Reengineering Ribosome Export (original) (raw)
Abstract
Large cargoes require multiple receptors for efficient transport through the nuclear pore complex. The 60S ribosomal subunit is one of the bulkiest transport cargoes, and in yeast three different receptors, Crm1, Mex67/Mtr2, and Arx1, collaborate in its export. However, only Crm1, recruited by the adapter Nmd3, appears to be conserved for 60S export in higher eukaryotes. We asked if export of the large subunit requires specific receptors. We made protein fusions between mutant Nmd3 and various export receptors. Surprisingly, fusions of Mex67, the tRNA exportin Los1, Mtr2, Cse1, or Msn5 to Nmd3, lacking its Crm1-dependent nuclear export signal (NES), all functioned in export. Furthermore, these chimeric proteins supported 60S export even in the presence of the Crm1 inhibitor leptomycin B, indicating that export was now independent of Crm1. These results suggest that there is not a requirement for a specific export receptor for the large subunit, as recruitment of any receptor will suffice. Finally we show that the addition of an NES directly to the 60S ribosomal subunit protein Rpl3 promotes export. These results imply remarkable flexibility in the export pathway for the 60S subunit and help explain how different export receptors could have evolved in different eukaryotic lineages.
INTRODUCTION
The nuclear export of most proteins and RNAs is mediated by karyopherins (Gorlich and Kutay, 1999; Fried and Kutay, 2003; Weis, 2003; Kohler and Hurt, 2007), a conserved group of soluble factors that facilitate the unidirectional transport of cargo molecules through the nuclear pore complex (NPC) in the nuclear envelope (Mattaj and Englmeier, 1998; Macara, 2001). Many nucleoporins, the proteins that constitute the NPC, contain natively unfolded domains (Denning et al., 2003) with short hydrophobic FG repeats, often in the context of GLFG or FXFG. These FG repeats are binding sites for karyopherins (reviewed in Tran and Wente, 2006). Four karyopherins in yeast are known to be involved in export. These are Crm1, which recognizes leucine-rich nuclear export sequences (NESs), Los1, the tRNA exporter, Cse1 which exports importin-α and Msn5, involved in the export of certain protein cargoes (reviewed in Pemberton and Paschal, 2005). The binding of these exportins to their substrates in the nucleus depends on the formation of a ternary complex with RanGTP that enhances the exportin-cargo interaction (reviewed in Lund and Dahlberg, 2001). On translocation to the cytoplasm, hydrolysis of GTP on Ran, stimulated by a cytoplasmic GTPase-activating protein (GAP) and Ran binding protein 1 (Yrb1), results in a conformational change necessary for disassembly of the export complex (Petosa et al., 2004). The directionality of export is controlled by the high concentration of Ran-GTP in the nucleus versus cytoplasm that promotes assembly of exportins with their cargo only in the nucleus. In contrast, the export of mRNA is driven by the heterodimer Mex67/Mtr2, unrelated to the karyopherins. Directionality of mRNA export appears to depend on the DEAD box helicase Dbp5 (Lund and Guthrie, 2005 and reviewed in Cole and Scarcelli, 2006; Stewart, 2007).
The actual mechanism of translocation through the NPC is not well understood, but the channel of the NPC is occupied by the unstructured domains of nucleoporins (Tran and Wente, 2006; Patel et al., 2007), held together in a meshwork by weak hydrophobic interactions between the FG repeats (Ribbeck and Gorlich, 2002) or extended as a polymer brush (Lim et al., 2006). Thus, translocation through the channel requires an export complex to partition into this hydrophobic environment (Ribbeck and Gorlich, 2002). Importin-β transport receptors, the group to which Crm1 belongs, contain multiple shallow hydrophobic pockets interspersed between each HEAT repeat that allow their interaction with the hydrophobic FG repeats of the nucleoporins (Bayliss et al., 2000; Fribourg et al., 2001). The multiple binding sites for FG repeats enable karyopherins with their associated cargo to pass through the hydrophobic channel. However, large cargoes require multiple export receptors for efficient transport (Ribbeck and Gorlich, 2002), probably to help cover the hydrophilic surface to facilitate their partitioning into the hydrophobic environment of the channel of the NPC.
Perhaps the bulkiest cargo to pass through the NPC in yeast is the large ribosomal subunit. Recent work has revealed that the large subunit utilizes three different export receptors: Crm1, which is recruited to the subunit via Nmd3 (Ho et al., 2000b; Gadal et al., 2001); the heterodimeric mRNA transporter Mex67/Mtr2 (Yao et al., 2007); and Arx1, a noncanonical receptor specific to the large subunit that binds directly to the subunit and to nucleoporins (Bradatsch et al., 2007; Hung et al., 2008). Whereas the function of Nmd3 as an export factor for the 60S subunit is conserved from yeast to humans (Thomas and Kutay, 2003; Trotta et al., 2003), Tap/p15, the human orthologues of Mex67/Mtr2, and Ebp1, the human ortholog of Arx1, do not appear to be involved in ribosome export in human cells (Bradatsch et al., 2007; Yao et al., 2007). Although we do not yet know what proteins act in conjunction with Crm1 for exporting 60S subunits in human cells, it appears that different proteins have evolved as export factors in different eukaryotic lineages. Considering that ribosome export is an essential pathway in eukaryotes and that many ribosome biogenesis factors are highly conserved, it is surprising that there appears to be such flexibility in the export receptors used for 60S subunit export. Here, we have asked if there is a particular requirement for utilizing specific export receptors in 60S export in yeast. We show that essentially any of the export receptors can replace Crm1 if they are directly recruited to the subunit as fusions to Nmd3. In addition, we have previously posed the question of why the Crm1-dependent NES for the 60S subunit is carried by an adapter protein, as it would seem more economical for the NES to be integral to the ribosome. We show that an NES can indeed work in cis to the ribosome when it is fused directly to a ribosomal protein.
MATERIALS AND METHODS
Strains, Plasmids, and Media
All Saccharomyces cerevisiae strains used in this study are listed in Table 1. All strains were grown at 30°C, unless otherwise indicated, in rich medium (yeast extract-peptone) or synthetic dropout medium, containing 2% glucose.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| PSY1687 | _MAT_α ade2 his3 leu2 trp1 ura3 mex67::HIS3 (pUN100: LEU2 mex67-5) | Segref et al. (1997) |
| AJY734 | _MAT_α ade2 ade3 leu2 lys3 ura3 his3 nmd3-4ts | Ho and Johnson (1999) |
| AJY1950 (SL348) | _MATa leu2 ura3 ade2 ade3 arx1_Δ nmd3_Δ_C14 (pAJ1029: ADE3 URA3 ARX1) | Hung et al. (2008) |
| AJY2110 | MATa ura3_Δ_0 his3_Δ_1 leu2_Δ_0 lys2_Δ_0 nmd3::KanMX (pAJ112: URA3 NMD3) | Hedges et al. (2006) |
| AJY2974 | MATa leu2 lys3 ura3 his3 nmd3-4ts crm1(T539C)-HA | This study |
Plasmids used in this work are listed in Table 2. pAJ1872 (MEX67) was made by moving a ClaI-SacI fragment from pAJ528 into pRS423. To create pAJ1882 (MEX67-nmd3_Δ_100-GFP) and pAJ2079 (mex67-5-nmd3_Δ_100-GFP), MEX67 was amplified with primers AJO994 (CCTGAATTCAGCGGATTTCACAATGTTGG) and AJO995 (GGAGAATTCGTTAATTAAGTTGTTGAACTGCACAAATGCTTC) from either wild-type or mex67-5 genome DNA. The products were digested with EcoRI and ligated into the EcoRI site at nucleotide 4 of NMD3 coding sequence in pAJ757 (nmd_Δ_100-GFP). pAJ1892 was made by moving the SstI to XmaI fragment from pAJ1882 to pAJ535. To make pAJ2066 (nmd3_Δ_100-MTR2-GFP), PCR was carried out with primers AJO1006 (CCTTTAATTAAAATTTTTAGCAGAGAATCCTCG) and AJO1073 (CCTCCCGGGATGAACACCAATAGTAATACTATG). The fragment was digested with XmaI and PacI and ligated into the same sites of pAJ757. pAJ2076, pAJ2077, and pAJ2078 were made in the same manner but with different primer pairs: AJO1106 (CTGCCCGGGAACAACTTAATTAACATGTCCGATTTGGAAACCGT) and AJO1107 (CAGCCCGGGATTACCAACTAATAATTGAT) for pAJ2076, AJO1108 (CTGCCCGGGAACAACTTAATTAACATGCTAGAACGGATTCAGCA) and AJO1109 (CAGCCCGGGTTGACCTTGCTTTAAAACAG) for pAJ2077, and AJO1110 (CTGCCCGGGAACAACTTAATTAACATGGATTCCACAGGCGCTTC) and AJO1111 (CAGCCCGGGGTTGTCATCAAAGAGATTAC) for pAJ2078. The PCR fragments were digested with XmaI and ligated into XmaI-cut pAJ757. pAJ2084, pAJ2085, pAJ2086, pAJ2087, and pAJ2088 were made similarly. The receptor containing fragments were moved as an EcoRI fragment (pAJ1882), an XmaI to PacI fragment (pAJ2066), or XmaI fragments (pAJ2076, pAJ2077, and pAJ2078) into the same site(s) of pAJ2083 (_nmd3[L263P F318I]Δ_100). pAJ2089 (RPL3-NES), pAJ2090 (RPL11B-NES), pAJ2091 (RPL12B-NES), and pAJ2094 (RPL25-NES) were made by PCR amplification of the NES of NMD3 using primers AJO329 (CTGCATCCAGTATACACACCCA) and AJO1118 (GTCTTAATTAACGATGAAGACGCTCCACAA) and ligating the PacI-HindIII NES-containing fragment into pAJ1090, pAJ1092, pAJ1093, and pAJ1127.
Table 2.
Plasmids used in this study
| Plasmids | Relevant markers | Source |
|---|---|---|
| pAJ123 | NMD3 LEU2 CEN | Ho and Johnson (1999) |
| pAJ1032 | ARX1 LEU2 CEN | Hung, unpublished |
| pAJ528 | MEX67 URA3 2μ | Ho et al. (2000b) |
| pAJ534 | nmd_Δ_50-myc LEU2 CEN | Ho et al. (2000b) |
| pAJ535 | nmd_Δ_100-myc LEU2 CEN | Ho et al. (2000b) |
| pAJ538 | NMD3-myc LEU2 CEN | Ho et al. (2000b) |
| pAJ755 | NMD3-GFP URA3 CEN | Hedges, unpublished data |
| pAJ757 | nmd3_Δ_100-GFP URA3 CEN | Hedges, unpublished data |
| pAJ908 | RPL25-GFP URA3 CEN | Kallstrom et al. (2003) |
| pAJ1025 | ARX1-GFP LEU2 CEN | Meyer et al. (2007) |
| pAJ1121 | GAL:LSG1 LEU2 CEN | West and Johnson, unpublished data |
| pAJ1129 | GAL:LSG1(K349T) LEU2 CEN | West and Johnson, unpublished data |
| pAJ1359 | nmd3_Δ_NLS-myc LEU2 CEN | Hedges et al. (2006) |
| pAJ1872 | MEX67 HIS3 2μ | This study |
| pAJ1882 | MEX67-nmd3_Δ_100-GFP URA3 CEN | This study |
| pAJ1892 | MEX67-nmd3_Δ_100-myc LEU2 CEN | This study |
| pAJ2066 | nmd3_Δ_100-MTR2 -GFP URA3 CEN | This study |
| pAJ2076 | nmd3_Δ_100-CSE1-GFP URA3 CEN | This study |
| pAJ2077 | nmd3_Δ_100-LOS1-GFP URA3 CEN | This study |
| pAJ2078 | nmd3_Δ_100-MSN5-GFP URA3 CEN | This study |
| pAJ2079 | (mex67-5)-nmd3_Δ_100-GFP URA3 CEN | This study |
| pAJ2084 | _MEX67-nmd3[L263P F318I]Δ_100-GFP URA3 CEN | This study |
| pAJ2085 | _nmd3[L263P F318I]Δ_100-MTR2-GFP URA3 CEN | This study |
| pAJ2086 | _nmd3[L263P F318I]Δ_100-CSE1-GFP URA3 CEN | This study |
| pAJ2087 | _nmd3[L263P F318I]Δ_100-LOS1-GFP URA3 CEN | This study |
| pAJ2088 | _nmd3[L263P F318I]Δ_100-MSN5-GFP URA3 CEN | This study |
| pAJ2089 | RPL3-NES LEU2 CEN | This study |
| pAJ2090 | RPL11B-NES LEU2 CEN | This study |
| pAJ2091 | RPL12B-NES LEU2 CEN | This study |
| pAJ2093 | RPL8B-NES LEU2 CEN | This study |
| pAJ2094 | RPL25-NES LEU2 CEN | This study |
| pAJ2095 | RPL32-NES LEU2 CEN | This study |
| pAJ2218 | MEX67-GFP LEU2 CEN | This study |
| pAJ2226 | nmd3_Δ_100-MSN5-myc LEU2 CEN | This study |
| pAJ2227 | nmd3_Δ_100-LOS1-myc LEU2 CEN | This study |
Microscopy
Overnight cultures of cells were diluted to an OD600 ∼0.1 in fresh media and then incubated for 5–6 h at appropriate temperatures. Hoechest dye was added at a final concentration of 4 μM to stain the nucleus. For leptomycin B (LMB) experiments, cells were concentrated 10-fold, and LMB (LC Laboratories, Cambridge, MA) was added at a final concentration of 0.1 μg/ml. Fluorescence was visualized on a Nikon E800 microscope fitted with an 100× objective (Melville, NY) and a Photometrics CoolSNAP ES digital camera (Tucson, AZ) controlled with the NIS-Elements AR 2.10 software (Nikon).
Sucrose Gradient Analysis
For polysome profile assays, cultures were collected at OD600 ∼0.2–0.3. Cycloheximide (200 μg/ml final concentration) was then added, and cells were immediately harvested by pouring onto ice and centrifugation. Extracts were prepared by glass bead extraction in polysome lysis buffer (10 mM Tris-HCl, pH 7.5, 100 mM KCl, 10 mM MgCl2, 6 mM BME, and 200 μg/ml cycloheximide). Nine OD260 nits of protein extract were loaded onto linear 7–47% sucrose gradients in polysome lysis. After a 2.5-h spin at 40,000 rpm in a Beckman SW40 rotor (Fullerton, CA), gradient fractions were collected on an ISCO density gradient fractionator (Lincoln, NE), continuously measuring absorbance at 254 nm.
To determine the ratio between 60S and 40S subunits, cell lysis buffer and sucrose solutions were made in low magnesium buffer (50 mM Tris, pH 7.4, 50 mM NaCl, and 1 mM DTT). Subsequent conditions for sucrose gradient sedimentation and analysis were exactly as described above.
RESULTS
High-copy Mex67 Can Partially Rescue the NES Function of Nmd3
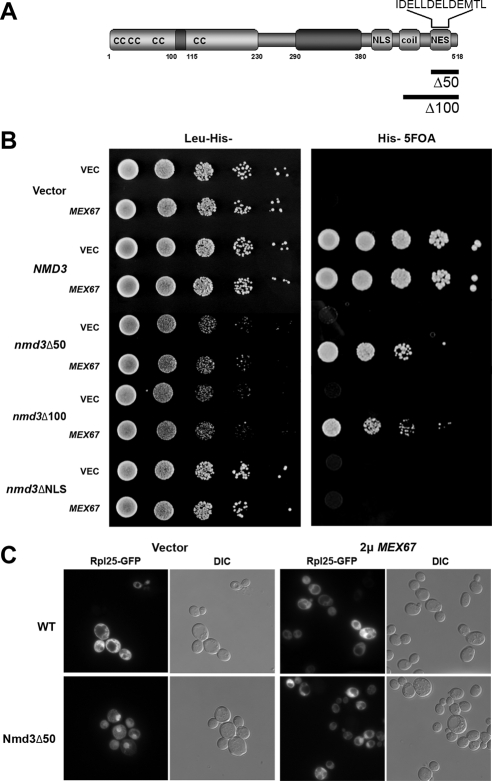
We previously identified MEX67 as a high-copy suppressor of an nmd3–1 mutant (Ho et al., 2000b). In the present work, we found that high-copy MEX67 improved the growth rate of cells containing truncated versions of Nmd3, Nmd3Δ50, or Nmd3Δ100 (C-terminal truncations of 50 or 100 amino acids, respectively; Figure 1, A and B). These truncations remove the Crm1-dependent leucine-rich NES of Nmd3. The suppression appeared to be specific for nmd3 mutants defective for export because MEX67 did not suppress the growth defect on an nmd3 mutant deleted of its nuclear localization signal (Figure 1B, _nmd3_ΔNLS). These results are consistent with the recent report that Mex67 acts as an export receptor for the 60S subunit in yeast (Yao et al., 2007) and suggest that recruitment of Mex67 to the large subunit is limiting in the export pathway. That Mex67 is limiting is supported by the finding that Mex67 and Mtr2 levels are increased on pre-60S subunits if their export is blocked by deletion of Arx1 or mutations in Nmd3 that disrupt its interaction with Crm1 (Hung et al., 2008). High-copy MEX67 could not replace NMD3 (Figure 1B, Vector, 5FOA), consistent with the idea that Nmd3 has a function in ribosome biogenesis beyond its role in export.
Figure 1.
High copy Mex67 rescues the growth defect and promotes 60S export in nmd3 NES mutants. (A) Schematic of Nmd3 primary structure. (B) Growth analysis of AJY2110 (nmd3::KanMX pAJ112 NMD3 URA3) expressing different mutant nmd3 alleles and high-copy MEX67 (pAJ1872). Tenfold serial dilutions of AJY2110 transformed with vector (pRS415), NMD3 (pAJ538), nmd3_Δ_50 (pAJ534), nmd3_Δ_100 (pAJ535), or nmd3_Δ_NLS (pAJ1359) in combination with vector (pRS423) or high-copy MEX67 were spotted onto His− Leu− dropout or His− 5FOA medium and incubated at 30°C. (C) AJY2110 (_nmd3_Δ) with wild-type NMD3 (WT) or nmd3_Δ_50 in combination with vector alone or high-copy (2μ) MEX67 were grown in selective medium at 30°C to OD600 ∼0.3 and GFP, and whole cells were visualized by fluorescence and DIC microscopy, respectively.
To confirm that suppression of mutant Nmd3 lacking its Crm1-dependent NES was due to bypassing the 60S export defect of the nmd3 mutant, we asked if we could detect enhanced export of 60S subunits using Rpl25-eGFP as a reporter for 60S localization. Indeed, we observed a modest increase in Rpl25-eGFP signal in the cytoplasm in nmd3_Δ_50 mutant cells when MEX67 was overexpressed (Figure 1C). However, we were unable to detect a change in the steady-state localization of Nmd3Δ50 (data not shown). This is explained by the fact that the mutant protein retains a strong NLS but lacks an NES. Thus, reimport is likely to exceed the rate of export, which is inefficient in the absence of Crm1 recruitment. Sucrose gradient analysis of polysomes and total subunit levels revealed no evident difference in 60S levels between nmd3_Δ_50 cells that contained high-copy MEX67 or control vector (data not shown). Thus, the level of suppression of 60S export by overexpression of MEX67 is modest.
A Chimeric Mex67-Nmd3Δ100 Fusion Protein Supports Export
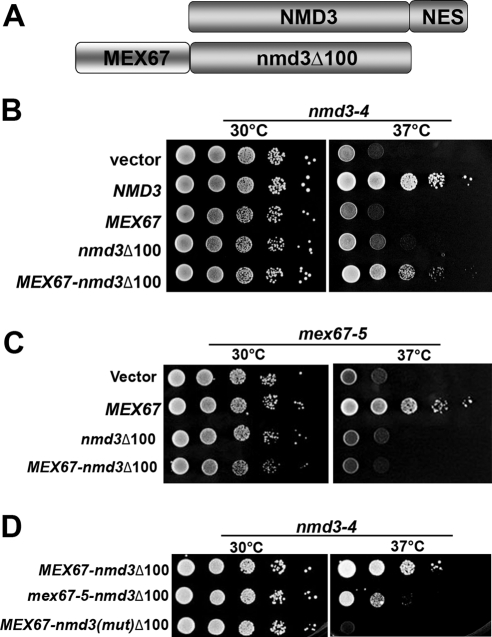
Export of the 60S subunit normally utilizes the three different receptors Crm1, Mex67/Mtr2, and Arx1 simultaneously. Because Mex67 appears to be limiting for 60S export, suppression of _nmd3_Δ50 by high-copy expression of MEX67 is probably the result of increasing the occupancy of Mex67 at its binding site on the subunit. To ask if we could bypass the export defect of an nmd3 mutant by recruiting an additional copy of Mex67 to the ribosome, we fused Mex67 to the amino terminus of Nmd3Δ100-GFP (Figure 2A). Nmd3Δ100 is completely lacking of any signals implicated in nuclear export to recruit Crm1 but retains the ability to bind to 60S subunits (Ho et al., 2000b; Gadal et al., 2001). The arrangement of this protein fusion was based on the fact that fusions to the C-terminus of Mex67 and the N-terminus of Nmd3 are functional (Ho et al., 2000a; Huh et al., 2003). Fusion of Mex67 to Nmd3Δ100 partially complemented the temperature sensitivity of nmd3-4 (Figure 2B), a temperature-sensitive lethal mutant that fails to export 60S subunits and shows a severe instability of 25S rRNA (Ho and Johnson, 1999; Ho et al., 2000b). On the other hand, this fusion did not complement a mex67-5 mutant (Figure 2C) and thus did not provide full Mex67 function. Because the mex67-5 mutant is defective for mRNA export (Santos-Rosa et al., 1998), the lack of complementation of mex67-5, but complementation of nmd3-4, suggests that this fusion was not functional for mRNA export but, remarkably, was able to support Nmd3-like function. As further controls to demonstrate that this fusion protein required both the 60S binding function of Nmd3 and the export function of Mex67, we introduced specific mutations into the fusion protein. We have shown previously that mutant Nmd3 containing two point mutations (L263P and F318I) is severely impaired for binding to the 60S subunit and is consequently nonfunctional (Hedges et al., 2006). The introduction of these point mutations into the Mex67-Nmd3Δ100 fusion protein abolished its ability to complement nmd3-4 (Figure 2D), indicating that the chimeric protein functions through binding to the 60S subunit. In addition, we introduced the mex67-5 mutation into the Mex67 moiety of the fusion protein. This mutation disrupts the interaction of Mex67 with Mtr2 (Santos-Rosa et al., 1998), and its introduction into the chimeric protein significantly reduced its activity. Thus, the Mex67 moiety also contributes to function of the chimeric protein.
Figure 2.
Fusion of Mex67 to NES-deficient nmd3 suppresses nmd3-4ts at nonpermissive temperature. (A) Schematic diagram showing the _MEX67-nmd3_Δ100 construct. (B) Tenfold serial dilutions of AJY734 (nmd3-4) transformed with empty vector, NMD3 (pAJ409), MEX67 (pAJ528), nmd3_Δ_100-GFP (pAJ757), and MEX67-nmd3_Δ_100-GFP (pAJ1882) were spotted onto selective medium and incubated for 3 d at the indicated temperature. (C) Growth test of AJY1231 (mex67-5) transformed with empty vector, MEX67 (pAJ528), nmd3_Δ_100-GFP (pAJ757), or MEX67-nmd3_Δ_100-GFP (pAJ1882) were spotted onto selective medium and incubated at 30 or 37°C. (D) MEX67-nmd3_Δ_100-GFP (pAJ1882), (mex67-5)-nmd3_Δ_100-GFP (pAJ2079), or _MEX67-nmd3[L263P F318I]Δ_100-GFP (pAJ2084) were transformed into AJY734 (nmd3-4) and tested to complementation as in B.
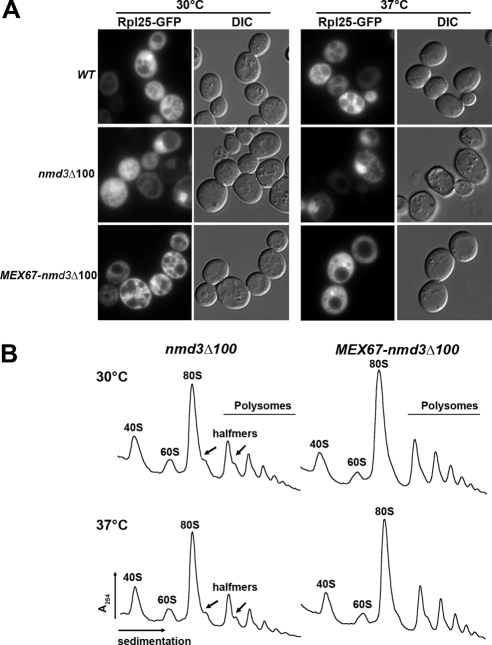
We next examined 60S subunit export in cells expressing Mex67-Nmd3Δ100 by monitoring Rpl25-eGFP. As expected, wild-type Nmd3 supported efficient export of 60S subunits: all cells showed cytoplasmic Rpl25-eGFP. Nmd3Δ100, without Mex67, conferred a strong block to 60S export (Figure 3A, middle panel), at both permissive and restrictive temperatures, because of its dominant negative nature and inability to complement nmd3-4 (Ho et al., 2000b). In contrast, the chimeric Mex67-Nmd3Δ100 fusion protein supported an intermediate level of export (Figure 3A, bottom panel), with 64 ± 7% cells showing cytoplasmic Rpl25-eGFP signal. This result is comparable to the growth test, implying that Mex67-Nmd3Δ100 fusion protein can partially complement Nmd3 export function. This was also reflected in increased 60S subunit levels detected by sucrose gradient analysis of free subunits and polysomes (Figure 3B). In the presence of Nmd3Δ100, halfmers (Figure 3B, arrows) are seen at both permissive and restrictive temperatures. Halfmers result from a deficiency in 60S subunits, causing initiation complexes with unjoined 40S subunits to stall on mRNAs. In the presence of the Mex67-Nmd3Δ100 fusion protein, the overall heights of the polysome peaks are increased and halfmers are reduced (Figure 3B). We quantified the ratio of polysomes to monosomes in these gradients. For nmd3Δ100 the ratio decreased from 0.88 to 0.82 when cells were shifted to restrictive temperature. The ratio remained constant at 0.94 when the Mex67-Nmd3Δ100 sample was shifted to restrictive temperature, supporting the conclusion that this fusion protein promotes export. Cells expressing wild-type Nmd3 gave a ratio of 0.97 (data not shown). Thus, this chimeric protein functions in export by replacing the Crm1-dependent NES of NMD3 with a direct protein fusion to Mex67. Under these conditions, two Mex67 molecules are recruited to the same subunit (Supplemental Figure S1). One wild-type Mex67 molecule would bind to its native binding site, whereas a second Mex67 is recruited as a fusion protein to the Nmd3-binding site, where it functionally replaces Crm1.
Figure 3.
The Mex67-Nmd3Δ100 fusion protein supports 60S subunit export in an nmd3-4 mutant. (A) AJY734 (nmd3-4) with NMD3-myc (pAJ538), nmd3_Δ_100-myc (pAJ535), and MEX67-nmd3_Δ_100-myc (pAJ1892) were grown to early log phase and then shifted to 37°C for 120 min. The localization of Rpl25-GFP (pAJ908) were detected by fluorescence microscopy (B) AJY734 (nmd3-4) with nmd3_Δ_100-myc (pAJ535) and MEX67-nmd3_Δ_100-myc (pAJ1892) were grown in selective medium to an OD600 ∼0.3 at 30°C or shift to 37°C for 30 min before harvest. Cell extracts were fractioned on 7–47% sucrose gradients containing 10 mM Tris, pH 7.5, 100 mM KCl, 6 mM βME, 10 mM MgCl2, and 200 μg/ml cycloheximide.
Wild-type Nmd3 recruits Crm1 for nuclear export. Consequently, its export is inhibited by the Crm1-specific inhibitor LMB. If the Mex67-Nmd3Δ100-GFP fusion acted independently of Crm1, its localization should not be sensitive to LMB. Because wild-type Crm1 in yeast is insensitive to LMB, we tested this assumption in an LMB-sensitive crm1(T539C) mutant (Neville and Rosbash, 1999). Indeed, in a time course of LMB treatment, the localization of Mex67-Nmd3Δ100-GFP was not affected by the addition of LMB, whereas wild-type Nmd3-GFP accumulated in the nucleus within 15 min of treatment with LMB (Supplemental Figure S2).
Other Export Receptors Can Also Substitute for Crm1
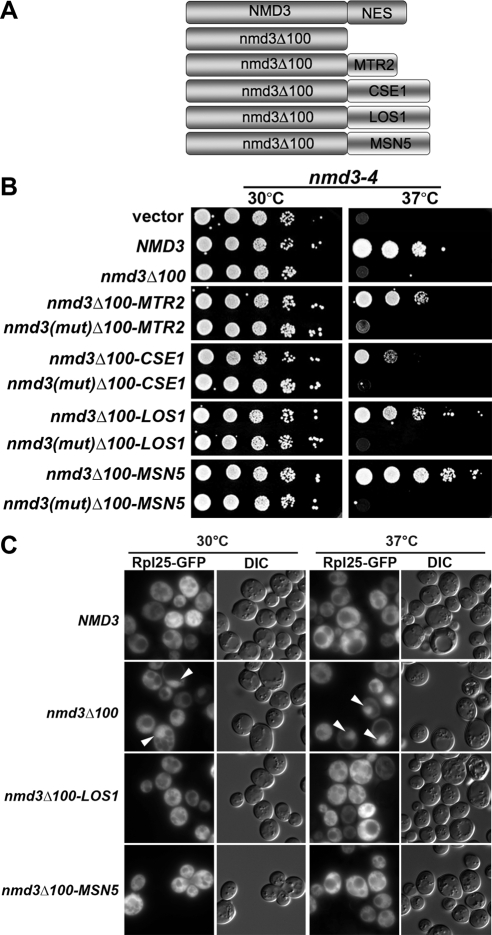
In view of the finding that Mex67 fused to an NES-deficient mutant Nmd3 protein promoted 60S export, we asked if other export receptors would suffice. We used Mtr2, Cse1, Los1, and Msn5. We fused each of these proteins separately to the C-terminus of Nmd3Δ100 (Figure 4A) and asked if they could support 60S export.
Figure 4.
Fusion of other receptors to NES-deficient Nmd3 supports ribosome export. (A) Schematic diagram showing the fusion protein constructs between different receptors and the NES deficient nmd3. (B) Tenfold serial dilutions of AJY734 (nmd3-4) containing empty vector, NMD3 (pAJ409), nmd3_Δ_100-GFP (pAJ757), nmd3_Δ_100-MTR2 -GFP (pAJ2066), _nmd3(mut)Δ_100-MTR2-GFP (pAJ2085), nmd3_Δ_100-CSE1 –GFP(pAJ2076), _nmd3 (mut)Δ_100-CSE1-GFP (pAJ2086), nmd3_Δ_100-LOS1 - GFP (pAJ2077), _nmd3(mut)Δ_100-LOS1-GFP (pAJ2087), nmd3_Δ_100-MSN5 -GFP (pAJ2078), and _nmd3(mut)Δ_100-MSN5-GFP (pAJ2088) were spotted onto selective medium and incubated for 6 d at the indicated temperature. nmd3(mut) is nmd3[L263P F318I]. (C) AJY734 (nmd3-4) expressing Rpl25-GFP (pAJ908) and wild-type NMD3-myc (pAJ538), nmd3_Δ_100-myc (pAJ535), nmd3_Δ_100-LOS1-myc (pAJ2227), or nmd3_Δ_100-MSN5-myc (pAJ2226) was grown in selective medium to early log phase at 30°C and then shifted to 37°C for 60 min before visualizing GFP.
Each fusion restored Nmd3-like function in nmd3-4 at nonpermissive temperature, but to varying degrees (Figure 4B). Msn5 and Los1 showed the best complementation, whereas the Mtr2 fusion was considerably weaker in activity and the Cse1 weaker yet. In each case, complementation was lost when the nmd3 double point mutation (L263P and F318I) was introduced into the fusion protein (Figure 4B), indicating that binding to the 60S subunit was required for function. In addition, each fusion protein was able to complement the lethality of an _nmd3_Δ mutant (data not shown), indicating that they are functional as the sole copies of Nmd3. Like the Mex67 fusion protein, the localization of these chimeric proteins also depends on the nature of the fusion receptors. The Cse1 and Msn5 fusion proteins were localized predominantly in the nucleus while the localization of the Los1 fusion protein was more like the Mex67 fusion protein, present in the nucleus, cytoplasm, and nuclear envelope (Supplemental Figure S3).
To demonstrate that these fusion proteins indeed supported export, we monitored Rpl25-eGFP localization in an nmd3-4 mutant containing the various fusion proteins at restrictive temperature. As seen in Figure 4C, both the Los1 and Msn5 fusion proteins supported efficient export of Rpl25-eGFP. Approximately 72 ± 1 and 79 ± 4% of the cells in Los1- and Msn5- fusion protein–containing strains, respectively, showed cytoplasmic Rpl25-eGFP localization. In conclusion, there is a correlation between complementation observed in the growth test (Figure 4A) and the efficiency of export, indicated by the percentage of cells with cytoplasmic Rpl25-GFP signal. Msn5-Nmd3Δ100 fusion protein showed the best complementation in nmd3-4 strain and the most efficient export compared with Mex67- or Los1-fusion proteins. We did not assay the Mtr2 or Cse1 fusions because of their poor complementation of nmd3-4. These results show that there is not a specific requirement for Crm1 in ribosome export and that multiple other receptors suffice, if recruited to the subunit.
For further evidence that the fusion proteins bound to the 60S subunit, we tested the Mex67-Nmd3Δ100 and the Nmd3Δ100-Msn5 fusion proteins for cosedimentation with the 60S subunits in sucrose density gradients. Indeed, both proteins were found exclusively at the position of free 60S (Supplemental Figure S4), as we have previously observed for Nmd3 (Ho et al., 2000a). We expect that all the fusion proteins would show cosedimentation with 60S subunits, reflecting the function of their Nmd3 domains, and because of the genetic evidence that a mutation that disrupts Nmd3 binding to 60S rendered these fusions nonfunctional (Figure 4B).
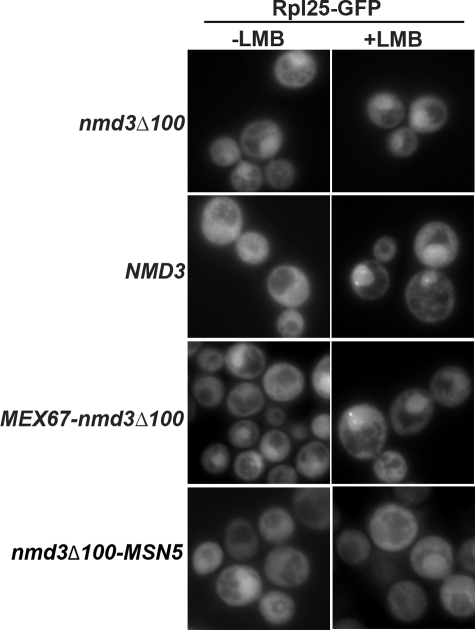
If export is driven by the chimeric receptors, it should be independent of Crm1. We tested this by examining the LMB sensitivity of 60S export in an nmd3-4ts crm1(T539C) mutant containing chimeric receptor fusion proteins. To detect 60S export in the absence of functional Nmd3, we shifted cells at early log phase to nonpermissive temperature to inactivate nmd3-4 so that 60S export would be dependent on the fusion protein. LMB was added thereafter. In wild-type cells Rpl25-GFP was cytoplasmic in the absence of LMB but was trapped in the nucleus after 30 min of LMB treatment (Figure 5). Consistent with previous studies (Ho et al., 2000b), Nmd3Δ100 severely blocked 60S export, regardless of LMB treatment. In the strains with Mex67-Nmd3Δ100 or Nmd3Δ100-Msn5 the percentage of cells showing nuclear localization of Rpl25-GFP did not change upon treatment with LMB (48 ± 4 to 48 ± 7 for the Mex67 fusion and 20 ± 1 to 18 ± 6 for the Msn5 fusion; Figure 5). The lower overall nuclear retention of Rpl25-GFP in the Nmd3Δ100-MSN case reflects the better complementation of growth by this construct compared with the Mex67 fusion (data not shown). Thus, 60S export by the chimeric fusion proteins was independent of Crm1.
Figure 5.
60S export by the chimeric Mex67-nmd3Δ100 fusion protein is insensitive to LMB. Rpl25-GFP (pAJ908) was monitored in AJY2974 (_nmd3-4 crm1_T539C) with pAJ535 (_nmd3_Δ100), pAJ538 (NMD3), or pAJ1892 (MEX67-nmd3_Δ_100) or pAJ2226 (nmd3_Δ_-MSN5). The cells were diluted into fresh medium from overnight cultures and incubate at 30°C for 30 min. After the temperature shift to 37°C for 1.5 h, LMB (0.1 μg/ml) was added and the cultures were incubated at 37°C for another 30 min before microscopy.
Release of the Chimeric Receptor Proteins Depends on Lsg1
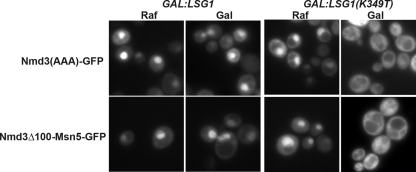
The directionality of export by receptors of the importin-β family, such as Crm1, is determined by their release from cargo in the cytoplasm upon dissociation from Ran (Dahlberg and Lund, 1998; Macara, 2001; Pemberton and Paschal, 2005). However, the chimeric receptor fusions that we generated are bound to the ribosome through the Nmd3 moiety and are not expected to be released in the same manner. Wild-type Nmd3 is released from ribosomes by the cytoplasmic GTPase Lsg1 (Hedges et al., 2005) and not by hydrolysis of GTP on Ran. Expression of the dominant negative LSG1(K349T) mutant traps wild-type Nmd3 on cytoplasmic 60S ribosomes (Hedges et al., 2005). We tested if this mutant could also trap the chimeric receptors in the cytoplasm. Because the Mex67-Nmd3Δ100 and Nmd3Δ100-Los1 fusion proteins were localized in both nucleus and cytoplasm with nuclear envelope decoration, their mislocalization may not be easily detected. Consequently, we took advantage of the nuclear localization of Nmd3Δ100-Msn5, and quantified the fraction of cells with cytoplasmic or nuclear signal upon LSG1(K349T) overexpression. Whereas 32 ± 3% of cells showed cytoplasmic localization of Nmd3Δ100-Msn5 before expression of the dominant negative LSG1 [Figure 6, bottom panel, LSG1(K349T) raf] after 2 h of induction in galactose, the fraction of cells showing cytoplasmic Nmd3Δ100-Msn5 rose to 89 ± 1% [Figure 6, bottom panel, LSG1(K349T) gal]. As a control, we observed similar results with nmd3(AAA), a mutant version of Nmd3 that displays a nuclear bias (Hedges et al., 2005). Although dissociation of export receptors from their cargo normally requires GTP hydrolysis on Ran or the RNA helicase Dbp5, in the case of Mex67 release, the mechanism of release of these chimeric receptors has been altered to depend on the GTPase Lsg1.
Figure 6.
Release of the chimeric fusion proteins from the ribosome in the cytoplasm requires the GTPase Lsg1. AJY734 (nmd3-4) with pAJ758 (nmd3AAA-GFP) or pAJ2078 (nmd3_Δ_-MSN5-GFP) were grown in the raffinose-containing medium to early log phase at 30°C. Either wild-type LSG1 (pAJ1121) or dominant negative mutant LSG1(K349T) (pAJ1129) was over expressed by adding 1% galactose. Then the cultures were shifted to 37°C for another 2 h.
The NES of Nmd3 Can Be Moved to Ribosomal Proteins
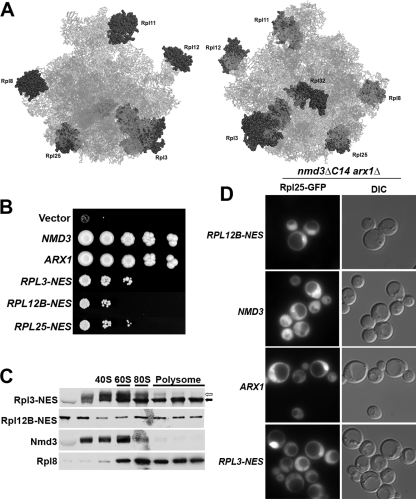
Nmd3 has evolved in eukaryotic cells as an adapter protein to provide the Crm1-dependent NES for the 60S subunit in trans to the ribosome. Although this could provide important mechanisms for regulating export of the pre-60S particle it would appear to be more economical for the NES to be provided by a ribosomal protein in cis. To ask if an NES can work in cis, we fused the NES of Nmd3 to the C-terminus of various ribosomal proteins (rproteins) of the 60S subunit. Fusions were made to Rpl3, Rpl8B, Rpl11B, Rpl12B, Rpl25, and Rpl32 (Figure 7A). We expected that not all these fusion proteins would function properly or efficiently for various reasons. The fusion could block efficient assembly of the rprotein into the subunit or its function post assembly. The NES could direct export of the rprotein before it was incorporated into the subunit or export of the subunit before it was correctly matured. Lastly, the NES may be inaccessible to Crm1. Consequently, to test these fusions we used a genetic background that would give us a sensitive readout of function. We used a strain deleted for the nonessential export receptor Arx1 that also expressed a mutant Nmd3 (Nmd3ΔC14) lacking the C-terminal 14 amino acids. This mutation removes the last leucine of the NES of Nmd3 (Hung et al., 2008) and, consequently, impairs export. The arx1 nmd3_Δ_C14 double mutant is synthetic lethal (Hung et al., 2008) and can be kept alive by the expression of a factor that provides nuclear export function, e.g., Arx1, Nmd3, or perhaps an rprotein bearing an NES. Among the proteins that we tested, we found that the Rpl3-NES construct gave the strongest complementation (Figure 7B). Rpl25 and Rpl12B complemented less well and Rpl11B was only slightly better than empty vector (Figure 7B and data not shown). Other fusions showed no difference from an empty vector control (data not shown). These rprotein-NES fusions were incorporated into ribosomes, indicated by their cosedimentation with ribosomal subunits in sucrose density gradients (Figure 7C). Only Rpl3 containing a functional NES was able to complement _arx1_Δ nmd3_Δ_C14 (Supplemental Figure S5A) although Rpl3 with either a functional or mutant NES was able to complement an rpl3Δ (Supplemental Figure S5B). In comparison to wild-type Nmd3 and Arx1, the rprotein-NES fusions complemented poorly (Figure 7B). Nevertheless, they did support cell growth. Furthermore, analysis of 60S export by monitoring Rpl25-eGFP localization confirmed that at least the Rpl3-NES fusion supported 60S export (Figure 7D). For technical reasons, it was not possible to make comparisons to an empty vector that was unable to complement the export defect of the double mutant. Consequently, we used cells expressing the poorly complementing Rpl12B-NES as a baseline for export because these cells show a very strong nuclear accumulation of Rpl25-eGFP in essentially all cells, reflecting their poor 60S export. In contrast, cells expressing the Rpl3-NES fusion showed reduced levels of nuclear retention, indicating export of Rpl25 out of the nucleus. Even greater levels of export were observed with expression of ARX1. However, only wild-type NMD3 supported efficient export, with essentially no nuclear retention of Rpl25-GFP. The lower efficiency of export by Rpl3-NES compared with Arx1 and Nmd3 is consistent with the lower growth rate supported by this fusion compared with NMD3 or ARX1. These results show that export of the 60S subunit can be supported by a Crm1-dependent NES in cis on the ribosome.
Figure 7.
Adding an NES to the ribosome in cis bypasses an nmd3 mutant. (A) Schematic of the 60S subunit showing the positions of the ribosomal proteins to which NESs were fused. Left, crown view of the joining face; right, rotated 180° around the vertical axis to show the solvent exposed surface. (B) Tenfold serial dilutions of cultures of AJY1950 with vector, NMD3 (pAJ123), ARX1 (pAJ1032), RPL3-NES (pAJ2089), RPL12B-NES (pAJ2091), and RPL25-NES (pAJ2094) on 5FOA plates, incubated at 30°C for 6 d. (C) Rpl3-NES (pAJ2089) and Rpl12b-NES (pAJ2091) were expressed in AJY1950, and their cosedimentation with ribosomes was analyzed using sucrose gradients. The NES fusions were detected by Western blotting using an antibody against Nmd3 that is specific for the NES of Nmd3. In the Rpl3-NES blot native Nmd3 and Rpl3-NES are indicated by white and black arrows, respectively. The distributions of Rpl8 and Nmd3 are shown for comparison. (D) The localization of Rpl25eGFP was detected in AJY1950 containing RPL12B-NES (pAJ2091), NMD3 (pAJ123), ARX1 (pAJ1032), or RPL3-NES (pAJ2089).
DISCUSSION
Requirements for Nuclear Export Receptors
The efficient transport of large cargoes through the NPC requires multiple receptors (Ribbeck and Gorlich, 2002). In yeast, nuclear export of the large ribosomal subunit depends on at least three receptors, Crm1, recruited to the subunit by Nmd3, the heterodimer of Mex67 and Mtr2, and the 60S-specific factor Arx1 (Ho et al., 2000b; Gadal et al., 2001; Bradatsch et al., 2007; Yao et al., 2007; Hung et al., 2008). Although Nmd3 is conserved throughout eukaryotes as an export adapter (Thomas and Kutay, 2003; Trotta et al., 2003), the functions of Mex67/Mtr2 and Arx1 in 60S export do not appear to be conserved in higher eukaryotes (Bradatsch et al., 2007; Yao et al., 2007), suggesting that other, as yet unidentified proteins, replace Mex67/Mtr2 and Arx1 as export receptors in higher eukaryotes. Ribosome export is a fundamental process in all eukaryotes, and many of the ribosomal proteins and ribosome biogenesis factors are highly conserved. Thus, it is surprising that different eukaryotic lineages have evolved the use of different receptors in the export process of the large subunit. Here, we asked if there are particular requirements for the receptors that are used for 60S export, i.e., must Crm1 be used or can other receptors substitute for Crm1. We found that Crm1 could be replaced by direct recruitment of any of the other known export receptors in yeast. Thus, there does not appear to be a specific requirement for Crm1 in 60S export. Not surprisingly, none of the chimeric receptors worked as efficiently as wild-type Nmd3 with Crm1. Several explanations could account for this reduced efficiency. The fusion of nmd3Δ100 to different receptors may impair their function preventing their proper interaction with nucleoporins or interaction of Nmd3 with 60S subunits. Alternatively, because Nmd3 is required for subunit biogenesis and the localization of the fusions differs from wild-type Nmd3, there may not be adequate amounts of Nmd3 with the proper localization in the nucleus for binding to the nascent subunits. Along these lines, there is a rough correlation between the expression level of the chimeric proteins and their degree of complementation (data not shown).
Nmd3 is an adapter protein: that is, it bridges the interaction between its cargo, the 60S subunit, and a receptor, in this case Crm1. In the work presented here, the fusions of export receptors to an NES-deficient Nmd3, bestow on Nmd3 the ability to bind both cargo and nucleoporins directly. Thus, these fusions themselves are not adapters but rather novel receptors. They are similar in function to the noncannonical 60S subunit receptor Arx1, which binds to cargo and interacts directly with nucleoporins, but does not belong to any family of known transport receptors (Bradatsch et al., 2007; Hung et al., 2008).
Do the Multiple Receptors Have to Be Different?
Whereas a single Mex67/Mtr2 dimer is probably recruited to a pre-60S particle in wild-type cells, in the case of the fusion of Mex67 to Nmd3, we suggest that two Mex67 molecules are recruited to the subunit; one to its normal binding site, possibly 5S (Yao et al., 2007), and the second to the Nmd3-binding site via fusion to Nmd3. Under these conditions, export would be driven by two molecules of Mex67 and Arx1, but not Crm1. This result suggests that multiple copies of the same receptor will suffice in export and that the receptors do not have to be different species. Nevertheless, utilization of different receptor species may have certain benefits. It may avoid competition between common receptors for the same binding sites. It may also provide cross talk between different cellular export pathways. The translational capacity of a cell is determined by its ribosome content. It would make etiological sense that mRNA export was regulated in response to translation capacity. Indeed, Mex67 and Mtr2 appear to toggle between utilization for 60S export and mRNA export (Yao et al., 2008).
Nuclear Export Signals in cis to the Ribosome
We previously posed the question of why the leucine-rich NES for 60S export is contained in the transacting factor, Nmd3, rather than in cis on a ribosomal protein (Johnson et al., 2002). After all, its presence on a ribosomal protein would seem more economical. We have suggested that providing the NES in trans on a shuttling factor may afford the cell a greater control of regulation of 60S export. For example, 60S export could be finely tuned to the cytoplasmic needs for free 60S subunits, regulated by the ratio of Nmd3 to free 60S subunits in the cytoplasm. Here we have demonstrated that the leucine-rich NES of Nmd3 can function when fused directly to a ribosomal protein. Thus, there is not an absolute requirement for the NES in trans to the ribosome. It should be noted that none of the ribosomal proteins containing an NES supported robust export. Various explanations could account for relatively weak function of the NES fusions to ribosomal proteins. The fusion protein must assemble into the subunit and not drive premature export of the pre-60S. The NES must be accessible to Crm1 once in the subunit. In addition, the position of the NES on the subunit may be important. Preliminary results suggest that Nmd3 binds to the joining face of the large subunit (Sengupta, Bussiere, Johnson, and Frank, unpublished data). Efficient export may require recruitment of a receptor to this large RNA surface to facilitate partitioning the ribosome into the hydrophobic channel of the NPC. As the joining face of the large subunit is highly constrained by its requirement to engage properly with the small subunit, the evolution of a transacting factor on this surface may have been favored over the acquisition of an NES on a ribosomal protein in the subunit interface. Furthermore, the Crm1 recruited position by rprotein-NES cannot compete with any native 60S export receptor.
Evolution of Ribosome Export Receptors
The origins of the nuclear envelope and the NPC are not well understood. It has been suggested that the NPC was already present, either in primitive or well-developed form, in the last common eukaryote ancestor (Mans et al., 2004; Bapteste et al., 2005). Regardless, at the time the nuclear envelope evolved as a barrier to separate the genome from the cytoplasm, transport mechanisms must have existed for ribosome export. Nmd3 is conserved from archaea to humans, suggesting a role for Nmd3 in 60S biogenesis that predates its role as a transport factor. Only the eukaryotic Nmd3 proteins contain nuclear shuttling sequences. Considering the occurrence of Nmd3 throughout eukaryotes, its conserved role in 60S export, and the lack of conservation of other export factors, Nmd3 may represent the first export adapter that evolved for the large subunit. Perhaps transport in an early eukaryote, with a less sophisticated NPC, could be driven by a single export receptor recruited by Nmd3. As eukaryotic lineages evolved and regulation of transport across the NPC became more highly regulated, additional proteins that associated with the pre-ribosomal particle acquired transport function to enhance the efficiency of export. The lack of a specific requirement for a given export receptor would allow ample flexibility for the evolution of different export factors in different eukaryotic lineages.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We thank E. Hurt (University of Heidelberg) and C. Dargemont (Institut Jacques Monod) for anti-Mtr2 and Mex67 antibodies, respectively, J. Hedges for preparation of plasmids, and E. Lund for helpful comments on this manuscript. This work was supported by National Institutes of Health Grant GM53655 to A.W.J.
Footnotes
REFERENCES
- Bapteste E., Charlebois R. L., MacLeod D., Brochier C. The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol. 2005;6:R85. doi: 10.1186/gb-2005-6-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R., Littlewood T., Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Bradatsch B., et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Scarcelli J. J. Unravelling mRNA export. Nat. Cell Biol. 2006;8:645–647. doi: 10.1038/ncb0706-645. [DOI] [PubMed] [Google Scholar]
- Dahlberg J. E., Lund E. Functions of the GTPase Ran in RNA export from the nucleus. Curr. Opin. Cell Biol. 1998;10:400–408. doi: 10.1016/s0955-0674(98)80017-3. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S., Braun I. C., Izaurralde E., Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Fried H., Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Hedges J., Chen Y. I., West M., Bussiere C., Johnson A. W. Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J. Biol. Chem. 2006;281:36579–36587. doi: 10.1074/jbc.M606798200. [DOI] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A. W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J., Johnson A. W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. H., Kallstrom G., Johnson A. W. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 2000a;6:1625–1634. doi: 10.1017/s1355838200001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H.N., Kallstrom G., Johnson A. W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000b;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hung N. J., Lo K. Y., Patel S. S., Helmke K., Johnson A. W. Arx1 Is a Nuclear Export Receptor for the 60S Ribosomal Subunit in Yeast. Mol. Biol. Cell. 2008;19:735–744. doi: 10.1091/mbc.E07-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. W., Lund E., Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 2002;11:580–585. doi: 10.1016/s0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- Kallstrom G., Hedges J., Johnson A. W. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Huang N. P., Koser J., Deng J., Lau K. H., Schwarz-Herion K., Fahrenkrog B., Aebi U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad Sci. USA. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. Direct and indirect roles of Ran GTP in nuclear export of RNAs in higher eukaryotes. In: Rush M., D'Eustachio P., editors. The Small GTPase Ran. Norwell, MA: Kluwer Academic Publishers; 2001. pp. 59–83. [Google Scholar]
- Lund M. K., Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Macara I. G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans B. J., Anantharaman V., Aravind L., Koonin E. V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Meyer A. E., Hung N. J., Yang P., Johnson A. W., Craig E. A. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad Sci. USA. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M., Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. S., Belmont B. J., Sante J. M., Rexach M. F. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Petosa C., Schoehn G., Askjaer P., Bauer U., Moulin M., Steuerwald U., Soler-Lopez M., Baudin F., Mattaj I. W., Muller C. W. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., Pante N., Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Luhrmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol. Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Thomas F., Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Trotta C. R., Lund E., Kahan L., Johnson A. W., Dahlberg J. E. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Yao W., Lutzmann M., Hurt E. A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J. 2008;27:6–16. doi: 10.1038/sj.emboj.7601947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., Roser D., Kohler A., Bradatsch B., Bassler J., Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]