Preserved glucose tolerance in high-fat-fed C57BL/6 mice transplanted with PPARγ−/−, PPARδ−/−, PPARγδ−/−, or LXRαβ−/− bone marrow (original) (raw)
Abstract
Macrophage lipid metabolism and inflammatory responses are both regulated by the nuclear receptors PPAR and LXR. Emerging links between inflammation and metabolic disease progression suggest that PPAR and LXR signaling may alter macrophage function and thereby impact systemic metabolism. In this study, the function of macrophage PPAR and LXR in Th1-biased C57BL/6 mice was tested using a bone marrow transplantation approach with PPARγ−/−, PPARδ−/−, PPARγδ−/−, and LXRαβ−/− cells. Despite their inhibitory effects on inflammatory gene expression, loss of PPARs or LXRs in macrophages did not exert major effects on obesity or glucose tolerance induced by a high-fat diet. Treatment with rosiglitazone effectively improved glucose tolerance in mice lacking macrophage PPARγ, suggesting that cell types other than macrophages are the primary mediators of the anti-diabetic effects of PPARγ agonists in our model system. C57BL/6 macrophages lacking PPARs or LXRs exhibited normal expression of most alternative activation gene markers, indicating that macrophage alternative activation is not absolutely dependent on these receptors in the C57BL/6 background under the conditions used here. These studies suggest that genetic background may be an important modifier of nuclear receptor effects in macrophages. Our results do not exclude a contribution of macrophage PPAR and LXR expression to systemic metabolism in certain contexts, but these factors do not appear to be dominant contributors to glucose tolerance in a high-fat-fed Th1-biased bone marrow transplant model.
Keywords: PPAR, LXR, macrophage, inflammation, insulin resistance, immune phenotype
Macrophages play an important role in innate and acquired immune responses. They can be directed to acquire distinct phenotypes through classical (M1) or alternative (M2) pathways of activation (1). Depending on the trigger for macrophage activation, different outcomes in the adaptive immune response, inflammation, and gene expression result. For instance, macrophages activated classically by lipopolysaccharide (LPS) (M1 phenotype) promote a Th1 T cell response, characterized by the release of inflammatory cytokines and the activation of cytotoxic CD8 T cells (2). The Th1 response is primarily geared toward the phagocytosis and killing of pathogens. Macrophages activated by alternative stimuli such as IL-4 (M2 phenotype) are less inflammatory and foster a Th2 response by directing the proliferation and differentiation of B cells (2). The biological impact of these distinct pathways of macrophage activation has been studied in mouse models that favor either Th1 or Th2 immune responses. The widely used C57BL/6 mouse strain preferentially mounts Th1-biased immune responses, including classical macrophage activation, leading to robust inflammatory responses (1). By contrast, the more Th2-permissive BALB/C strain is more susceptible to alternative macrophage activation and does not exhibit such a dominant inflammatory reaction (1).
Recent work suggests that macrophage-dependent inflammatory pathways are linked to the development and possibly treatment of insulin resistance. In particular, adipose tissue inflammation has attracted widespread interest as a potential causal factor in insulin resistance (3). Type 2 diabetes is associated with low-grade or chronic inflammation (4), and adipose tissue is now recognized to be a major producer of bioactive signaling molecules, known as adipokines, some of which may contribute to chronic inflammation (5). In addition, macrophages have been shown to accumulate in adipose tissue of obese mice and humans, where they are postulated to contribute to inflammatory signaling (6, 7). In fact, studies have consistently revealed elevated expression of inflammatory genes such as TNFα, IL-6, and iNOS in adipose tissue of obese mice, and tissue macrophages appear to be a prime source of these factors (6, 8, 9).
Several lines of evidence support a causal role for macrophage inflammatory signaling in insulin resistance. For example, inhibition of TNFα signaling in obese mice improves insulin sensitivity, whereas exogenously administering TNFα worsens insulin resistance (4, 10, 11). Furthermore, blocking macrophage inflammatory gene expression through genetic deletion of IKKβ, an essential kinase in the NF-κB activation pathway, improves insulin sensitivity in obese mice (12, 13). Inhibiting macrophage infiltration into adipose tissue by deletion of MCP-1 expression in fat also improves insulin resistance (14–16).
The observation that inflammatory gene expression in adipose tissue is suppressed by systemic treatment with PPARγ agonists such as rosiglitazone has led several groups to investigate the impact of anti-inflammatory nuclear receptor signaling on diabetes (17, 18). Members of both the peroxisome proliferator-activated receptor (PPAR) and liver X receptor (LXR) subfamilies have emerged as important negative regulators of macrophage-mediated inflammation. PPARγ, PPARδ, LXRα, and LXRβ are each expressed at high levels in murine macrophages, and numerous studies have addressed the potential for these receptors to alter macrophage inflammatory gene expression (19–21). Recent studies in Th2-biased BALB/C mice have suggested that PPARγ is important for M2 macrophage activation and that genetic deletion of PPARγ in myeloid cells alters the balance between M1 and M2 macrophages (22). Moreover, BALB/C mice lacking PPARγ expression in macrophages are more susceptible to the development of diet-induced insulin resistance (22). Hevener et al. (18) have also examined the consequence of loss of bone marrow (BM) PPARγ expression on a mixed genetic background. Similar to the previous study, loss of PPARγ was associated with increased expression of inflammatory genes as well as defective insulin signaling in target tissues such as adipose tissue, liver, and muscle. The contribution of macrophage PPARδ or LXR expression to systemic glucose metabolism has not been addressed previously.
C57BL/6 is the genetic background most widely studied in the context of metabolism; however, the importance of anti-inflammatory nuclear receptor function for obesity-linked insulin resistance in this strain remains unclear. Here we assess the impact of loss of BM expression of four different nuclear receptors on systemic glucose metabolism in C57BL/6 mice. BM genetically deleted for PPARγ, PPARδ, both PPARγ and PPARδ, or both LXRα and LXRβ was isolated from C57BL/6 donors and transplanted into wild-type C57BL/6 recipients. Mice were fed a high-fat diet to induce obesity and insulin resistance. None of these nuclear receptors significantly altered obesity or glucose tolerance of transplanted mice in our study. In addition, treatment with rosiglitazone improved glucose tolerance in mice receiving PPARγ-null BM. Although our results do not exclude a contribution of macrophage PPAR and LXR expression to systemic metabolism in certain contexts, these factors do not appear to be dominant contributors to glucose tolerance in a high-fat-fed Th1-biased bone marrow transplant (BMT) model. Our data suggest that genetic background and diet may influence the metabolic consequences of nuclear receptor signaling in macrophages.
METHODS
Reagents and cell culture
The synthetic ligands for LXR (GW3965 and T1317), PPARα (GW9544), PPARδ (GW742), and PPARγ (GW7845) were provided by GlaxoSmithKline. All ligands were dissolved in DMSO before use in cell culture. The LXR ligand was used at a concentration of 1 μM, whereas the PPAR ligands were all used at 100 nM. LPS from Salmonella typhimurium and poly(I:C) were purchased from Sigma and used at 100 ng/ml and 2.5 mg/ml, respectively. IL-13 was purchased from Peprotech and used at 10 ng/ml. Thioglycollate-elicted primary murine macrophages were maintained in DMEM containing 10% FBS. BM-derived macrophages were differentiated in DMEM containing 20% FBS and 30% L929-conditioned media for 7 days. After differentiation, macrophages were cultured in DMEM containing 10% FBS. Cells were harvested from wild-type, PPARγ−/−, PPARδ−/−, or PPARγδ−/− mice. The Mx Cre mice were purchased from Jackson Laboratories. The Mx Cre, PPARγfl/fl, and PPARδ−/− mice are all on the C57BL/6 background (more than nine generations backcrossed).
Animal studies
The recipient wild-type mice used for the BMT studies were irradiated with 900 rads the night prior to reconstitution. Each of the four groups of recipient mice contained 12 mice. BM cells from recipient mice were harvested and injected into tail veins of the recipient mice. The irradiated mice were kept in sterile cages with autoclaved food and trimethoprim-sulfamethoxazole-treated water for 2 weeks. Mice were challenged with a 60% caloric fat diet (Research Diets) for 16 weeks. Mice were fasted the night prior to glucose tolerance tests, and glucose levels were monitored after intraperitoneal injections of glucose (2 g/kg; Sigma). For gavage experiments, mice were gavaged with either vehicle or rosiglitazone (30 mg/kg; Cayman Chemicals) for 8 days.
Insulin ELISA
Wild-type, PPARγ−/−, and PPARγδ−/− BMT mice were fasted overnight, and blood was collected in heparin tubes. Samples were then spun at 8,000 rpm for 5 min to isolate serum. An ultrasensitive mouse insulin ELISA kit (Crystal Chem, Inc.) was used to perform an insulin ELISA on the serum to determine insulin levels.
RNA and quantitative PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) and was reverse transcribed to obtain cDNA (Applied Biosystems). Real-time quantitative PCR assays were performed using an Applied Biosystems 7900 sequence detector as previously described (23). Data were normalized to housekeeping gene 36B4.
Statistical analysis
Statistical significance was determined using Student's _t_-test for in vitro assays. A one-way or two-way ANOVA test was used to determine statistical significance for all glucose tolerance tests.
RESULTS
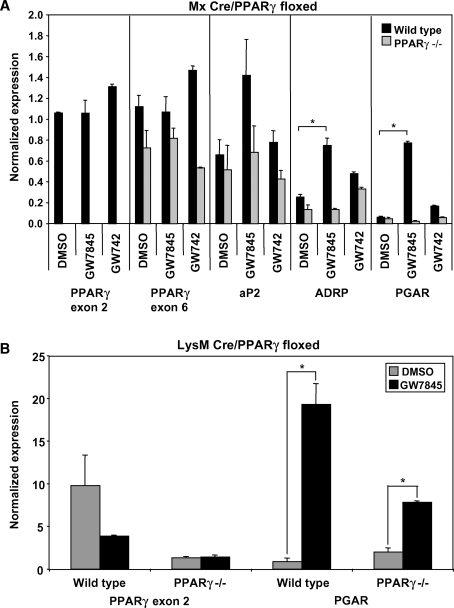
To investigate the function of macrophage PPARγ, we used a Cre recombinase system to disrupt the PPARγ gene. Specifically, we crossed C57BL/6 mice with loxP sites flanking the second exon of PPARγ (PPARγfl/fl) with C57BL/6 mice expressing Cre recombinase downstream of the α/β interferon-inducible (Mx) promoter. Cre recombinase activity was initiated by intraperitoneal injection of the Mx activator poly IC 3 to 4 weeks after birth. Interestingly, injection of poly IC at weaning age caused a permanent disruption of PPARγ in myeloid precursor cells; new myeloid cells generated postinjection remain PPARγ-null for the life of the mouse. Typically, we analyzed macrophages from these mice at least 4 weeks after inducing recombination to reduce any potential effects of the poly IC. Quantitative real-time PCR confirmed that the Mx Cre system was extremely efficient, yielding nearly complete (>95%) deletion of PPARγ exon 2 in vivo (Fig. 1A). As expected, induction of PPAR target genes (aP2, ADRP, PGAR) by PPARγ but not PPARδ agonists was lost in Mx Cre PPARγ−/− macrophages (Fig. 1A).
Fig. 1.
Induction of the Mx promoter effectively disrupts PPARγ, whereas LysM Cre cannot efficiently delete PPARγ exon 2. Cells were harvested and pooled from three to five mice per group. The presented data are representative of at least three independent experiments. A: Thioglycollate-elicited wild-type and Mx Cre PPARγ−/− peritoneal macrophages were treated with PPARγ or PPARδ ligands (GW7845 or GD742, respectively; 100 nM each) overnight. PPARγ exon 2 could not be detected in PPARγ−/− cells (P < 0.005), whereas PPARγ exon 6 expression was unaffected. Known PPARγ target genes (aP2, ADRP, and PGAR) were tested to confirm loss of ligand response in PPARγ−/− macrophages. B: Thioglycollate-elicited wild-type and LysM Cre PPARγ−/− peritoneal macrophages were treated with PPARγ ligand (GW7845; 100 nM) overnight. PPARγ exon 2 could still be detected in PPARγ−/− macrophages, and PGAR, a known PPARγ target gene, could still be induced by PPARγ ligand in these cells (P < 0.02). Error bars represent SEM.
In addition to using the Mx Cre system, we also generated mice with macrophage-specific PPARγ deletion using the LysM Cre system. Unlike Mx Cre, LysM Cre is constitutively active. However, compared with the Mx Cre system, the LysM Cre system was not as efficient in recombining PPARγ to create a disrupted gene in our hands. After harvesting peritoneal macrophages from wild-type and LysM Cre PPARγ−/− mice, we treated the cells with PPARγ ligand to test whether disruption of PPARγ was able to prevent PPARγ target gene regulation. We found that PPARγ exon 2 could still be detected by quantitative real-time PCR and that PGAR, a PPARγ target gene, could be regulated by ligand even in PPARγ−/− macrophages (Fig. 1B). In contrast, the Mx Cre system yields cell populations with complete deletion of PPARγ exon 2 expression, and more importantly, these cells are unable to respond to PPARγ ligand and do not regulate target genes. Consequently, we opted to use the Mx Cre system for our studies.
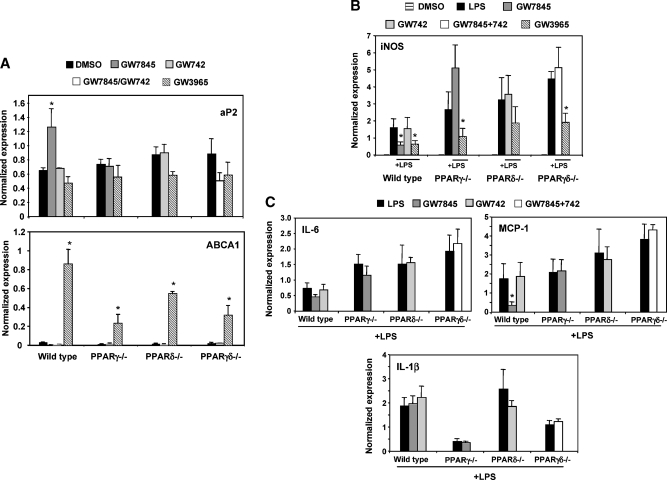
Recent work suggests that PPARs mediate inflammatory signaling pathways in macrophages and may affect inflammation associated with insulin resistance (18, 22). To address this issue in our genetic loss-of-function system, thioglycollate-elicited PPARγ−/−, PPARδ−/−, or PPARγδ−/− peritoneal macrophages were isolated and pretreated with either PPAR ligand or LXR ligand overnight. Cells were then stimulated with LPS (10 ng/ml) for 6 h, and inflammatory and receptor target gene was measured by real-time PCR. As shown in Fig. 2A, the PPAR target gene aP2, and the LXR target gene ABCA1, were upregulated in wild-type cells by ligand, confirming proper ligand response. As expected, LXR agonist (GW3965) efficiently suppressed inflammatory gene expression (iNOS). Furthermore, PPARγ ligand (GW7845) repressed IL-6, MCP-1, and iNOS expression in wild-type cells but not in PPARγ-null cells (Fig. 2B, C). PPARδ agonist (GW742) had no significant effect on expression of these genes in any genotype.
Fig. 2.
PPARγ−/−, PPARδ−/−, and PPARγδ−/− macrophages exhibited heightened inflammatory gene expression compared with wild-type cells in vitro_._ Cells were harvested and pooled from three to five mice per group. The presented data are representative of at least three independent experiments. A: Thioglycollate-elicited wild-type, PPARγ−/−, PPARδ−/−, and PPARγδ−/− peritoneal macrophages were harvested and pretreated with GW7845 (100 nM), GW742 (100 nM), or GW3965 (1 μM) overnight. The cells were then stimulated with LPS (10 ng/ml) for 6 h. PPAR and LXR target genes (aP2 and ABCA1, respectively) were tested to confirm ligand response. B: Macrophages were analyzed for inflammatory gene expression. iNOS was markedly upregulated in macrophages with deleted PPARs; in wild-type cells, treatment with LXR ligands consistently suppressed inflammatory genes, whereas treatment only with PPARγ and not PPARδ ligand resulted in repression. C: Additional inflammatory genes were tested in response to LPS and PPAR ligand treatments. Similar to iNOS, both IL-6 and MCP-1 expression was repressed by PPARγ ligand. However, IL-1β has a different expression profile compared with the other genes and highlights inconsistencies in PPAR signaling in inflammation. Error bars represent SEM.
We also noted a significant effect of loss of PPAR expression on LPS-inducible inflammatory gene expression in the absence of receptor ligands. Loss of either PPARγ or PPARδ alone led to enhanced induction of iNOS and IL-6 by LPS (Fig. 2B, C). Moreover, the combined deletion of both receptors had an additive effect, i.e., inflammatory gene expression was most responsive in the double knockouts. Interestingly, the combined loss of PPARγ and PPARδ affected the expression of some inflammatory markers more than others. For example, in contrast to IL-6, MCP-1, and iNOS, expression of IL-1β was not altered in the wild-type cells (Fig. 2C).
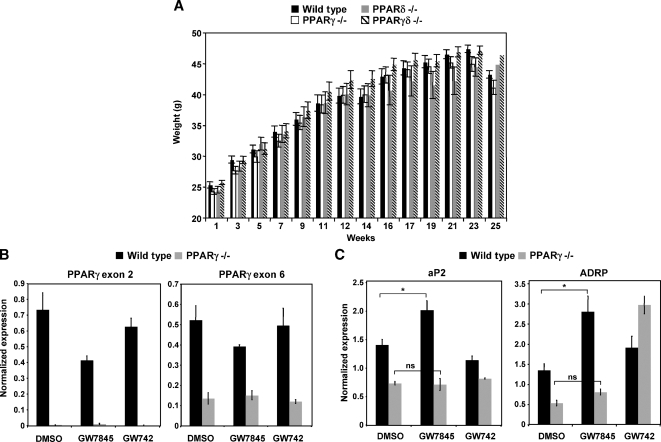
To investigate the impact of BM PPAR expression on systemic glucose metabolism in C57BL/6 mice, we used wild-type, PPARγ−/−, PPARδ−/−, or PPARγδ−/− BM (from mice more than 10 generations backcrossed to C57/BL6) to reconstitute irradiated wild-type mice. After recovery for 4 weeks, the mice were challenged with a 60% fat diet for an additional 14 weeks to cause obesity. All four groups of mice gained weight at a similar rate and had equivalent food intake during the course of the feeding (Fig. 3A; data not shown). To confirm the degree of BM reconstitution after 14 weeks of high-fat diet, we harvested thioglycollate-elicted peritoneal macrophages from PPARγ−/− and wild-type BMT-recipient mice. Real-time PCR verified that virtually all of the macrophages from PPARγ−/− BMT mice were derived from the donor cells, because PPARγ exon 2 was not expressed (Fig. 3B). We also tested whether the macrophages were responsive to ligand by examining PPAR target gene expression. As shown in Fig. 3C, expression of ADRP was induced by PPARγ ligand (GW7845) in cells from wild-type but not PPARγ−/− BMT mice. Basal expression of ADRP was also reduced in PPARγ-null cells.
Fig. 3.
All PPAR mice had equivalent weight gain and exhibited reconstitution with myeloid cells derived from donor mice. A: Irradiated mice transplanted with wild-type, PPARγ−/−, PPARδ−/−, or PPARγδ−/− myeloid cells were challenged with a 60% fat diet. All groups of mice gained weight at similar rates. B: Thioglycollate-elicted wild-type and PPARγ−/− peritoneal macrophages were harvested from recipient mice to determine degree of reconstitution. PPARγ exon 2 could not be detected in PPARγ−/− macrophages (P < 0.05), whereas PPARγ exon 6 could be detected. Cells were harvested and pooled from four wild-type and three PPARγ−/− bone marrow transplant (BMT) mice. C: Wild-type and PPARγ−/− macrophages were treated with GW7845 overnight. PPARγ target genes (aP2 and ADRP) did not increase in expression as a result of ligand treatment in PPARγ−/− cells. Cells were harvested and pooled from four wild-type and three PPARγ−/− BMT mice. Error bars represent SEM.
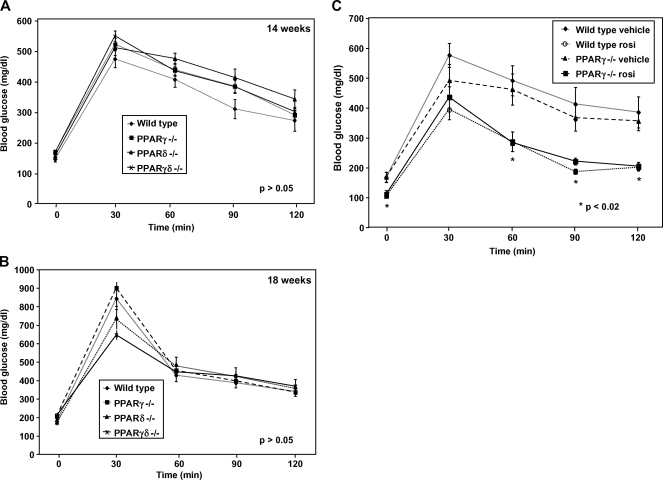
After mice were on the diet for 14 weeks, we performed glucose tolerance tests to determine whether reconstitution with differing PPAR knockout macrophages would alter systemic metabolism. Mice clearly became glucose intolerant as a result of high-fat feeding; however, high-fat-fed mice lacking BM expression of PPARγ, PPARδ, or both on the C57BL/6 background showed no significant difference from controls (Fig. 4A). A second set of glucose tolerance tests performed 4 weeks later yielded similar results (Fig. 4B). Fasting serum insulin levels were also not significantly different between groups, as determined by ELISA (P > 0.05; data not shown).
Fig. 4.
Glucose tolerance tests and insulin levels exhibited no significant differences in insulin sensitivity. A two-way ANOVA test was used to determine significance. A: After 14 weeks on the diet, the four groups of PPAR mice were equally diabetic and similarly insulin resistant (P > 0.05). After the mice were fasted overnight, basal blood glucose levels were measured (0 min) before intraperitoneal administration of 2 g/kg glucose. Blood glucose was assessed every 30 min after the glucose bolus. B: A second glucose tolerance test performed after an additional 4 weeks yielded similar results (P > 0.05). C: Wild-type and PPARγ−/− mice were treated with rosiglitazone (30 mg/kg) for 8 days. A glucose tolerance test was performed as previously described. Rosiglitazone improved insulin sensitivity in both wild-type and PPARγ−/− mice, whereas vehicle treatment did not demonstrate a decrease in blood glucose levels in either group (P < 0.02). Error bars represent SEM.
We also assessed the influence of BM PPARγ expression for the anti-diabetic effects of PPARγ agonists on a C57BL/6 background. We treated wild-type and PPARγ−/− BMT-recipient mice with rosiglitazone for 8 days. As shown in Fig. 4C, both groups treated with rosiglitazone showed markedly and equivalently improved glucose tolerance, compared with vehicle controls. This observation suggests that tissues other than BM are the primary mediators of the anti-diabetic effects of PPARγ ligand in mice on a C57BL/6 background under the experimental conditions employed here.
Macrophage infiltration into adipose tissue is a well-characterized feature in obese mouse models (6). Furthermore, administration of thiazoladinediones reverses macrophage infiltration, presumably by activating PPARγ. We therefore investigated whether the lack of macrophage PPARγ would affect macrophage infiltration into adipose tissue. To our surprise, we found that there was no change in the numbers of macrophages present in adipose tissue when we compared wild-type and PPARγ BMT mice (data not shown). Additionally, the size of adipocytes was similar in wild-type and PPARγ BMT mice and we did not observe any obvious differences in body composition (data not shown).
Previous work has established that both LXRα and LXRβ are expressed at high levels in macrophages, and both isotypes have strong anti-inflammatory effects (19). As shown in Fig. 2B, the anti-inflammatory effects of LXRs are especially prominent in vitro. We hypothesized that loss of LXR receptor expression in BM cells might exacerbate inflammatory signaling in vivo and impact glucose metabolism. We therefore transplanted BM cells from wild-type and LXRαβ−/− mice into irradiated wild-type recipients. As with the mice used for PPAR studies, the animals used for these experiments were also more than 10 generations backcrossed to C57BL/6 mice. After 4 weeks of recovery, the BMT mice were challenged with a 60% fat diet to induce obesity and insulin resistance. All groups consumed similar amounts of food and gained weight at similar rates (Fig. 5A). We confirmed that the reconstituted animals had been repopulated with LXRαβ−/− BM by harvesting thioglycollate-elicted peritoneal macrophages and performing real-time PCR to examine the expression of LXRs and their downstream target genes. As expected, LXR ligands (GW3965 and T1317) induced expression of ABCA1 in wild-type but not in LXRαβ−/− BMT cells (Fig. 5B). We also verified that LXRs had maintained their anti-inflammatory abilities after reconstitution by stimulating macrophages with LPS in the presence or absence of LXR ligand. Indeed, LXR ligand repressed expression of TNFα in wild-type but not LXR-null BM recipients (Fig. 5B).
Fig. 5.
Recipient mice showed equivalent weight gain and exhibited efficient reconstitution with wild-type or LXR-null donor bone marrow (BM). A: Wild-type and LXRαβ−/− mice gained weight at similar rates on a 60% fat diet. B: Thioglycollate-elicted wild-type and LXRαβ−/− peritoneal macrophages were harvested from recipient mice to determine degree of reconstitution. Wild-type and LXRαβ−/− macrophages were pretreated with GW3965 or T1317 (1 μM) and then stimulated with LPS (10 ng/ml) for 6 h. The LXRαβ target gene, ABCA1, did not increase in expression as a result of ligand treatment in LXRαβ−/− cells. Furthermore, LXRαβ−/− cells failed to repress TNFα gene expression. Wild-type cells treated with LXR ligand were able to exert anti-inflammatory control. Error bars represent SEM.
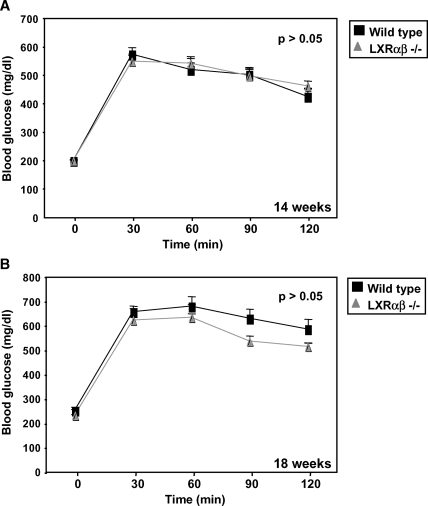
After mice were on on a high-fat diet for 14 weeks, a glucose tolerance test was performed. Similar to the results obtained with the PPAR BM transplants, we observed no difference in glucose tolerance or insulin levels between mice transplanted with wild-type or LXRαβ−/− BM (Fig. 6A; data not shown). A second independent test performed 4 weeks later showed similar results (Fig. 6B). These observations indicate that despite their anti-inflammatory effects, loss of LXR expression in the BM does not exert a major influence on glucose tolerance in high-fat-fed mice.
Fig. 6.
Glucose tolerance tests in wild-type and LXRαβ−/− mice revealed no difference in blood glucose levels. A two-way ANOVA test was used to determine significance. A: Wild-type and LXRαβ−/− mice were challenged with a 60% fat diet for 14 weeks. After mice were fasted overnight, basal blood glucose levels were measured. Glucose (2 g/kg) was administered, then blood glucose was assessed every 30 min for 2 h. Both groups of mice were equally insulin resistant (P > 0.05). B: A second glucose tolerance test was performed after an additional 4 weeks as previously described. Similar results were obtained (P > 0.05). Error bars represent SEM.
It has been previously reported that alternative activation of BALB/C macrophages relies significantly on appropriate PPAR signaling (22, 24). Studies have demonstrated that PPARγ-deficient macrophages express low levels of M2 marker genes and are not effectively alternatively activated (22). As a result, these cells may be more inflammatory and may act as contributors to worsened insulin sensitivity. To test whether PPARγ−/− or PPARδ−/− macrophages from C57BL/6 mice also expressed reduced levels of alternative activation marker genes, we stimulated BM-derived macrophages with cytokines that upregulate Th-2 response, such as IL-4 or IL-13 (25). We found that the absence of PPARs did not compromise M2 inflammatory responses under our experimental conditions. Expression of classical M2 marker genes such as Chi3l3, Pdcd1lg2, and Clec7a (26, 27) in response to IL-4 or IL-13 in PPARγ−/−, PPARδ−/−, or PPARγδ−/− macrophages was not significantly different from wild-type macrophages (Fig. 7). However, the loss of PPARγ in macrophages resulted in lower expression of arginase I (ArgI), a primary M2 marker gene (28), in response to IL-4 or IL-13 stimulation. ArgI has been previously described as a direct PPARγ and PPARδ target gene, and this may explain the reduced ArgI expression in PPARγ-deficient macrophages (22, 29). Interestingly, ArgI expression was unique in its dependence on PPARγ for induction; the expression of the remaining M2 marker genes we tested was not altered by the loss of PPARs.
Fig. 7.
Classical M2 gene expression is not absolutely dependent on PPAR expression in BM-derived macrophages. Wild-type, PPARγ−/−, PPARδ−/−, and PPARγδ−/− BM cells were harvested, differentiated into macrophages, and treated with IL-4 or IL-13 (10 ng/ml each) overnight. Expression of M2 genes was analyzed to determine impact of PPARγ, PPARδ, or both PPARγ and PPARδ deficiencies. The presented data are representative of two independent experiments. Error bars represent SEM.
DISCUSSION
In the present study, we report that loss of BM expression of anti-inflammatory nuclear receptors is not a dominant contributor to the development of diet-induced obesity or glucose intolerance in high-fat-fed C57BL/6 mice. This was true not only for PPARγ, which has been reported to play a role in insulin resistance in BALB/C mice, but also for PPARδ, LXRα, and LXRβ, which have not been previously studied in this context. Using a BMT approach, we found that loss of PPARγ, PPARδ, both PPARγ and PPARδ, or both LXRα and LXRβ did not alter weight gain in mice challenged with a 60% fat diet or have a significant effect on glucose tolerance. We also found that administration of rosiglitazone to PPARγ−/− BMT mice improved glucose tolerance, suggesting that BM expression of this receptor is not absolutely required for the therapeutic effects of thiazolidinediones in C57BL/6 mice.
Previous work has reported that macrophage PPARγ expression affects the development of obesity and insulin resistance (18, 22). It is important to point out that our studies were conducted in parallel to those of Glass and Chawla (18, 22) and were not designed to replicate the experimental conditions used in either of these reports. Therefore, our results are not necessarily in conflict with those of prior work, and there are a number of factors that may have contributed to the differences in experimental outcomes.
One possible contributor is the influence of genetic background. The macrophages in prior work were derived from either BALB/C or mixed-background mice. Genetic background in mice contributes to the dominance of M1 or M2 activation of macrophages and can bias how mouse models respond to specific stimuli. For example, the BALB/C mouse strain is Th2-permissive and is more susceptible to diseases that require an aggressive immune response (1). Conversely, C57BL/6 mice are dominant for Th1 signaling and display more-robust inflammatory signaling and the upregulation of pro-inflammatory genes. The C57BL/6 strain is most widely used for the study of metabolic disorders because of its susceptibility to diet-induced insulin resistance and atherosclerosis.
Another plausible explanation for the difference between our results and those of the Chawla and Glass groups (18, 22) is environment and the immune status of the mice. All animal facilities are different, and mice at different sites may therefore be exposed to a range of different pathogens and commensal bacteria. It is conceivable that such differences alter the immune systems of experimental mice in ways that may affect macrophage responses.
Another notable difference between our study and prior work is the diet employed. Hevener et al. (18) reported more-significant differences in blood glucose levels between wild-type and PPARγ−/− BMT mice fed a chow diet rather than a high-fat diet. Odegaard et al. (22) employed a 36% fat diet. We chose to use a 60% fat diet because our aim was to study PPAR/LXR function in obese models. However, it is possible that our high-fat diet may have been too robust a diabetic stimulus and therefore masked differences otherwise seen on a chow diet.
Finally, the BMT approach employed in our study may have contributed to differences in results. Prior studies employed a LysM Cre approach to delete PPARγ expression from macrophages. We chose to use Mx Cre-mediated deletion coupled with BMT because of incomplete deletion by LysM Cre in our hands (Fig. 1). However, it is possible that the BMT procedure itself has a confounding effect on systemic metabolism. Furthermore, although we confirmed reconstitution of recipient mice with the appropriate donor BM cells, there is a possibility that the tissue macrophage reconstitution was not complete at the time we performed our studies. Prior work has shown that adipose tissue macrophages and hepatic Kupffer cells are effectively replaced by BM cells in the time frame employed in our study (more than 4 months). However, the possibility that some wild-type macrophages may have remained and affected our results cannot be excluded.
While this paper was under review, two additional studies reported an impact of loss of PPARδ expression on glucose tolerance and alternative macrophage activation (30, 31). Some of the findings reported in these studies also differ from the results we report here. As outlined above for PPARγ, it is likely that a number of factors may contribute to the prominence of PPARδ signaling in different experimental contexts. Additional studies will be required to more precisely define the role of both PPARγ and PPARδ in macrophage inflammatory signaling in the context of metabolic disease.
In conclusion, our studies suggest that the in vitro anti-inflammatory activities of nuclear receptors alone are not predictive of in vivo metabolic effects. Macrophage LXRs show profound anti-inflammatory effects in vitro but do not dramatically influence systemic glucose metabolism under the conditions used here. Furthermore, our results suggest that careful consideration of the immunological differences between mouse strains is warranted in the context of metabolic disease studies. Inherent biases in mouse strains can steer immune responses and affect the outcomes of various disease models in different ways. Finally, our data indicate that the influence of BM nuclear expression on systemic metabolism may differ between experimental context and genetic background and that these factors may not be obligatory components of disease pathogenesis or thiazolidinedione therapeutic action in all contexts.
Acknowledgments
The authors thank Jon Collins and Tim Willson of GlaxoSmithKline for the synthetic ligands for LXR (GW3965 and T1317), PPARα (GW9544), PPARδ (GW742), and PPARγ (GW7845). The PPARγfl/fl and PPARδ−/− mice were provided by Ronald Evans of the Salk Institute and Frank Gonzalez of NCI, respectively.
Published, JLR Papers in Press, September 4, 2008.
Footnotes
*
This work was supported by National Institutes of Health grant HL030568.
References
- 1.Mills C. D., K. Kincaid, J. M. Alt, M. J. Heilman, and A. M. Hill. 2000. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164 6166–6173.10843666 [Google Scholar]
- 2.Abbas A. K., K. M. Murphy, and A. Sher. 1996. Functional diversity of helper T lymphocytes. Nature. 383 787–793. [DOI] [PubMed] [Google Scholar]
- 3.Ferrante A. W., Jr. 2007. Obesity-induced inflammation: a metabolic dialogue in the language of inflammation. J. Intern. Med. 262 408–414. [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil G. S. 2006. Inflammation and metabolic disorders. Nature. 444 860–867. [DOI] [PubMed] [Google Scholar]
- 5.Wellen K. E., and G. S. Hotamisligil. 2005. Inflammation, stress, and diabetes. J. Clin. Invest. 115 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisberg S. P., D. McCann, M. Desai, M. Rosenbaum, R. L. Leibel, and A. W. Ferrante, Jr. 2003. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H., G. T. Barnes, Q. Yang, G. Tan, D. Yang, C. J. Chou, J. Sole, A. Nichols, J. S. Ross, L. A. Tartaglia, et al. 2003. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 112 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotamisligil G. S., N. S. Shargill, and B. M. Spiegelman. 1993. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 259 87–91. [DOI] [PubMed] [Google Scholar]
- 9.Lumeng C. N., S. M. Deyoung, J. L. Bodzin, and A. R. Saltiel. 2007. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 56 16–23. [DOI] [PubMed] [Google Scholar]
- 10.Uysal K. T., S. M. Wiesbrock, M. W. Marino, and G. S. Hotamisligil. 1997. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature. 389 610–614. [DOI] [PubMed] [Google Scholar]
- 11.Hotamisligil G. S., P. Arner, J. F. Caro, R. L. Atkinson, and B. M. Spiegelman. 1995. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Invest. 95 2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arkan M. C., A. L. Hevener, F. R. Greten, S. Maeda, Z. W. Li, J. M. Long, A. Wynshaw-Boris, G. Poli, J. Olefsky, and M. Karin. 2005. IKK-beta links inflammation to obesity-induced insulin resistance. Nat. Med. 11 191–198. [DOI] [PubMed] [Google Scholar]
- 13.Yuan M., N. Konstantopoulos, J. Lee, L. Hansen, Z. W. Li, M. Karin, and S. E. Shoelson. 2001. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science. 293 1673–1677. [DOI] [PubMed] [Google Scholar]
- 14.Sartipy P., and D. J. Loskutoff. 2003. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc. Natl. Acad. Sci. USA. 100 7265–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanda H., S. Tateya, Y. Tamori, K. Kotani, K. Hiasa, R. Kitazawa, S. Kitazawa, H. Miyachi, S. Maeda, K. Egashira, et al. 2006. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Invest. 116 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamei N., K. Tobe, R. Suzuki, M. Ohsugi, T. Watanabe, N. Kubota, N. Ohtsuka-Kowatari, K. Kumagai, K. Sakamoto, M. Kobayashi, et al. 2006. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem. 281 26602–26614. [DOI] [PubMed] [Google Scholar]
- 17.Jiang C., A. T. Ting, and B. Seed. 1998. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature. 391 82–86. [DOI] [PubMed] [Google Scholar]
- 18.Hevener A. L., J. M. Olefsky, D. Reichart, M. T. Nguyen, G. Bandyopadyhay, H. Y. Leung, M. J. Watt, C. Benner, M. A. Febbraio, A. K. Nguyen, et al. 2007. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Invest. 117 1658–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph S. B., A. Castrillo, B. A. Laffitte, D. J. Mangelsdorf, and P. Tontonoz. 2003. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 9 213–219. [DOI] [PubMed] [Google Scholar]
- 20.Ricote M., A. C. Li, T. M. Willson, C. J. Kelly, and C. K. Glass. 1998. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 391 79–82. [DOI] [PubMed] [Google Scholar]
- 21.Lee C. H., A. Chawla, N. Urbiztondo, D. Liao, W. A. Boisvert, R. M. Evans, and L. K. Curtiss. 2003. Transcriptional repression of atherogenic inflammation: modulation by PPARdelta. Science. 302 453–457. [DOI] [PubMed] [Google Scholar]
- 22.Odegaard J. I., R. R. Ricardo-Gonzalez, M. H. Goforth, C. R. Morel, V. Subramanian, L. Mukundan, A. R. Eagle, D. Vats, F. Brombacher, A. W. Ferrante, et al. 2007. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 447 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laffitte B. A., S. B. Joseph, R. Walczak, L. Pei, D. C. Wilpitz, J. L. Collins, and P. Tontonoz. 2001. Autoregulation of the human liver X receptor alpha promoter. Mol. Cell. Biol. 21 7558–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouhlel M. A., B. Derudas, E. Rigamonti, R. Dievart, J. Brozek, S. Haulon, C. Zawadzki, B. Jude, G. Torpier, N. Marx, et al. 2007. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6 137–143. [DOI] [PubMed] [Google Scholar]
- 25.Gordon S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3 23–35. [DOI] [PubMed] [Google Scholar]
- 26.Loke P., and J. P. Allison. 2003. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc. Natl. Acad. Sci. USA. 100 5336–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willment J. A., H. H. Lin, D. M. Reid, P. R. Taylor, D. L. Williams, S. Y. Wong, S. Gordon, and G. D. Brown. 2003. Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide. J. Immunol. 171 4569–4573. [DOI] [PubMed] [Google Scholar]
- 28.Loke P., M. G. Nair, J. Parkinson, D. Guiliano, M. Blaxter, and J. E. Allen. 2002. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallardo-Soler A., C. Gomez-Nieto, M. L. Campo, C. Marathe, P. Tontonoz, A. Castrillo, and I. Corraliza. 2008. Arginase I induction by modified lipoproteins in macrophages: a peroxisome proliferator-activated receptor-gamma/delta-mediated effect that links lipid metabolism and immunity. Mol. Endocrinol. 22 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang K., S. M. Reilly, V. Karabacak, M. R. Gangl, K. Fitzgerald, B. Hatano, and C. H. Lee. 2008. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 7 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odegaard J. I., R. R. Ricardo-Gonzalez, A. Red Eagle, D. Vats, C. R. Morel, M. H. Goforth, V. Subramanian, L. Mukundan, A. W. Ferrante, and A. Chawla. 2008. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 7 496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]