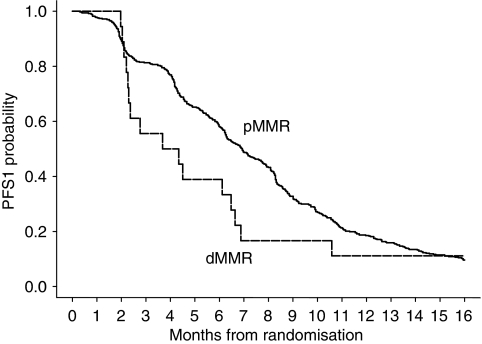
The median PFS for patients with a pMMR tumour, dMMR tumours caused by hypermethylation of the MLH1 promoter, and the total group of patients with a dMMR tumour, was 6.9 months (95% CI 6.3–7.7), 4.3 months (95% CI 2.4–6.6) and 4.0 months (95% CI 2.3–6.5), respectively (Figure 3). In the sequential treatment arm the median PFS for these groups of patients was 5.8 months (95% CI 4.9–6.3), 6.6 months (95% CI 2.2–->) and 4.2 months (95% CI 2.2–10.6), respectively, and for the combination treatment arm 8.3 months (95% CI 7.6–8.7), 4.0 months (95% CI 2.3–6.5) and 8.2 months (95% CI 7.4–8.5), respectively. The pair-wise comparison with pMMR and the two groups of dMMR patients was not significant in the first comparison (_P_=0.06), and significant in the second comparison (_P_=0.02).
Figure 3.
PFS by MMR status.
Tumour response in relation to MMR status
A total of 511 patients received first-line therapy. Deficient mismatch repair was observed in 18 patients, of whom 13 patients showed hypermethylation of the MLH1 promoter. Of these 511 patients, dMMR was detected in 16 out of 469 patients evaluable for response on first-line therapy, in 8 out of 288 patients evaluable for response on second-line therapy and in none of 96 patients evaluable for response on third-line therapy. The response and PFS analyses are only shown for first-line due to the low number of patients with dMMR tumours in second- and third-line treatment (Tables 2 and 3). The overall response rate in first-line treatment for pMMR, dMMR caused by hypermethylation of the MLH1 promoter and the total group of patients with a dMMR tumour was 31, 33 and 25%, respectively. Results on disease control in first-line treatment in the pMMR group, the dMMR group caused by hypermethylation of the MLH1 promoter and the total group of patients with a dMMR tumour, were 83, 58 and 56%, respectively (Table 3).
Table 3. Best overall response in first-line treatment according to MMR status.
| Patients evaluable for response | pMMR (_n_=453) | dMMR MLH1-hypermethylation (_n_=12) | dMMR total (_n_=16) | _P_-value# pMMR vs dMMR hypermethylation | _P_-value# pMMR vs dMMR total |
|---|---|---|---|---|---|
| Best overall response | |||||
| Complete response | 16 (4%) | — | — | ||
| Partial response | 123 (27%) | 4 (33%) | 4 (25%) | ||
| Stable disease | 235 (52%) | 3 (25%) | 5 (31%) | ||
| Progressive disease | 79 (17%) | 5 (42%) | 7 (44%) | ||
| Response rate (95% CI) | 31% (27–35%) | 33% (10–65%) | 25% (7–52%) | 0.84 | 0.63 |
| _n_=139 | _n_=4 | _n_=4 | |||
| Disease control (95% CI) | 83% (79–86%) | 58% (28–85%) | 56% (30–80%) | 0.031 | 0.008 |
| _n_=374 | _n_=7 | _n_=9 | |||
| Sequential treatment | _n_=239 | _n_=5 | _n_=8 | ||
| Response rate (95% CI) | 17% (13–23%) | 40% (5–85%) | 25% (3–65%) | 0.19 | 0.57 |
| _n_=41 | _n_=2 | _n_=2 | |||
| Disease control (95% CI) | 76% (70–81%) | 60% (15–95%) | 50% (16–84%) | 0.40 | 0.09 |
| _n_=182 | _n_=3 | _n_=4 | |||
| Combination treatment | _n_=214 | _n_=7 | _n_=8 | ||
| Response rate (95% CI) | 46% (39–53%) | 29% (4–71%) | 25% (3–65%) | 0.46 | 0.30 |
| _n_=98 | _n_=1 | _n_=2 | |||
| Disease control (95% CI) | 90% (85–93%) | 57% (18–90%) | 63% (25–92%) | 0.033 | 0.048 |
| _n_=193 | _n_=4 | _n_=5 |
Discussion
This is the first large randomised study in advanced CRC patients in which the incidence of dMMR and the correlation between sporadic dMMR and the outcome of chemotherapy is investigated. The incidence of dMMR was low (18 out of 515 patients, 3.5%). In 13 of these patients (2.5%) dMMR was caused by MLH1 promoter hypermethylation.
The incidence is lower as the previously reported incidence of 10–20% dMMR in sporadic CRC's (Lothe et al, 1993; Peltomaki, 2003). We ruled out the possibility of an underestimation of dMMR due to technical failure for several reasons. Previously it has been shown that IHC is an accurate, highly specific and sensitive method for detecting dMMR (Ward et al, 2001; Lindor et al, 2002; Overbeek et al, 2008). We performed IHC staining on TMA's (Hendriks et al, 2003), and confirmed the results on whole tissue slides and by MSI analysis. Furthermore, MSI analysis in a random sample of patients with pMMR as determined by IHC did confirm the results.
The low incidence in our series may be explained by a reduced potential in stages I–III dMMR patients to develop metastases (Lim et al, 2004; Malesci et al, 2007), as most of the previously reported incidences were observed in these earlier stages of CRC (Popat et al, 2005). Thus, as a consequence the incidence of dMMR in advanced CRC studies will be lower than 10–20%. Data on the incidence in stage IV patients are scarce and are mostly derived from small, non-randomised studies, with a reported incidence between 0.5 and 21% (Kochhar et al, 1997; Samowitz et al, 2001; Liang et al, 2002; Ricciardiello et al, 2005; Malesci et al, 2007; Muller et al, 2008). A low dMMR incidence of 0 and 2.7% was found in advanced CRC patients with resected liver (Haddad et al, 2004) and lung metastases (Melloni et al, 2006), respectively. An incidence of 4.4% of MLH1/MSH2 loss was reported in a large cohort of advanced CRC patients (Braun et al, 2008).
Our results on the correlation of dMMR with clinical outcome should be interpreted with caution due to the low number of dMMR patients. We found a non-significant decrease in median OS of 17.9 vs 10.2 months, and in median PFS of 6.9 vs 4.0 months in pMMR vs dMMR patients treated with chemotherapy, with a decreased disease control rate of 83 vs 56%, respectively. These results are difficult to compare with previously reported results for several reasons. First, dMMR has been tested as a predictive marker in advanced CRC in only one randomised study (Muller et al, 2008). In this study with 474 patients, tumour tissue was available from 104 patients of whom only 4 patients treated with 5FU and oxaliplatin tested positive for dMMR. Second, we used capecitabine as a fluoropyrimidine. It is yet unknown whether the outcome of 5FU and capecitabine differs in respect to the MMR status of patients. Third, in contrast to most studies we differentiated between dMMR caused by hypermethylation of the MLH1 promoter, dMMR without MLH1 promoter hypermethylation, and pMMR. Hereditary and sporadic dMMR tumours may differ in terms of pathologic features, underlying molecular alterations, and prognosis, and it is known that patients with dMMR tumours with overlapping hypermethylator phenotype have a worse clinical outcome compared to dMMR tumours without promoter hypermethylation (Hawkins et al, 2002; Ward et al, 2003; McGivern et al, 2004; Benatti et al, 2005; Johnson et al, 2005; Jass, 2007).
A reduced chemosensitivity was observed in CRC cell lines with MLH1 promoter hypermethylation treated with fluoropyrimidines (Arnold et al, 2003). Although we found a decreased disease control rate in dMMR patients, our numbers are too small to draw meaningful conclusions. Definite proof for a reduced chemosensitivity of advanced dMMR tumours can only be obtained from prospective randomised studies with chemotherapy vs observation. However studies with this design are considered unethical given the benefit of chemotherapy. We considered a control group within the same prospective trial which therefore was selected, treated and monitored in exactly the same way as the most appropriate alternative.
In conclusion, dMMR caused by hypermethylation of the _MLH1 –_promoter is a rare event in advanced CRC patients. This supports the hypothesis that dMMR tumours have a reduced metastatic potential. Given the low incidence of dMMR in advanced CRC patients, our results do not allow any meaningful conclusions on the correlation between dMMR status and clinical outcome. This topic should be addressed by a pooled analysis of multiple trials. The low incidence of dMMR does not justify the need for standard dMMR testing in advanced CRC patients.
Acknowledgments
This study was supported by the Dutch Colorectal Cancer Group (DCCG) and the Cornelis Visser Foundation.
References
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR (1993) Clues to the pathogenesis of familial colorectal cancer. Science 260: 812–816 [DOI] [PubMed] [Google Scholar]
- Arnold CN, Goel A, Boland CR (2003) Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. Int J Cancer 106: 66–73 [DOI] [PubMed] [Google Scholar]
- Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M, Roncari B, Maffei S, Rossi G, Ponti G, Santini A, Losi L, Di GC, Oliani C, Ponz De LM, Lanza G (2005) Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res 11: 8332–8340 [DOI] [PubMed] [Google Scholar]
- Bettstetter M, Dechant S, Ruemmele P, Grabowski M, Keller G, Holinski-Feder E, Hartmann A, Hofstaedter F, Dietmaier W (2007) Distinction of hereditary nonpolyposis colorectal cancer and sporadic microsatellite-unstable colorectal cancer through quantification of MLH1 methylation by real-time PCR. Clin Cancer Res 13: 3221–3228 [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257 [PubMed] [Google Scholar]
- Braun MS, Richman SD, Quirke P, Daly C, Adlard JW, Elliott F, Barrett JH, Selby P, Meade AM, Stephens RJ, Parmar MK, Seymour MT (2008) Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol 26: 2690–2698 [DOI] [PubMed] [Google Scholar]
- Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR (1999) Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology 117: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR (2004) Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology 126: 394–401 [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN (1998) Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58: 3455–3460 [PubMed] [Google Scholar]
- Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, Halling KC, McDonnell SK, Schaid DJ, Walsh VC, Kubly V, Nelson H, Michels VV, Thibodeau SN (2001) The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet 69: 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, van DA, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer 43: 1348–1360 [DOI] [PubMed] [Google Scholar]
- Elsaleh H, Iacopetta B (2001) Microsatellite instability is a predictive marker for survival benefit from adjuvant chemotherapy in a population-based series of stage III colorectal carcinoma. Clin Colorectal Cancer 1: 104–109 [DOI] [PubMed] [Google Scholar]
- Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB (1996) The role of DNA mismatch repair in platinum drug resistance. Cancer Res 56: 4881–4886 [PubMed] [Google Scholar]
- Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S (2000) Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med 342: 69–77 [DOI] [PubMed] [Google Scholar]
- Haddad R, Ogilvie RT, Croitoru M, Muniz V, Gryfe R, Pollet A, Shanmugathasan P, Fitzgerald T, Law CH, Hanna SS, Jothy S, Redston M, Gallinger S, Smith AJ (2004) Microsatellite instability as a prognostic factor in resected colorectal cancer liver metastases. Ann Surg Oncol 11: 977–982 [DOI] [PubMed] [Google Scholar]
- Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R (2002) CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology 122: 1376–1387 [DOI] [PubMed] [Google Scholar]
- Hendriks Y, Franken P, Dierssen JW, De LW, Wijnen J, Dreef E, Tops C, Breuning M, Brocker-Vriends A, Vasen H, Fodde R, Morreau H (2003) Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 162: 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363: 558–561 [DOI] [PubMed] [Google Scholar]
- Jass JR, Sobin LH, Wanatabe H (1990) The WHO classification of gastrointestinal tumors: a commentary on the second edition. Cancer 66: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Jass JR (2007) Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 50: 113–130 [DOI] [PubMed] [Google Scholar]
- Johnson V, Volikos E, Halford SE, Eftekhar Sadat ET, Popat S, Talbot I, Truninger K, Martin J, Jass J, Houlston R, Atkin W, Tomlinson IP, Silver AR (2005) Exon 3 beta-catenin mutations are specifically associated with colorectal carcinomas in hereditary non-polyposis colorectal cancer syndrome. Gut 54: 264–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R (1997) Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57: 808–811 [PubMed] [Google Scholar]
- Kochhar R, Halling KC, McDonnell S, Schaid DJ, French AJ, O'Connell MJ, Nagorney DM, Thibodeau SN (1997) Allelic imbalance and microsatellite instability in resected Duke's D colorectal cancer. Diagn Mol Pathol 6: 78–84 [DOI] [PubMed] [Google Scholar]
- Koopman M, Antonini NF, Douma J, Wals J, Honkoop AH, Erdkamp FL, de Jong RS, Rodenburg CJ, Vreugdenhil G, Loosveld OJ, van BA, Sinnige HA, Creemers GJ, Tesselaar ME, Slee PH, Werter MJ, Mol L, Dalesio O, Punt CJ (2007) Sequential vs combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO): a phase III randomised controlled trial. Lancet 370: 135–142 [DOI] [PubMed] [Google Scholar]
- Liang JT, Huang KC, Lai HS, Lee PH, Cheng YM, Hsu HC, Cheng AL, Hsu CH, Yeh KH, Wang SM, Tang C, Chang KJ (2002) High-frequency microsatellite instability predicts better chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV sporadic colorectal cancer after palliative bowel resection. Int J Cancer 101: 519–525 [DOI] [PubMed] [Google Scholar]
- Lim SB, Jeong SY, Lee MR, Ku JL, Shin YK, Kim WH, Park JG (2004) Prognostic significance of microsatellite instability in sporadic colorectal cancer. Int J Colorectal Dis 19: 533–537 [DOI] [PubMed] [Google Scholar]
- Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN (2002) Immunohistochemistry vs microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 20: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 24: 5313–5327 [DOI] [PubMed] [Google Scholar]
- Lothe RA, Peltomaki P, Meling GI, Aaltonen LA, Nystrom-Lahti M, Pylkkanen L, Heimdal K, Andersen TI, Moller P, Rognum TO (1993) Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res 53: 5849–5852 [PubMed] [Google Scholar]
- Magrini R, Bhonde MR, Hanski ML, Notter M, Scherubl H, Boland CR, Zeitz M, Hanski C (2002) Cellular effects of CPT-11 on colon carcinoma cells: dependence on p53 and hMLH1 status. Int J Cancer 101: 23–31 [DOI] [PubMed] [Google Scholar]
- Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, Roncalli M, Gennari L, Santoro A (2007) Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res 13: 3831–3839 [DOI] [PubMed] [Google Scholar]
- McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, Barker MA, Arnold S, Simms LA, Leggett BA, Young J, Jass JR (2004) Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer 3: 101–107 [DOI] [PubMed] [Google Scholar]
- Mecklin JP (1987) Frequency of hereditary colorectal carcinoma. Gastroenterology 93: 1021–1025 [DOI] [PubMed] [Google Scholar]
- Melloni G, Doglioni C, Bandiera A, Carretta A, Ciriaco P, Arrigoni G, Zannini P (2006) Prognostic factors and analysis of microsatellite instability in resected pulmonary metastases from colorectal carcinoma. Ann Thorac Surg 81: 2008–2013 [DOI] [PubMed] [Google Scholar]
- Muller CI, Schulmann K, Reinacher-Schick A, Andre N, Arnold D, Tannapfel A, Arkenau H, Hahn SA, Schmoll SH, Porschen R, Schmiegel W, Graeven U (2008) Predictive and prognostic value of microsatellite instability in patients with advanced colorectal cancer treated with a fluoropyrimidine and oxaliplatin containing first-line chemotherapy. A report of the AIO Colorectal Study Group. Int J Colorectal Dis 23: 1033–1039 [DOI] [PubMed] [Google Scholar]
- Overbeek LI, Kets CM, Hebeda KM, Bodmer D, van der LE, Willems R, Goossens M, Arts N, Brunner HG, van Krieken JH, Hoogerbrugge N, Ligtenberg MJ (2007) Patients with an unexplained microsatellite instable tumour have a low risk of familial cancer. Br J Cancer 96: 1605–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek LI, Ligtenberg MJ, Willems RW, Hermens RP, Blokx WA, Dubois SV, van der LH, Meijer JW, Mlynek-Kersjes ML, Hoogerbrugge N, Hebeda KM, van Krieken JH (2008) Interpretation of immunohistochemistry for mismatch repair proteins is only reliable in a specialized setting. Am J Surg Pathol 32: 1246–1251 [DOI] [PubMed] [Google Scholar]
- Peltomaki P (2003) Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol 21: 1174–1179 [DOI] [PubMed] [Google Scholar]
- Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23: 609–618 [DOI] [PubMed] [Google Scholar]
- Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, Young J, Jenkins MA, Hopper JL, Baron JA, Buchanan D, Casey G, Levine AJ, Le ML, Gallinger S, Bapat B, Potter JD, Newcomb PA, Haile RW, Laird PW (2008) Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev 17: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt CJ (2004) New options and old dilemmas in the treatment of patients with advanced colorectal cancer. Ann Oncol 15: 1453–1459 [DOI] [PubMed] [Google Scholar]
- Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S (2003) Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 349: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardiello L, Ceccarelli C, Angiolini G, Pariali M, Chieco P, Paterini P, Biasco G, Martinelli GN, Roda E, Bazzoli F (2005) High thymidylate synthase expression in colorectal cancer with microsatellite instability: implications for chemotherapeutic strategies. Clin Cancer Res 11: 4234–4240 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S (1997) A national cancer institute workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting hightlights and Bethesda guidelines. J Natl Cancer Inst 89: 1758–1762. Ref Type: Generic [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Curtin K, Ma KN, Schaffer D, Coleman LW, Leppert M, Slattery ML (2001) Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev 10: 917–923 [PubMed] [Google Scholar]
- Samowitz WS, Slattery ML, Potter JD, Leppert MF (1999) BAT-26 and BAT-40 instability in colorectal adenomas and carcinomas and germline polymorphisms. Am J Pathol 154: 1637–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankila R, Aaltonen LA, Jarvinen HJ, Mecklin JP (1996) Better survival rates in patients with MLH1-associated hereditary colorectal cancer. Gastroenterology 110: 682–687 [DOI] [PubMed] [Google Scholar]
- Sergent C, Franco N, Chapusot C, Lizard-Nacol S, Isambert N, Correia M, Chauffert B (2002) Human colon cancer cells surviving high doses of cisplatin or oxaliplatin in vitro are not defective in DNA mismatch repair proteins. Cancer Chemother Pharmacol 49: 445–452 [DOI] [PubMed] [Google Scholar]
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van GM, van Oosterom AT, Christian MC, Gwyther SG (2000) New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 92: 205–216 [DOI] [PubMed] [Google Scholar]
- Ward R, Meagher A, Tomlinson I, O'Connor T, Norrie M, Wu R, Hawkins N (2001) Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut 48: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ (2003) Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol 21: 3729–3736 [DOI] [PubMed] [Google Scholar]