Widespread balancing selection and pathogen-driven selection at blood group antigen genes (original) (raw)
Abstract
Historically, allelic variations in blood group antigen (BGA) genes have been regarded as possible susceptibility factors for infectious diseases. Since host–pathogen interactions are major determinants in evolution, BGAs can be thought of as selection targets. In order to verify this hypothesis, we obtained an estimate of pathogen richness for geographic locations corresponding to 52 populations distributed worldwide; after correction for multiple tests and for variables different from selective forces, significant correlations with pathogen richness were obtained for multiple variants at 11 BGA loci out of 26. In line with this finding, we demonstrate that three BGA genes, namely CD55, CD151, and SLC14A1, have been subjected to balancing selection, a process, rare outside MHC genes, which maintains variability at a locus. Moreover, we identified a gene region immediately upstream the transcription start site of FUT2 which has undergone non-neutral evolution independently from the coding region. Finally, in the case of BSG, we describe the presence of a highly divergent haplotype clade and the possible reasons for its maintenance, including frequency-dependent balancing selection, are discussed. These data indicate that BGAs have been playing a central role in the host–pathogen arms race during human evolutionary history and no other gene category shows similar levels of widespread selection, with the only exception of loci involved in antigen recognition.
Since the discovery of the ABO blood group in 1900 by Karl Landsteiner, as many as 29 blood group (BG) systems have been identified in humans (Blood Group Antigen Gene Mutation Database, BGMUT [Blumenfeld and Patnaik 2004]). Each system is specified by a blood group antigen (BGA) constituted by a protein or carbohydrate molecule which is expressed on the erythrocyte membrane and is polymorphic in human populations.
The molecular basis of all blood group systems (except for P1) has been clarified, with one or more polymorphic loci accounting for BG phenotypes. BGA genes belong to different functional categories, including receptors, transporters, channels, adhesion molecules, and enzymes; among the latter, the great majority of loci code for glycotransferases. While a few BGAs are confined to the erythrocyte membrane, others are expressed at the surface of different cell types or secreted in body fluids (Reid and Lomas-Frances 1997).
The number of different alleles is highly variable among BGA genes and ranges from two to >100 (Blumenfeld and Patnaik 2004) with the most common form of variation being accounted for by missense or nonsense single nucleotide polymorphisms (SNPs). BGA polymorphisms have attracted considerable attention over recent years not only with respect to erythrocyte physiology per se, but also due to the possibility that variations in BGAs might underlie different susceptibility to diseases. In particular, the association between infections and BGA polymorphisms has been extensively investigated, although conclusive results have been obtained in a minority of cases. For example, specific BGA alleles have been shown to alter susceptibility to malaria (Moulds and Moulds 2000), while FUT2 variants (Lewis system) influence the predisposition to Norwalk virus (Lindesmith et al. 2003) and Campylobacter (Ruiz-Palacios et al. 2003) infection, as well as to vulvovaginal candidisis (Hurd and Domino 2004) and urinary tract infections (Schaeffer et al. 2001).
Such findings are in line with the vision whereby different BGAs serve as “incidental receptors for viruses and bacteria” (Moulds et al. 1996), but also function as modulators of innate immune response (Ruiz-Palacios et al. 2003; Linden et al. 2008) and possibly as “decoy-sink” molecules targeting pathogens to macrophages (Gagneux and Varki 1999).
Given this premise and the conundrum whereby host–pathogen interactions are major determinants in evolution, BGAs can be thought of as possible targets of diverse selective pressures. This view is in agreement with the geographic differentiation pattern observed for BGAs and with previous reports of non-neutral evolution at the ABO, DARC, GYPA, and FUT2 loci (Saitou and Yamamoto 1997; Koda et al. 2001; Baum et al. 2002; Hamblin et al. 2002; Calafell et al. 2008).
Here we exploited the availability of extensive resequencing data, as well as of SNP genotyping in world-wide populations, to investigate the evolutionary forces underlying the evolution of BGA genes: Our data provide evidence that balancing and pathogen-driven selections have acted at multiple BGA loci.
Results and Discussion
Pathogen richness and BGA gene polymorphisms
As a first step we wished to verify whether allele frequencies of SNPs in BGA genes varied with pathogen richness, in terms of different species in a geographic location. Similar approaches have been applied to test this same hypothesis for HLA genes (Prugnolle et al. 2005) and for other gene–environment interactions (Thompson et al. 2004; Young et al. 2005; Hancock et al. 2008). To this aim we exploited the fact that a set of over 650,000 tag SNPs has been typed in 52 populations (HGDP-CEPH panel) distributed world wide (Li et al. 2008). As for pathogen richness, we gathered information concerning the number of different micropathogen species from the Gideon database; pathogen richness was calculated on a country basis by pooling together viruses, bacteria, fungi and protozoa (see Methods for further details). A total of 262 SNPs in BGA genes had been typed in the HGDP-CEPH panel allowing analysis of the following loci: RHCE, ERMAP, DARC, CD55, CR1, GYPC, GYPA, GCNT2, RHAG, C1GALT1, AQP1, KEL, AQP3, ABO, CD44, ART4 (also known as DO), SEMA7A, SLC4A1, SLC14A1, FUT3, BCAM (also known as LU), FUT2, FUT1, A4GALT, XG, and XK.
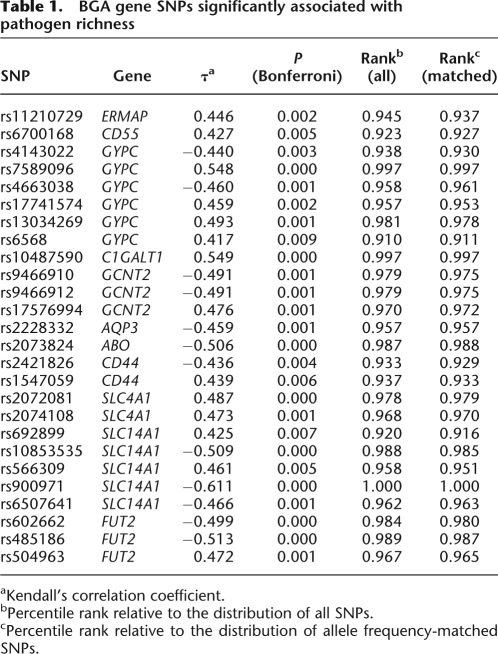
For all 262 BGA SNPs in the data set we calculated Kendall's rank correlation coefficient (τ) between pathogen richness and allele frequencies in HGDP-CEPH populations; a normal approximation with continuity correction to account for ties was used for _P_-value calculations (Kendall 1976). We verified that, after Bonferroni correction for multiple tests, 26 BGA gene SNPs were significantly associated with pathogen richness (Table 1). Since variables different from selective forces (e.g., colonization routes; Handley et al. 2007) are expected to affect allele frequency spectra across populations, we compared the strength of BGA gene SNP correlations to control sets of SNPs extracted from the data set. In particular, for each BGA SNP in Table 1 we extracted from the full data set all SNPs having an overall minor allele frequency (averaged over all populations) differing less than 0.01 from its frequency; for all SNPs in the 26 frequency-matched groups we calculated Kendall's τ between pathogen richness and allele frequencies. Next, we calculated the percentile rank of BGA gene SNPs in the distribution of Kendall's τ obtained for the control sets and in the distribution of all SNPs in the data set. Data are reported in Table 1 and indicate that all SNPs ranked above the 90th percentile of τ-values, with 19 of them ranking above the 95th (data for all 262 SNPs are available in Supplemental File 1). By performing 30,000 simulations using samples of 262 SNPs we verified that the probability of obtaining 19 SNPs with a correlation value above the 95th percentile amounted to 0.045; a similar result is obtained by considering the probability to obtain n SNPs with a τ higher than the 95th percentile in a sample of 262 to be Poisson-distributed (P = 0.043). These data therefore indicate that the fraction of BGA SNPs that correlate with pathogen richness is higher than expected; yet, these calculations also suggest that a portion of SNPs in Table 1 might represent false positive associations in that the retrieval of 13 variants with a percentile rank above the 95th would be expected by chance. An estimation of the magnitude of selective effects exerted by pathogens on human genes would be required to accurately estimate the expected fraction of truly correlated SNPs.
Table 1.
BGA gene SNPs significantly associated with pathogen richness
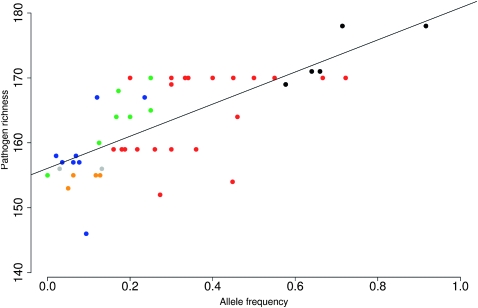
The strongest correlation between BGA SNP allele frequency and pathogen richness was obtained for rs900971 in SLC14A1 (Fig. 1; similar representations for all SNPs in Table 1 are available as Supplemental Fig. 1). In order to verify that environmental variables correlating with pathogen richness (Guernier et al. 2004) did not determine the association signal with BGA genes, we calculated the mean temperature and maximum precipitation rate for geographic locations corresponding to HGDP-CEPH populations; none of the SNPs reported in Table 1 significantly correlated with either variable (data not shown).
Figure 1.
Correlation between pathogen richness and allele frequency for rs900971 in SLC14A1. Populations from different broad geographic areas are coded by different colors: (green) Sub-Saharan Africa, (black) America, (red) Asia, (blue) Europe, (orange) Middle East, and (gray) Oceania.
The identification of correlations between specific environmental variables and allele frequencies has been regarded as a strategy complementary to common population genetic approaches for the detection of selection signatures (Hancock et al. 2008). All such analyses rely on the assumption that the environmental variable we measure nowadays has changed little over human history and that gene flow due to recent admixture has had a minor impact on human genetic diversity. In this case, we implicitly assumed that the number of different pathogen species per country has been maintained proportionally unchanged along human evolutionary history. Although an oversimplification, this might not be so different from the reality, given that climatic variables have been shown to be of primary importance in driving the distribution of human pathogens (Guernier et al. 2004). As for gene flow, the influence of recent admixture in most populations is considered to be modest (Li et al. 2008), as also demonstrated by the good relationship between population genetic diversity and distance from Africa (Handley et al. 2007).
Our data therefore indicate that the allele frequencies of a subset of BGA genes vary with pathogen richness, supporting the vision whereby these loci affect the susceptibility to infectious diseases. This hypothesis had previously been formulated for ABO and FUT2 (Greenwell 1997; Hill 2006; Casanova and Abel 2007), while in the case of GYPC, DARC, and SLC4A1 the ability of specific alleles to modulate infection susceptibility has been demonstrated for malaria (Moulds and Moulds 2000). Also, in the case of AQP3, modulation of malaria severity can be hypothesized since AQP3 represents the major channel for glycerol transport in human erythrocytes (Roudier et al. 2002), and mice knockout for Aqp9, a related glycerol transporting aquaporin, display increased survival to P. berghei (Liu et al. 2007). One possibility to explain the observed associations is that pathogen richness has co-varied with malaria prevalence and that nucleotide variations, even different from those previously reported to confer resistance in SLC4A1 and GYPC, affect Plasmodium entry, spread, or rosetting. Conversely, no association with infectious disease predisposition has ever been reported for SLC14A1, ERMAP, C1GALT1, and GCNT2; yet these genes encode either glycotransferases or surface glycosylated proteins, suggesting that carbohydrate determinants might affect pathogen attachment and entry.
Another possibility involving SLC14A1 variants is that the association with pathogen richness reflects some important aspect of urea metabolism during infection; indeed the gene codes for an urea transporter and the intracellular availability of urea has been shown to be a limiting factor for the ability of Mycobacterim bovis to attenuate expression of MHC class II molecules during macrophage infection through urease-induced alkalinization of intracellular compartments (Sendide et al. 2004).
As for CD44, it has been shown to act as a receptor for group A Streptococcus (Cywes et al. 2000), Mycobacterium tuberculosis (Leemans et al. 2003), and Escherichia coli in urinary tract infections (Rouschop et al. 2006).
Population genetics analysis of BGA genes
Given the results obtained above and the premise whereby BGA genes might represent selective targets, we wished to verify whether selection signatures could be identified at BGA genes. To this aim we exploited the fact that 22 out of 38 loci involved in BGA specification have been included in the SeattleSNPs program so that resequencing data (although with some gaps) in at least two populations are available; in particular, all data refer to one population with European ancestry (EA) and one with African ancestry (either Yorubans [YRI] or African American [AA]). From the SeattleSNP gene list we excluded ABO, which has been previously studied (Saitou and Yamamoto 1997; Calafell et al. 2008), A4GALT, XK, and ART4 due to poor resequencing coverage, and KEL, because of its being located in a region subjected to a selective sweep possibly driven by the nearby TRPV6 locus (Akey et al. 2006). The following genes were left for analysis: AQP1, AQP3, ACHE, BSG, B3GALNT1 (previously B3GALT3), CD55 (previously DAF), CD151, SLC4A1, ICAM4, FUT3, FUT2, FUT1, BCAM, ERMAP, GYPC, SEMA7A, and SLC14A1.
With the aim of identifying loci that have been subjected to natural selection, and following the conundrum whereby selection signatures might extend over relatively short gene regions (due to the action of mutation and recombination; Wiuf et al. 2004; Bubb et al. 2006), we applied a sliding window approach to all BGA genes (except for ACHE, due to its small size and FUT2, as detailed below) and calculated population genetic differentiation, measured as _F_ST. Under the assumption of neutrality, _F_ST is determined by demographic history (i.e., genetic drift and gene flow), which affects all loci similarly. We therefore calculated the 2.5th and 97.5th percentiles in the distribution of _F_ST-values obtained for sliding windows across SeattleSNPs genes (see Methods for details) and searched for BGA gene regions that display unusually high or low population differentiation. Overall, 8.3% of sliding windows deriving from the 17 BGA genes displayed exceedingly high or low _F_ST-values; estimation of an empirical probability (see Methods) to obtain an equal or higher fraction of outliers in windows deriving from SeattleSNP genes yielded a _P_-value of 0.19. These data indicate that an excess of unusual _F_ST-values can be observed for BGA genes, with the failure to reach statistical significance being likely due to the presence of other non-neutrally evolving genes in the SeattleSNP data set (which mainly gathers genes involved in inflammatory processes).
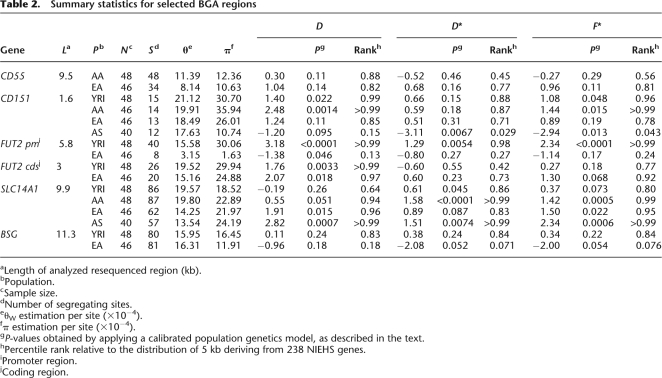
BGA gene regions displaying unusual _F_ST-values were further studied by application of population genetics statistics. In particular, widely used test include Tajima's D (Tajima 1989) and Fu and Li's D* and F* (Fu and Li 1993). Tajima's D (_D_T) tests the departure from neutrality by comparing two nucleotide diversity indexes: θW (Watterson 1975), an estimate of the expected per site heterozygosity, and π (Nei and Li 1979), the average number of pairwise sequence nucleotide differences. Positive values of _D_T indicate an excess of intermediate frequency variants and are a hallmark of balancing selection; negative _D_T-values indicate either purifying selection or a high representation of rare variants as a result of a selective sweep. Fu and Li's F* and D* are also based on SNP frequency spectra and differ from _D_T in that they also take into account whether mutations occur in external or internal branches of a genealogy. Since population history, in addition to selective processes, is known to affect frequency spectra and all related statistics; we performed coalescent simulations using a calibrated population genetics model that incorporates demographic scenarios (Schaffner et al. 2005). Also, in order to disentangle the effects of selection and population history, we exploited the conundrum whereby selection acts on a single locus while demography affects the whole genome: As a control data set we therefore calculated diversity parameters and test statistics for 5 kb windows deriving from 238 genes resequenced by the NIEHS program (see Methods for details). A similar comparison with SeattleSNPs gene data is provided in Supplementary File 2 (Supplemental Table 2). Sliding window analyses identified gene regions showing unusual _F_ST-values (Supplemental Fig. 3), which were selected for further study, as reported in the following paragraphs. In addition, for the remaining BGA loci we calculated summary statistics (_D_T, D*, and F*) for the entire gene region and unusual values were found for BSG (detailed below) and FUT1. The latter was not further analyzed as high homology with other gene family members and a pseudogene suggested that gene conversion events might affect the result (this was not the case for FUT2 since we focused on the promoter region, as detailed below).
CD55 (Cromer system)
A sliding-window analysis along the CD55 gene (OMIM no. +125240; referred to as DAF in the SeattleSNPs database) revealed the presence of a region encompassing nucleotides ∼9000–19,000 showing exceedingly low _F_ST-values (Supplemental Fig. 3). Both nucleotide diversity estimates and test statistics (Table 2) revealed no significant departure from neutrality for both, AA and EA, yet _D_T and Fu and Li's D* and F* ranked relatively high in the distribution of 5 kb windows from NIEHS genes (see Supplemental Table 2 for a comparison with SeattleSNPs genes).
Table 2.
Summary statistics for selected BGA regions
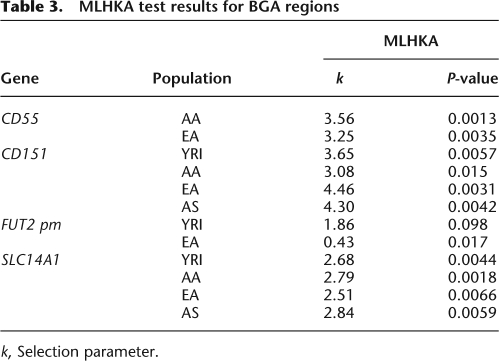
Under neutral evolution, the amount of within-species diversity is predicted to correlate with levels of between-species divergence, since both depend on the neutral mutation rate (Kimura 1983). The HKA test (Hudson et al. 1987) is commonly used to verify whether this expectation is verified. Here we performed a maximum likelihood HKA test (MLHKA) by comparing the CD55 region to 16 neutrally evolving genes (see Methods for details): A significant result was obtained for both AA and EA (Table 3).
Table 3.
MLHKA test results for BGA regions
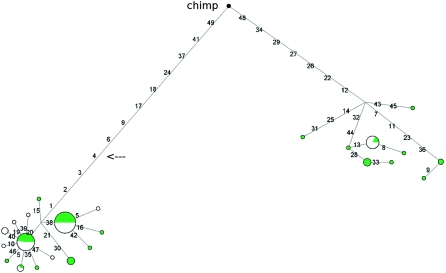
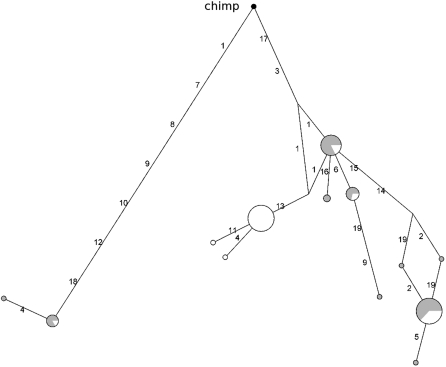
Therefore, we wished to study the genealogy of CD55 haplotypes in the region and to this aim a neighbor-joining network was constructed. Two major clades separated by long branch lengths are evident (Fig. 2), each containing common haplotypes. In order to estimate the TMRCA (time to the most recent common ancestor) of the two haplotype clades, we applied a phylogeny-based method (Bandelt et al. 1999) based on the measure ρ, the average pairwise difference between the two haplotype clusters. ρ resulted in a value equal to 13.28, so that, using a mutation rate based on 50 fixed differences between chimpanzee and humans, and a separation time of 6 million years (Myr) (Glazko and Nei 2003), we estimated a TMRCA of 3.19 Myr (SD = 673 Kyr). Given the low recombination rate in the region, we wished to verify this result using GENETREE, which is based on a maximum-likelihood coalescent analysis (Griffiths and Tavare 1994, 1995). The method assumes an infinite-site model without recombination and, therefore, haplotypes and sites that violate these assumptions need to be removed: in this case, only three single segregating sites had to be removed. The resulting gene tree, rooted using the chimpanzee sequence, is partitioned into two deep branches (Supplemental Fig. 4). A maximum-likelihood estimate of θ (θML) of 9.2 was obtained, resulting in an estimated effective population size (_N_e) of 22,000, a value comparable to most figures reported in the literature (Tishkoff and Verrelli 2003). Using this method, the TMRCA of the CD55 haplotype lineages amounted to 2.61 Myr (SD = 552 Kyr). Such deep coalescent time is unusual, as estimates for neutrally evolving autosomal loci range between 0.8 Myr and 1.5 Myr (Tishkoff and Verrelli 2003).
Figure 2.
Genealogy of CD55 haplotypes reconstructed through a median-joining network. The analysis corresponds to the gene region spanning nucleotides ∼9500–18,300 (as described in the text). Each node represents a different haplotype, with the size of the circle proportional to the haplotype frequency. Nucleotide differences between haplotypes are indicated on the branches of the network. Circles are color-coded according to population (green, AA; white, EA). The chimpanzee sequence is also shown. The arrow shows the position of rs6700168 (Table 1). Note that the relative position of mutations along a branch is arbitrary.
Overall these data strongly support the idea that the CD55 region we analyzed has evolved under long-standing balancing selection. This gene portion covers roughly 10 kb surrounding exon 6–7 and contains four DNase I hypersensitive sites in CD4+ T cells (Boyle et al. 2008); five intermediate frequency SNPs (rs6700079, rs2184476, rs1507760, rs10746462, and rs10746463) located along the branch separating the two haplogroups lie within DNase I hypersensitive sites. Since DNase I hyperaccessibilty is thought to be a hallmark of active _cis_-regulatory regions (Gross and Garrard 1988; Felsenfeld and Groudine 2003), these variants might represent good candidates as functional SNPs with a role in transcriptional regulation of CD55. Importantly, another variant (rs6700168) located in this genomic portion was found to correlate with pathogen richness (Table 1) and it lies along the branch separating the two haplotype clusters (Fig. 2). In order to verify whether heterozygote advantage might underlie the action of balancing selection we calculated the observed over expected heterozygosity for rs6700168 and verified whether this ratio varied with pathogen richness. Since this was not the case, we suggest that the maintenance of the two haplotype lineages is not due to overdominance but possibly to antagonistic selection (see below).
CD55 (also known as DAF, decay-accelerating factor) is a complement-regulatory protein expressed by most cell types, which protects host tissues from damage by the autologous complement system (Nicholson-Weller and Wang 1994). Previous studies have indicated that the membrane-anchored form of CD55 serves as a receptor for very common human pathogens, such as Dr+ E. coli (Nowicki et al. 1993), coxsackieviruses B1, B3, and B5 (Shafren et al. 1995), and echovirus 7 (Clarkson et al. 1995), suggesting that decreased or abolished DAF expression might confer decreased susceptibility to these infectious agents. Total absence of CD55 (Inab phenotype) is very rare in humans (Blumenfeld and Patnaik 2004) and associates with no overt phenotype. Yet, other observations point to a possible role of the gene in fertility and pregnancy: CD55 is dynamically regulated during the menstrual cycle (Young et al. 2002) and it is highly expressed at the feto–maternal interface (Sood et al. 2006); moreover, reduced DAF expression has been associated with luteal phase defect of the endometrium associated with infertility or pregnancy loss. Also, mice lacking DAF are more susceptible to autoimmune manifestations (Kaul et al. 1995).
These evidences might therefore suggest that regulation of CD55 expression levels, either in a cell-type- or stage-dependent fashion might affect vital processes, such as reproduction and immunity. Also, the lack of evidence supporting heterozygote advantage and the phenotype of cd55 −/− mice possibly suggest that balancing selection ensues from antagonist selection trading-off resistance to infection with autoimmune phenomena. Obviously, other hypotheses are possible (e.g., adaptation to variable environmental conditions with special reference to different environmental pathogens) and further studies on the biological function of CD55 will be instrumental in addressing this issue.
CD151 (RAPH system)
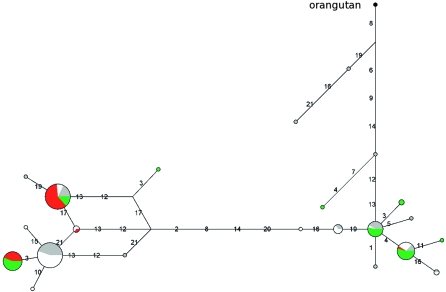
A sliding window analysis along CD151 (OMIM no. *602243) indicated the 3′ gene region displays reduced population differentiation and exceedingly low _F_ST-values are observed in a region roughly corresponding to the terminal region extending from exon 6 to the 3′ UTR (Supplemental Fig. 3). Nucleotide diversity (both _θ_W and π), in this restricted region, ranked above the 97.5th percentile in the distribution of 5 kb windows deriving from NIEHS genes (Supplemental Table 1) for both EA and YRI. Summary statistics revealed significantly positive values for _D_T, D*, and F* in YRI, but not in EA. In order to further investigate the possible departure from neutrality in other human populations, the same region was resequenced in two additional samples: Asians (AS) and AA. As shown in Table 2, significantly positive test statistics were obtained for populations of African ancestry but not for Asians and Europeans. In the case of AS, negative values of summary statistics are due to the presence of a single highly divergent haplotype (Fig. 3).
Figure 3.
Genealogy of CD151 haplotypes reconstructed through a median-joining network. The analysis corresponds to the gene region spanning nucleotides 9100–10,400. Population color codes are as follows: (green) AA; (white) EA; (red) AS; (gray) YRI.
Application of the MLHKA test, using 16 neutrally evolving genes (see Methods), rejected the hypothesis of neutrality for all populations (Table 3).
Next, we wished to examine haplotype genealogy for the terminal CD151 gene region and, to this aim, a median-joining network was constructed (Fig. 3). The topology of this network was relatively unambiguous showing two major clades, each containing common haplotypes, separated by long branch lengths. Calculation of the TMRCA (ρ = 8.55; fixed differences = 58 using Pongo pygmaeus) yielded an estimate of 3.83 Myr (SD = 1.06 Myr), again a deep coalescent time compared to neutral loci. As reported above for CD55, this result was verified using GENETREE and an estimated TMRCA of 2.14 Myr was obtained (SD = 643 Kyr, _θ_ML = 1.9, _N_e = 13,722; Supplemental Fig. 5). In analogy to CD55, these features suggest the action of long-standing balancing selection in African populations.
The analyzed CD151 region covers the last four coding exons and the 3′ UTR; most variants are located in noncoding regions, with the majority of intermediate frequency SNPs falling within the UTR. Analysis of known functional elements in the 3′ UTR was performed using UTRscan and no SNPs were found to affect predicted motifs. Conversely, a search for microRNA target sites (miRBase) indicated that one variant, namely rs1130698, falls within the highest scoring predicted target site. In particular the T allele changes a G–C pairing between the CD151 UTR sequence and hsa-miR-940 to a G–U wobble; unfortunately, little is known about the expression pattern of hsa-miR-940, except for the fact that it was cloned from cervical cell lines (Lui et al. 2007).
CD151, a member of the tetraspanin protein family involved in cell adhesion and motility, is expressed in most human tissues (Fitter et al. 1995). Mutations of CD151 in humans result in nephropathy with epidermolysis bullosa and deafness (Karamatic Crew et al. 2004), while different phenotypes have been reported for cd151 −/− mice, including abnormal hemostasis (Wright et al. 2004), defective wound healing (Cowin et al. 2006), and renal defects (Sachs et al. 2006). Members of the tetraspanin family have been implicated in virus infection in animals and humans; in particular, different tetraspanins have been shown to act as receptors for HCV (Pileri et al. 1998), HIV (von Lindern et al. 2003), canine distemper virus (Loffler et al. 1997), feline leukemia virus (Willett et al. 1994), and porcine reproductive and respiratory syndrome virus (Shanmukhappa et al. 2007); yet a recent report has also shown that members of the tetraspanin family, including CD151, protect human macrophages from HIV-1 and vescicular stomatitis virus infection, possibly by blocking virion binding/uptake (Ho et al. 2006).
Whether the maintenance of balancing selection at the CD151 locus is pathogen-driven remains to be elucidated, and unfortunately no SNP mapping to the gene has been typed in the HGDP-CEPH panel; it is worth noting that besides its possible direct role in predisposing to infections (by acting as a viral receptor/binding factor), its function in wound healing (Cowin et al. 2006) might also be regarded as linked to pathogens and their prevalence, in that the risk of wound infection likely depends on how long the healing process takes to completion.
FUT2 (Lewis system)
In humans two alpha (1,2)-fucosyltransferases, encoded by the paralogous FUT1 and FUT2 genes, determine expression of the human H antigen, a precursor of blood group A and B antigens.
The two genes differ in substrate specificities and tissue expression (Costache et al. 1997): FUT1 (H enzyme, H/h system) is responsible for the expression of H antigen in red cells and vascular endothelia, whereas the Se enzyme (encoded by FUT2, Lewis system, OMIM no. +182100) is responsible for the synthesis of the same antigen in secretory glands and the intestinal mucosa; individuals referred to as “secretors” (Se) have at least one functional FUT2 allele.
Common FUT2 null alleles are present in many populations; in particular a frequent null allele (se 428) is responsible for most nonsecretor phenotypes in Europe and Africa, while a missense mutation (_se_385) is widespread in East Asians (Kelly et al. 1995; Koda et al. 1996; Liu et al. 1998). Interestingly, the coding region of FUT2 has previously been hypothesized to be subjected to balancing selection, possibly under an overdominance regime (Koda et al. 2001).
In the case of FUT2 we did not perform a sliding window analysis as described for the above genes due to extensive resequencing gaps. Rather, we divided the gene in three major portions: coding exon, intron, and 5′ upstream region (10 kb upstream the transcription start site, thereafter referred to as putative promoter). In line with previous findings, the coding exon displayed high nucleotide diversity and positive statistics (Table 2), while we verified that low levels of nucleotide variation characterize the only intron (not shown). Interestingly, an unusual pattern was observed at the putative promoter region: as shown in Table 2, YRI displayed high values of θW, while EA presented low nucleotide diversity (percentile rank of θW = 0.18). Calculation of _F_ST yielded a high value of 0.45, corresponding to a percentile rank of 0.977 in the distribution of SeattleSNPs gene windows and being 20-fold greater than population differentiation calculated for the coding region (_F_ST = 0.022).
Summary statistics for the putative promoter revealed deviation from neutrality in YRI, since all tests yielded significantly positive values (Table 2); conversely, statistics for EA resulted in negative values, although significance was only obtained for _D_T. Human/chimpanzee divergence in this gene region amounted to 1.35%, a value higher than the genome average (average = 1.06%, SD = 0.25%; Chimpanzee Sequencing and Analysis Consortium 2005) and greater than that of control loci used in the MLHKA test; the latter gave no significant results for YRI, while a reduction of polymorphism compared to intraspecific divergence was evidenced for EA (Table 3). The greater than average divergence and high polymorphism level observed for YRI might be consistent with the region having low sequence constraints, resulting in an increase of both divergence and diversity; yet, this hypothesis does not fit the EA data whereby low diversity is observed; moreover, the high population differentiation we observed can hardly be reconciled with a neutral pattern of evolution.
Low diversity values and negative statistics are consistent with both purifying and directional selection; Fay and Wu's H (Fay and Wu 2000) is usually applied to distinguish between these possibilities, since significantly negative _H_-values indicate an excess of high-frequency derived alleles, consistent with directional selection. H equaled −4.66 in EA with a borderline _P_-value of 0.062 (calculated using the calibrated model). It should be noted that the interpretation of H can be complicated by the fact that the power of this statistic to detect selection is poor when the sweep is relatively old (Przeworski 2002) and population structure can result in significantly negative H statistics (Przeworski 2002).
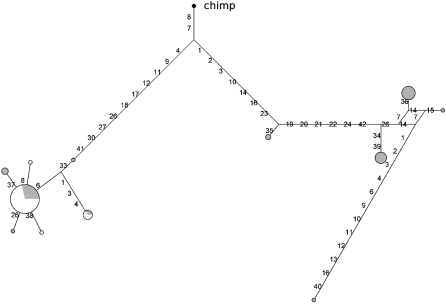
Reconstruction of haplotype genealogy for the FUT2 putative promoter using yielded a topology with two major clades separated by long branch lengths (Fig. 4); consistent with the high degree of geographic structure, all European haplotypes cluster with the same haplogroup while African chromosomes are divided in the two clades. Calculation of the TMRCA (ρ = 13.10; fixed differences = 79, using chimpanzee) yielded an estimate of 1.99 Myr (SD = 410 Kyr). A similar TMRCA was estimated with the use of GENETREE (TMRCA = 1.70 Myr, SD = 375 Kyr, _θ_ML = 3.4, _N_e = 8681; Supplemental Fig. 6). Construction of a haplotype genealogy for the coding region (data not shown) resulted in a TMRCA of 3 Myr, in agreement with previous findings (Koda et al. 2001).
Figure 4.
Median-joining network for the putative promoter region of FUT2. The analysis corresponds to the gene region spanning nucleotides 7400–17,800. Population color codes are as follows: (white) EA; (gray) YRI.
Overall, the data presented above are consistent with the presence of a selected variant/haplotype in the promoter region of FUT2; this is in line with a recent report indicating that distinct promoter haplotypes have an effect on the gene transcription levels (Soejima and Koda 2008). In the case of EA, the statistics we performed did not allow a firm rejection of the neutral model; in part this might be due to the small number of SNPs in the region (only eight) which reduces the power of all tests; also, failure to reject neutrality might be accounted for by the pattern being a relic of older selective events.
In the case of YRI, we consider that our observations might be consistent with the presence of a balanced polymorphism. This raises the possibility that the signatures we obtained at the promoter region are due to hitchhiking and linkage disequilibrium (LD) with the coding exon. Nonetheless, different observations suggest that this is not the case. First, calculation of _D_′ between the _se_428 variant and common SNPs in the putative promoter revealed a maximum value of 0.27, indicating low LD, in agreement with a previous report (Soejima and Koda 2008). Second, summary statistics yielded stronger results for the putative promoter compared to the coding exon. Third, although hitchhiking has the potential to affect large genomic regions, the signatures of balancing selection are predicted to extend over relatively short distances (Wiuf et al. 2004; Bubb et al. 2006); as an example, the high nucleotide diversity that characterizes the second exon of MHC loci decays rapidly in flanking intronic sequences (Cereb et al. 1997; Fu et al. 2003) and neighboring exons (Takahata and Sata 1998). This suggests that the departure from neutrality and the high level of nucleotide diversity we observe in the FUT2 putative promoter region is not merely a result of hitchhiking with the coding exon, given the 7 kb separating the transcription start site from the second exon.
As reported above, a recent study (Soejima and Koda 2008) of the FUT2 proximal promoter region indicated that nucleotide diversity patterns differ between African and non-African populations, and the authors identified two common haplotypes with different cell-type specific activities. These observations raise the interesting possibility that balancing selection at the FUT2 promoter region might result from overdominance due to differential activity of the two promoter haplotypes in different tissues. The “secretor” status has been associated with increased susceptibility to infection by caliciviruses (Lindesmith et al. 2003), HIV (Ali et al. 2000), and respiratory viruses (Raza et al. 1991); yet secretor subjects also display advantages compared to nonsecretors, such as lower susceptibility to urinary tract and Candida infections, and increased protection against Neisseria meningitis and Streptococcus (Haverkorn and Goslings 1969; Blackwell et al. 1990). Also, situations exist where the secretor status might underlie a double-faceted situation. One example involves Campylobacter jejuni infection (the most common cause of bacterial diarrhea): the pathogen exploits H antigens for tethering to the intestinal mucosa, but at the same time alpha (1,2)-linked fucosyloligosaccharides in human milk inhibit Campylobacter infection by competing with intestinal cell surface receptors (Ruiz-Palacios et al. 2003). As a result, a breast-fed infant is expected to be at variable risk of infectious diarrhea depending on his/her intestinal expression of H antigens and his/her mother secretion of the same molecule in milk; in this scenario, maximization of FUT2 expression in lactating epithelia might be extremely important in providing immunization to newborns. Indeed, different oligosaccharide species in human milk form part of the innate immune system with activity against different pathogens (Newburg et al. 2005), and fucosyloligosaccharides containing alpha (1,2)-linked fucose are prevalent (Chaturvedi et al. 2001). Women who are nonsecretors do not express measurable 2-linked fucosyloligosaccharides and the amount of milk fucosyloligosaccharides varies even among secretors (Chaturvedi et al. 2001), possibly suggesting the presence of genotype differences responsible for such variation (Chaturvedi et al. 2001). Since diarrhea represents a very common cause of mortality in newborns throughout the world, the adaptive significance of decreasing the chance of infection in breast-fed infants is evident. Therefore, maintenance of the advantages conferred by the secretor status, while modulating the levels of glycotransferase activity in a cell-type-dependent fashion, might represent a beneficial strategy in specific circumstances. Obviously, other explanations for the maintenance of different FUT2 promoter haplotypes are possible, and further studies will be required to analyze the activity of FUT2 promoter haplotypes. Unfortunately, no SNP located in the putative promoter region of FUT2 was available to test association with pathogen richness or verify whether a heterozygote excess could be observed in specific geographic locations. Conversely, significantly associated SNPs are located in the coding exon (rs602662 and rs485186) or 3′ UTR (rs504963) and are in full LD with the null _se_428 allele in both EA and AA (in all cases, _D_′ = 1, P < 0.001). No correlation was observed between the observed/expected heterozygosity ratio for these SNPs and pathogen richness, suggesting that, although pathogens have exerted a selective pressure on the gene and balancing selection has been operating, the underlying explanation is not accounted for by overdominance. This might be expected if the null allele is thought of as the selected variant: Heterozygotes are secretors and they are not expected to have an advantage compared to subjects carrying two active alleles. One possibility is that secretors and nonsecretors experience advantages or disadvantages depending on variable environmental conditions in terms of pathogen prevalence, since they display different susceptibility to diverse pathogen types. Alternatively, as suggested above, more complex scenarios can be envisaged that also take into account promoter variants.
SLC14A1 (Kidd system)
Sliding window analysis of SLC14A1 (OMIM no. *111000) revealed an extended region of about 6.3 kb showing high levels of population differentiation (Supplemental Fig. 3). The single variant (Asp280Asn) responsible for the common JK*A/JK*B antigens (Blumenfeld and Patnaik 2004) is located ∼6 kb downstream and, with the aim of analyzing the evolutionary history of the gene, we decided to resequence the entire region in YRI and AS with the exception of a small central gap of 2 kb (Supplemental Fig. 3). Two novel nonsynonymous variants were identified, Val10Met and Val76Ile, and both were present in the same three AA subjects.
Summary statistics and diversity parameters for the four populations (Table 2) revealed high levels of polymorphism and allowed rejection of neutrality for AA, AS, and EA, while borderline values were obtained for YRI.
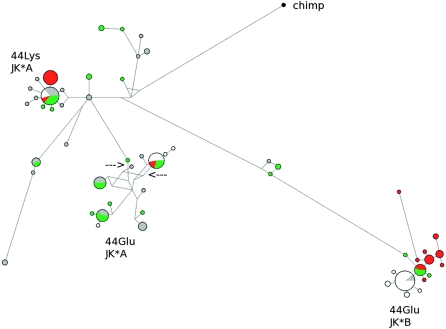
Application of the MLHKA test, as described above, rejected the hypothesis of neutrality for all populations (Table 3). Construction of the median-joining network is recommended when regions displaying low recombination are being analyzed; in the case of SLC14A1, the gene region carrying the Asp280Asn polymorphism displays low LD with the more 5′ region (Supplemental Fig. 7); yet, we decided to calculate the network over the entire region so that the relative distribution of chromosomes carrying the JK*A/JK*B variants could be visualized (Fig. 5); conversely, TMRCA estimate was performed using GENETREE and for this analysis only variants in linkage disequilibrium were included (Supplemental Fig. 7). In both cases, three major haplotype clades are evident and TMRCA estimated equaled 2.28 Myr (SD = 283 Kyr, _θ_ML = 12, _N_e = 28,800). The median joining network shows that a long branch separates haplotypes carrying the JK*B allele from JK*A, while a nonsynonymous Glu44Lys SNP might be regarded as the selected variant maintaining the two closer clusters carrying JK*A (Fig. 5). It is interesting to notice that two variants located in this gene region (rs10853535 and rs692899) correlate with pathogen richness (Table 1); one of them lies on the branch leading to the haplotype cluster carrying the JK*A and 44Glu alleles, while the second is internal to this same cluster and defines a smaller haplotype group (Fig. 5).
Figure 5.
Genealogy of SLC14A1 haplotypes reconstructed through a median-joining network. The analysis corresponds to the gene region spanning nucleotides 4887–17,350. Population color codes are as follows: (green) AA; (white) EA; (red) AS; (gray) YRI. The allelic status at amino acid position 44 and 280 (JK*A/JK*B) is reported for the three major clusters. The two arrows denote the position of rs10853535 and rs692899, which correlate with pathogen richness (Table 1).
Interestingly, for both these variants the observed over expected heterozygosity ratio significantly correlated with pathogen richness (rs692899, τ = 0.327, P = 0.0012; rs10853535, τ = 0.311, P = 0.0019, Supplemental Fig. 2), possibly suggesting that the two subclades carrying the JK*A allele are maintained by overdominance. Conversely, we found no heterozygote excess for rs900971, which showed the strongest correlation with pathogens among all BGA SNPs (Table 1). This variant is located further downstream the Asp280Asn SNP and displays low linkage disequilibrium with the balancing selection region. It is therefore tempting to speculate that different variants in SLC14A1 have been subjected to pathogen-driven selection under different regimes that might include heterozygote advantage and, possibly, directional selection.
Altogether, the data reported above concur with the idea that multiallelic balancing selection has shaped the evolutionary history of SLC14A1, although _s_everal issues remain to be clarified. In particular, the possible role of urea metabolism in relation to pathogen resistance has been briefly mentioned above as a possible explanation for selection at this locus, but current knowledge on this issue is too limited to warrant extensive speculation. Moreover, consistent with the biological function of SLC14A1, Kidd-null subjects and knockout mice display mild urinary concentrating defects and greater urine output (Sands et al. 1992; Yang et al. 2002). This observation raises the possibility that, together with pathogen-driven selection, the transporter might also have adapted to climatic variables, possibly driven, for example, by the necessity to spare water in hot dry climates. In fact, we did not find any SNP in SLC14A1 to correlate with climatic variables, such as mean temperature and maximum precipitation rate. Yet, the effect might be confounded by pathogen-driven selection or the power to detect a correlation might vary depending on the environmental variable, as previously suggested (Hancock et al. 2008).
BSG (OK system)
Calculation of nucleotide diversity parameters and summary statistics for the whole BSG gene (OMIM no. *109480) revealed an unusual pattern in EA. In both this population and in YRI we observed a θW of 16 × 10−4, a value higher than the 97.5th percentile in EA (Supplemental Table 1). Yet, while in YRI relatively high values for Fu and Li's D* and F* were obtained, all statistics were negative in EA with borderline significance (Table 2). Closer examination indicated that the negative statistics in Europeans are due to the presence of a single highly divergent haplotype carrying 24 singletons. We therefore verified whether this haplotype was present in the African sample and identified six additional chromosomes carrying closely related haplotypes. We next constructed a median-joining network of a 2-kb gene region showing low recombination (Supplemental Fig. 8): The topology indicated the presence of two distantly related haplotype clusters (Fig. 6) with an estimated TMRCA of 1.76 Myr (SD = 576 Kyr, ρ = 4.99; fixed differences with chimpanzee = 34). Calculation of the TMRCA using GENETREE resulted in a comparable estimate (TMRCA = 1.53 Myr, SD = 443 Kyr, _θ_ML = 2.5, _N_e = 10,714; Supplemental Fig. 8). Such divergent haplotype clades can be expected under two different circumstances, namely balancing selection and ancient population structure. Yet, some difference exists in that symmetric balancing selection is expected to elongate the entire neutral genealogy, while the effects of ancient population structure are reflected in an increase in the genealogical time occupied by two single lineages (Takahata 1990; Wall 2000). A possibility to discriminate between these scenarios is to calculate the percentage of congruent mutations, meaning those that occur on the basal branches of a genealogy (Wall 2000). When we applied this approach to the two major BSG clades, a percentage of congruent mutations equal to 34% was obtained; this is lower than previous estimates under a model of ancient population structure, which ranged from 42% to 45% (Barreiro et al. 2005; Garrigan et al. 2005); also, the TMRCA we estimated for the BSG gene is not unusual (Tishkoff and Verrelli 2003; Garrigan and Hammer 2006), while deep coalescent times are expected when ancient population subdivision is involved. The asymmetric structure of the haplotype genealogy whereby most chromosomes cluster in one clade with a relatively deep coalescent, while a minor branch is accounted for by a small number of less diverged chromosomes is difficult to interpret within a theoretical framework. Different explanations might account for the BSG genealogy, one appealing possibility being frequency-dependent balancing selection, accounting for the maintenance of a distantly related haplotype with a low frequency in the population. Another possibility is that different selective events have being acting on the BSG locus or, else, that complex demographic scenarios account for the pattern we observe nowadays.
Figure 6.
Genealogy of BSG haplotypes reconstructed through a median-joining network. The analysis corresponds to the gene region spanning nucleotides 8500–10,300. Population color codes are as follows: (white) EA; (gray) YRI.
Basigin (also known as CD147) has been involved in different biologic and pathologic processes, such as amyloid-beta production, thymocyte maturation, cellular invasion, and rheumatoid arthritis (Iacono et al. 2007). Moreover, functioning as a receptor for cyclophilin A makes CD147 a facilitator of HIV-1 infection (Pushkarsky et al. 2001). This property derives from the ability of HIV-1 to incorporate cyclophilin A into virions, a feature which is common to other viruses (Castro et al. 2003; Lin and Emerman 2006). Additional studies aimed at clarifying the evolutionary history of BSG and its role in infections might benefit from this initial description.
Conclusions
Haldane's hypothesis as formulated in 1932 posits that infectious diseases have been a major threat to human populations and have therefore exerted strong selective pressures throughout human history (Haldane 1932). A few years later he also presciently proposed that antigens constituted of protein-carbohydrates molecules account for “surprising biochemical diversity by serological tests” and possibly play a role in resistance/predisposition to pathogen infection (Haldane 1949). These lines seem to perfectly fit BGA genes, as demonstrated by both this study and previous descriptions (Saitou and Yamamoto 1997; Koda et al. 2001; Baum et al. 2002; Hamblin et al. 2002).
On the one hand, despite medical advances in treatment and prevention, infectious diseases represent a major selective pressure in humans and account for about 48% of deaths in people younger than 45 yr worldwide (Kapp 1999). On the other hand, different BGAs have been shown to act as receptors for one or more pathogens and differential disease susceptibility has been substantiated in some cases depending on BG phenotype. In this scenario, it is not surprising that BGA genes have been the target of selective pressures and associations between pathogen richness and BGA alleles can be identified.
Indeed, here we show that four BGA genes have been subjected to balancing selection (the underlying selective pressure possibly being an infectious agent) and that pathogen richness has shaped allele frequencies in 11 genes. These data, together with a previous description of non neutral evolution for ABO (Saitou and Yamamoto 1997; Calafell et al. 2008), FUT2 (Koda et al. 2001), GYPA (Baum et al. 2002), and DARC (Hamblin et al. 2002), indicate that BGAs played a central role in the host–pathogen arms race during human evolutionary history.
Methods
DNA samples and sequencing
Human genomic DNA was obtained from the Coriell Institute for Medical Research. All analyzed regions were PCR amplified and directly sequenced; primer sequences are available upon request. PCR products were treated with ExoSAP-IT (USB Corporation), directly sequenced on both strands with a Big Dye Terminator sequencing Kit (v3.1 Applied Biosystem) and run on an Applied Biosystems ABI 3130 XL Genetic Analyzer (Applied Biosystem). Sequences were assembled using AutoAssembler version 1.4.0 (Applied Biosystems), inspected manually by two distinct operators, and singletons were re-amplified and resequenced.
Data retrieval and haplotype construction
Genotype data for two populations, one of African ancestry and one of Caucasian ancestry, were retrieved from the SeattleSNPs website (http://pga.mbt.washington.edu). Nucleotide positions for all analyzed genes correspond to those of SeattleSNPs, which in turn are derived from the following GenBank accession nos.: AY942196 (BSG), AY851161 (CD55), DQ074789 (CD151), AY937240 (FUT2), and AY942197 (SLC14A1).
Genotype data for 238 resequenced human genes were derived from the NIEHS SNPs Program website (http://egp.gs.washington.edu). In particular we selected genes that had been resequenced in populations of defined ethnicity including African American (AA), Caucasians (European ancestry, EA), Yoruba (YRI), and Asians (AS) (NIEHS panel 2). Similarly, genotype data from 304 resequenced genes were derived from the SeattleSNPs Web site. In particular, 201 and 103 genes have been resequenced across panels 1 and 2, respectively, the former containing African American and European American, the latter Yoruban and European subjects.
Haplotypes were inferred using PHASE version 2.1 (Stephens et al. 2001; Stephens and Scheet 2005), a program for reconstructing haplotypes from unrelated genotype data through a Bayesian statistical method. Haplotypes for individuals resequenced in this study are available as supplementary material (Supplemental File 3).
Linkage disequilibrium analyses were performed using Haploview (Barrett et al. 2005), and haplotype blocks were identified through an implemented method (Gabriel et al. 2002).
Data concerning HGDP-CEPH SNPs derive from a previous work (Li et al. 2008). A SNP was ascribed to a specific gene if it was located within the transcribed region or no more than 700 bp upstream from the transcription start site.
Statistical analysis
The correlation between pathogen richness and BGA allele frequencies was assessed by Kendall's rank correlation coefficient (τ), a non-parametric statistic used to measure the degree of correspondence between two rankings. The reason for using this test is that even in the presence of ties, the sampling distribution of τ satisfactorily converges to a normal distribution for values of n larger than 10 (Salkind 2007).
In order to evaluate the probability of obtaining 19 SNPs out of 262 with a τ higher than the 95th percentile, we performed 30,000 simulations. In particular, samples of 262 SNPs were extracted from the full data set by searching for each BGA SNP, one with an allele frequency matched at the 0.001 level. For each sample, the number of SNPs with a percentile rank higher than the 95th percentile (calculated over all SNPs) was counted. By this procedure, the empirical probability of obtaining 19 or more SNPs was estimated equal to 0.045. A theoretic approach can also be applied by considering that the probability to obtain n SNPs with a τ higher than the 95th percentile in a sample of 262 is Poisson-distributed with lambda = 13 (5% of 262). For such a distribution the probability of obtaining 19 or more SNPs equals 0.043.
The _F_ST statistic (Wright 1950) estimates genetic differentiation among populations and was calculated as previously proposed (Hudson et al. 1992). In order to identify gene regions showing extreme _F_ST-values, sliding windows of 5 kb moving along BGA genes with a step of 150 bp were used; the same procedure was applied to all genes resequenced by the SeattleSNPs program. Values deriving from sliding windows obtained from all genes resequenced in panels 1 and 2 were used to identify the 2.5th and 97.5th percentiles that represented the threshold to define unusually high or low _F_ST-values in BGA genes. It is worth noting that negative _F_ST should be interpreted as 0 and the 2.5th percentile value of _F_ST from SeattleSNPs gene sliding windows resulted extremely close to 0. Therefore, BGA windows displaying an _F_ST-value negative or equal to 0 were considered to display exceedingly low population differentiation. In order to evaluate the probability of obtaining 8.3% of windows showing _F_ST-values, either below the 2.5th or above the 97.5th percentiles, we used a simulation-based approach. In particular, 17 genes were randomly selected from the SeattleSNPs database and for each group the fraction of sliding windows showing exceedingly low or high values was counted. Ten thousand simulations were performed and the probability of obtaining a fraction of outliers equal to or higher than 8.3% was calculated.
Tajima's D (Tajima 1989), Fu and Li's D* and F* (Fu and Li 1993) statistics, as well as diversity parameters θW (Watterson 1975) and π (Nei and Li 1979) and Fay and Wu's H (Fay and Wu 2000) were calculated using libsequence (Thornton 2003), a C++ class library providing an object-oriented framework for the analysis of molecular population genetic data. Calibrated coalescent simulations were performed using the cosi package (Schaffner et al. 2005) and its best-fit parameters for YRI, AA, EA, and AS populations with 10,000 iterations. As a further control, summary statistics were calculated for 5 kb windows deriving from NIEHS genes and the values obtained for BGA gene regions compared to their distribution. In particular, for each gene a 5 kb region was randomly selected; the only requirement was that it did not contain any long (>500 bp) resequencing gap; if the gene did not fulfill this requirement it was discarded, as were 5 kb regions displaying less than five SNPs. The numbers of analyzed windows for AA, YRI, EA, and AS were 209, 203, 177, and 172, respectively. The same procedure was applied to SeattleSNPs genes and a total of 103, 201, and 298 windows were obtained for YRI, AA, and subjects with European ancestry, respectively.
The maximum-likelihood-ratio HKA test was performed using the MLHKA software (Wright and Charlesworth 2004) using multilocus data of 16 genes and Pan troglodytes (NCBI panTro2) as an outgroup. The 16 reference genes were randomly selected among NIEHS loci shorter than 20 kb that have been resequenced in the four populations (YRI, AA, EA, and AS; panel 2); the only criterion was that Tajima's D did not suggest the action of natural selection (i.e., _D_T is higher than the 2.5th and lower than the 97.5th percentiles in the distribution of NIEHS genes; see Supplemental Table 3). The reference set was accounted for by the following genes: VNN3, PLA2G2D, MB, MAD2L2, HRAS, CYP17A1, ATOX1, BNIP3, CDC20, NGB, TUBA1, MT3, NUDT1, PRDX5, RETN, and JUND.
We evaluated the likelihood of the model under two different assumptions: that all loci evolved neutrally and that only the region under analysis was subjected to natural selection; statistical significance was assessed by a likelihood ratio test. We used a chain length (the number of cycles of the Markov chain) of 2 × 105 and, as suggested by Wright and Charlesworth (2004), we ran the program several times with different seeds to ensure stability of results.
Median-joining networks to infer haplotype genealogy was constructed using NETWORK 4.5 (Bandelt et al. 1999). Estimate of the time to the most common ancestor (TMRCA) was obtained using a phylogeny based approach implemented in NETWORK using a mutation rate based on the number of fixed differences between human and chimpanzee or orangutan and assuming a separation time from humans of 6 Myr and 13 Myr ago, respectively (Glazko and Nei 2003). In all cases, a second TMRCA estimate derived from application of a maximum-likelihood coalescent method implemented in GENETREE (Griffiths and Tavare 1994, 1995). Again, the mutation rate μ was obtained on the basis of the divergence between human and a primate, assuming a generation time of 25 yr. In using this μ and the estimated maximum likelihood θ (θML), we estimated the effective population size parameter (_N_e). With these assumptions, the coalescence time, scaled in 2_N_e units, was converted into years. For the coalescence process, 106 simulations were performed. All calculations were performed in the R environment (www.r-project.org).
Environmental variables
Pathogen absence/presence matrices for the 21 countries where HGDP-CEPH populations are located were derived from the Gideon database (http://www.gideononline.com) following previous indications (Prugnolle et al. 2005). Briefly, only species that are transmitted in the countries were included, meaning that cases of transmission due to tourism and immigration were not taken into account; also, species that have recently been eradicated as a result, for example, of vaccination campaigns, were recorded as present in the matrix. It should be noted that the final number of different pathogen species per country differ from those calculated by Prugnolle et al. (2005), since these authors only took into account intracellular disease agents. Precipitation rate and mean temperature were derived for the geographic coordinates corresponding to HGDP-CEPH populations from the NCEP/NCAR database (Kistler et al. 2001).
Sequence annotation
Data concerning DNase I hypersensitive sites in CD4+ T cells derive from a previous work (Boyle et al. 2008) and were retrieved from the UCSC annotation tables (http://genome.ucsc.edu, Duke DNase I HS track). MicroRNA binding sites were identified through the dedicated utility at miRBase, which relies on the miRanda algorithm (John et al. 2004) and requires a target site to be conserved in at least two species. Functional elements in 3′UTR were searched for using UTRscan (Pesole and Liuni 1999).
Acknowledgments
We thank Dr. Roberto Giorda for helpful comments and discussion about the manuscript. M.S. and R.C. are part of the Doctorate School of Molecular Medicine, University of Milan.
Footnotes
References
- Akey J.M., Swanson W.J., Madeoy J., Eberle M., Shriver M.D. TRPV6 exhibits unusual patterns of polymorphism and divergence in worldwide populations. Hum. Mol. Genet. 2006;15:2106–2113. doi: 10.1093/hmg/ddl134. [DOI] [PubMed] [Google Scholar]
- Ali S., Niang M.A., N'doye I., Critchlow C.W., Hawes S.E., Hill A.V., Kiviat N.B. Secretor polymorphism and human immunodeficiency virus infection in Senegalese women. J. Infect. Dis. 2000;181:737–739. doi: 10.1086/315234. [DOI] [PubMed] [Google Scholar]
- Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Patin E., Neyrolles O., Cann H.M., Gicquel B., Quintana-Murci L. The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region. Am. J. Hum. Genet. 2005;77:869–886. doi: 10.1086/497613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baum J., Ward R.H., Conway D.J. Natural selection on the erythrocyte surface. Mol. Biol. Evol. 2002;19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- Blackwell C.C., Weir D.M., James V.S., Todd W.T., Banatvala N., Chaudhuri A.K., Gray H.G., Thomson E.J., Fallon R.J. Secretor status, smoking and carriage of Neisseria meningitidis. Epidemiol. Infect. 1990;104:203–209. doi: 10.1017/s0950268800059367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld O.O., Patnaik S.K. Allelic genes of blood group antigens: A source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Hum. Mutat. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb K.L., Bovee D., Buckley D., Haugen E., Kibukawa M., Paddock M., Palmieri A., Subramanian S., Zhou Y., Kaul R., et al. Scan of human genome reveals no new loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafell F., Roubinet F., Ramírez-Soriano A., Saitou N., Bertranpetit J., Blancher A. Evolutionary dynamics of the human ABO gene. Hum. Genet. 2008;124:123–135. doi: 10.1007/s00439-008-0530-8. [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Abel L. Human genetics of infectious diseases: A unified theory. EMBO J. 2007;26:915–922. doi: 10.1038/sj.emboj.7601558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A.P., Carvalho T.M., Moussatche N., Damaso C.R. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 2003;77:9052–9068. doi: 10.1128/JVI.77.16.9052-9068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereb N., Hughes A.L., Yang S.Y. Locus-specific conservation of the HLA class I introns by intra-locus homogenization. Immunogenetics. 1997;47:30–36. doi: 10.1007/s002510050323. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P., Warren C.D., Altaye M., Morrow A.L., Ruiz-Palacios G., Pickering L.K., Newburg D.S. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology. 2001;11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clarkson N.A., Kaufman R., Lublin D.M., Ward T., Pipkin P.A., Minor P.D., Evans D.J., Almond J.W. Characterization of the echovirus 7 receptor: Domains of CD55 critical for virus binding. J. Virol. 1995;69:5497–5501. doi: 10.1128/jvi.69.9.5497-5501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costache M., Apoil P.A., Cailleau A., Elmgren A., Larson G., Henry S., Blancher A., Iordachescu D., Oriol R., Mollicone R. Evolution of fucosyltransferase genes in vertebrates. J. Biol. Chem. 1997;272:29721–29728. doi: 10.1074/jbc.272.47.29721. [DOI] [PubMed] [Google Scholar]
- Cowin A.J., Adams D., Geary S.M., Wright M.D., Jones J.C., Ashman L.K. Wound healing is defective in mice lacking tetraspanin CD151. J. Invest. Dermatol. 2006;126:680–689. doi: 10.1038/sj.jid.5700142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywes C., Stamenkovic I., Wessels M.R. CD44 as a receptor for colonization of the pharynx by group A Streptococcus. J. Clin. Invest. 2000;106:995–1002. doi: 10.1172/JCI10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J.C., Wu C.I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Fitter S., Tetaz T.J., Berndt M.C., Ashman L.K. Molecular cloning of cDNA encoding a novel platelet-endothelial cell tetra-span antigen, PETA-3. Blood. 1995;86:1348–1355. [PubMed] [Google Scholar]
- Fu Y.X., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Liu Z., Lin J., Chen W., Jia Z., Pan D., Xu A. Extensive polymorphism and different evolutionary patterns of intron 2 were identified in the HLA-DQB1 gene. Immunogenetics. 2003;54:761–766. doi: 10.1007/s00251-002-0523-z. [DOI] [PubMed] [Google Scholar]
- Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gagneux P., Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- Garrigan D., Hammer M.F. Reconstructing human origins in the genomic era. Nat. Rev. Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- Garrigan D., Mobasher Z., Kingan S.B., Wilder J.A., Hammer M.F. Deep haplotype divergence and long-range linkage disequilibrium at xp21.1 provide evidence that humans descend from a structured ancestral population. Genetics. 2005;170:1849–1856. doi: 10.1534/genetics.105.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G.V., Nei M. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Greenwell P. Blood group antigens: Molecules seeking a function? Glycoconj. J. 1997;14:159–173. doi: 10.1023/a:1018581503164. [DOI] [PubMed] [Google Scholar]
- Griffiths R.C., Tavare S. Sampling theory for neutral alleles in a varying environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994;344:403–410. doi: 10.1098/rstb.1994.0079. [DOI] [PubMed] [Google Scholar]
- Griffiths R.C., Tavare S. Unrooted genealogical tree probabilities in the infinitely-many-sites model. Math. Biosci. 1995;127:77–98. doi: 10.1016/0025-5564(94)00044-z. [DOI] [PubMed] [Google Scholar]
- Gross D.S., Garrard W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Guernier V., Hochberg M.E., Guegan J.F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2:e141. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J.B.S. The causes of evolution. Longmans, Green & Co; London, UK: 1932. [Google Scholar]
- Haldane J.B.S. Garland Publishing Inc; New York/London: 1949. Disease and evolution. Symposium sui fattori ecologici e genetici della speciazione negli animali; Selected genetic papers of J.B.S. Haldane (Anonymous) pp. 325–334. [Google Scholar]
- Hamblin M.T., Thompson E.E., Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A.M., Witonsky D.B., Gordon A.S., Eshel G., Pritchard J.K., Coop G., Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley L.J., Manica A., Goudet J., Balloux F. Going the distance: Human population genetics in a clinal world. Trends Genet. 2007;23:432–439. doi: 10.1016/j.tig.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Haverkorn M.J., Goslings W.R. Streptococci, ABO blood groups, and secretor status. Am. J. Hum. Genet. 1969;21:360–375. [PMC free article] [PubMed] [Google Scholar]
- Hill A.V. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 2006;40:469–486. doi: 10.1146/annurev.genet.40.110405.090546. [DOI] [PubMed] [Google Scholar]
- Ho S.H., Martin F., Higginbottom A., Partridge L.J., Parthasarathy V., Moseley G.W., Lopez P., Cheng-Mayer C., Monk P.N. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J. Virol. 2006;80:6487–6496. doi: 10.1128/JVI.02539-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R., Kreitman M., Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R., Slatkin M., Maddison W.P. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd E.A., Domino S.E. Increased susceptibility of secretor factor gene Fut2-null mice to experimental vaginal candidiasis. Infect. Immun. 2004;72:4279–4281. doi: 10.1128/IAI.72.7.4279-4281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono K.T., Brown A.L., Greene M.I., Saouaf S.J. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp. Mol. Pathol. 2007;83:283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp C. WHO warns of microbial threat. Lancet. 1999;353:2222. doi: 10.1016/S0140-6736(05)76281-4. [DOI] [PubMed] [Google Scholar]
- Karamatic Crew V., Burton N., Kagan A., Green C.A., Levene C., Flinter F., Brady R.L., Daniels G., Anstee D.J. CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood. 2004;104:2217–2223. doi: 10.1182/blood-2004-04-1512. [DOI] [PubMed] [Google Scholar]
- Kaul A., Nagamani M., Nowicki B. Decreased expression of endometrial decay accelerating factor (DAF), a complement regulatory protein, in patients with luteal phase defect. Am. J. Reprod. Immunol. 1995;34:236–240. doi: 10.1111/j.1600-0897.1995.tb00947.x. [DOI] [PubMed] [Google Scholar]
- Kelly R.J., Rouquier S., Giorgi D., Lennon G.G., Lowe J.B. Sequence and expression of a candidate for the human secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- Kendall M.G. Rank correlation methods. Griffin; London: 1976. [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Kistler R., Kalnay E., Collins W., Saha S., White G., Woollen J., Chelliah M., Ebisuzaki W., Kanamitsu M., Kousky V., et al. The NCEP-NCAR 50-year reanalysis: Monthly means CD-ROM and documentation. Bull. Am. Meteorol. Soc. 2001;82:247–268. [Google Scholar]
- Koda Y., Soejima M., Liu Y., Kimura H. Molecular basis for secretor type alpha(1,2)-fucosyltransferase gene deficiency in a Japanese population: A fusion gene generated by unequal crossover responsible for the enzyme deficiency. Am. J. Hum. Genet. 1996;59:343–350. [PMC free article] [PubMed] [Google Scholar]
- Koda Y., Tachida H., Pang H., Liu Y., Soejima M., Ghaderi A.A., Takenaka O., Kimura H. Contrasting patterns of polymorphisms at the ABO-secretor gene (FUT2) and plasma alpha(1,3)fucosyltransferase gene (FUT6) in human populations. Genetics. 2001;158:747–756. doi: 10.1093/genetics/158.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans J.C., Florquin S., Heikens M., Pals S.T., van der Neut R., Van Der Poll T. CD44 is a macrophage binding site for Mycobacterium tuberculosis that mediates macrophage recruitment and protective immunity against tuberculosis. J. Clin. Invest. 2003;111:681–689. doi: 10.1172/JCI16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Lin T.Y., Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden S., Mahdavi J., Semino-Mora C., Olsen C., Carlstedt I., Boren T., Dubois A. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4:e2. doi: 10.1371/journal.ppat.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., LePendu J., Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Liu Y., Koda Y., Soejima M., Pang H., Schlaphoff T., du Toit E.D., Kimura H. Extensive polymorphism of the FUT2 gene in an African (Xhosa) population of South Africa. Hum. Genet. 1998;103:204–210. doi: 10.1007/s004390050808. [DOI] [PubMed] [Google Scholar]
- Liu Y., Promeneur D., Rojek A., Kumar N., Frokiaer J., Nielsen S., King L.S., Agre P., Carbrey J.M. Aquaporin 9 is the major pathway for glycerol uptake by mouse erythrocytes, with implications for malarial virulence. Proc. Natl. Acad. Sci. 2007;104:12560–12564. doi: 10.1073/pnas.0705313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler S., Lottspeich F., Lanza F., Azorsa D.O., ter Meulen V., Schneider-Schaulies J. CD9, a tetraspan transmembrane protein, renders cells susceptible to canine distemper virus. J. Virol. 1997;71:42–49. doi: 10.1128/jvi.71.1.42-49.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui W.O., Pourmand N., Patterson B.K., Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Moulds J.M., Moulds J.J. Blood group associations with parasites, bacteria, and viruses. Transfus. Med. Rev. 2000;14:302–311. doi: 10.1053/tmrv.2000.16227. [DOI] [PubMed] [Google Scholar]
- Moulds J.M., Nowicki S., Moulds J.J., Nowicki B.J. Human blood groups: Incidental receptors for viruses and bacteria. Transfusion. 1996;36:362–374. doi: 10.1046/j.1537-2995.1996.36496226154.x. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg D.S., Ruiz-Palacios G.M., Morrow A.L. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- Nicholson-Weller A., Wang C.E. Structure and function of decay accelerating factor CD55. J. Lab. Clin. Med. 1994;123:485–491. [PubMed] [Google Scholar]
- Nowicki B., Hart A., Coyne K.E., Lublin D.M., Nowicki S. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J. Exp. Med. 1993;178:2115–2121. doi: 10.1084/jem.178.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G., Liuni S. Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Trends Genet. 1999;15:378. doi: 10.1016/S0168-9525(99)01795-3. [DOI] [PubMed] [Google Scholar]
- Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A.J., Houghton M., Rosa D., Grandi G., et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Prugnolle F., Manica A., Charpentier M., Guegan J.F., Guernier V., Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 2005;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarsky T., Zybarth G., Dubrovsky L., Yurchenko V., Tang H., Guo H., Toole B., Sherry B., Bukrinsky M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza M.W., Blackwell C.C., Molyneaux P., James V.S., Ogilvie M.M., Inglis J.M., Weir D.M. Association between secretor status and respiratory viral illness. BMJ. 1991;303:815–818. doi: 10.1136/bmj.303.6806.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid M.E., Lomas-Frances C. The blood group antigen facts book. Academic; San Diego: 1997. [Google Scholar]
- Roudier N., Bailly P., Gane P., Lucien N., Gobin R., Cartron J.P., Ripoche P. Erythroid expression and oligomeric state of the AQP3 protein. J. Biol. Chem. 2002;277:7664–7669. doi: 10.1074/jbc.M105411200. [DOI] [PubMed] [Google Scholar]
- Rouschop K.M., Sylva M., Teske G.J., Hoedemaeker I., Pals S.T., Weening J.J., van der Poll T., Florquin S. Urothelial CD44 facilitates Escherichia coli infection of the murine urinary tract. J. Immunol. 2006;177:7225–7232. doi: 10.4049/jimmunol.177.10.7225. [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios G.M., Cervantes L.E., Ramos P., Chavez-Munguia B., Newburg D.S. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- Sachs N., Kreft M., van den Bergh Weerman M.A., Beynon A.J., Peters T.A., Weening J.J., Sonnenberg A. Kidney failure in mice lacking the tetraspanin CD151. J. Cell Biol. 2006;175:33–39. doi: 10.1083/jcb.200603073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Yamamoto F. Evolution of primate ABO blood group genes and their homologous genes. Mol. Biol. Evol. 1997;14:399–411. doi: 10.1093/oxfordjournals.molbev.a025776. [DOI] [PubMed] [Google Scholar]
- Salkind N.J. Encyclopedia of measurement and statistics. Sage Publications; Thousand Oaks, CA: 2007. [Google Scholar]
- Sands J.M., Gargus J.J., Frohlich O., Gunn R.B., Kokko J.P. Urinary concentrating ability in patients with Jk(a-b-) blood type who lack carrier-mediated urea transport. J. Am. Soc. Nephrol. 1992;2:1689–1696. doi: 10.1681/ASN.V2121689. [DOI] [PubMed] [Google Scholar]
- Schaeffer A.J., Rajan N., Cao Q., Anderson B.E., Pruden D.L., Sensibar J., Duncan J.L. Host pathogenesis in urinary tract infections. Int. J. Antimicrob. Agents. 2001;17:245–251. doi: 10.1016/s0924-8579(01)00302-8. [DOI] [PubMed] [Google Scholar]
- Schaffner S.F., Foo C., Gabriel S., Reich D., Daly M.J., Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendide K., Deghmane A.E., Reyrat J.M., Talal A., Hmama Z. Mycobacterium bovis BCG urease attenuates major histocompatibility complex class II trafficking to the macrophage cell surface. Infect. Immun. 2004;72:4200–4209. doi: 10.1128/IAI.72.7.4200-4209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafren D.R., Bates R.C., Agrez M.V., Herd R.L., Burns G.F., Barry R.D. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J. Virol. 1995;69:3873–3877. doi: 10.1128/jvi.69.6.3873-3877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmukhappa K., Kim J.K., Kapil S. Role of CD151, A tetraspanin, in porcine reproductive and respiratory syndrome virus infection. Virol. J. 2007;4:62. doi: 10.1186/1743-422X-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima M., Koda Y. Distinct single nucleotide polymorphism pattern at the FUT2 promoter among human populations. Ann. Hematol. 2008;87:19–25. doi: 10.1007/s00277-007-0362-y. [DOI] [PubMed] [Google Scholar]
- Sood R., Zehnder J.L., Druzin M.L., Brown P.O. Gene expression patterns in human placenta. Proc. Natl. Acad. Sci. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. 1990;87:2419–2423. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Satta Y. Footprints of intragenic recombination at HLA loci. Immunogenetics. 1998;47:430–441. doi: 10.1007/s002510050380. [DOI] [PubMed] [Google Scholar]
- Thompson E.E., Kuttab-Boulos H., Witonsky D., Yang L., Roe B.A., Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am. J. Hum. Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. libsequence: A C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Tishkoff S.A., Verrelli B.C. Patterns of human genetic diversity: Implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- von Lindern J.J., Rojo D., Grovit-Ferbas K., Yeramian C., Deng C., Herbein G., Ferguson M.R., Pappas T.C., Decker J.M., Singh A., et al. Potential role for CD63 in CCR5-mediated human immunodeficiency virus type 1 infection of macrophages. J. Virol. 2003;77:3624–3633. doi: 10.1128/JVI.77.6.3624-3633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J.D. Detecting ancient admixture in humans using sequence polymorphism data. Genetics. 2000;154:1271–1279. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Willett B.J., Hosie M.J., Jarrett O., Neil J.C. Identification of a putative cellular receptor for feline immunodeficiency virus as the feline homologue of CD9. Immunology. 1994;81:228–233. [PMC free article] [PubMed] [Google Scholar]
- Wiuf C., Zhao K., Innan H., Nordborg M. The probability and chromosomal extent of trans-specific polymorphism. Genetics. 2004;168:2363–2372. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Genetical structure of populations. Nature. 1950;166:247–249. doi: 10.1038/166247a0. [DOI] [PubMed] [Google Scholar]
- Wright S.I., Charlesworth B. The HKA test revisited: A maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M.D., Geary S.M., Fitter S., Moseley G.W., Lau L.M., Sheng K.C., Apostolopoulos V., Stanley E.G., Jackson D.E., Ashman L.K. Characterization of mice lacking the tetraspanin superfamily member CD151. Mol. Cell. Biol. 2004;24:5978–5988. doi: 10.1128/MCB.24.13.5978-5988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Bankir L., Gillespie A., Epstein C.J., Verkman A.S. Urea-selective concentrating defect in transgenic mice lacking urea transporter UT-B. J. Biol. Chem. 2002;277:10633–10637. doi: 10.1074/jbc.M200207200. [DOI] [PubMed] [Google Scholar]
- Young S.L., Lessey B.A., Fritz M.A., Meyer W.R., Murray M.J., Speckman P.L., Nowicki B.J. In vivo and in vitro evidence suggest that HB-EGF regulates endometrial expression of human decay-accelerating factor. J. Clin. Endocrinol. Metab. 2002;87:1368–1375. doi: 10.1210/jcem.87.3.8350. [DOI] [PubMed] [Google Scholar]
- Young J.H., Chang Y.P., Kim J.D., Chretien J.P., Klag M.J., Levine M.A., Ruff C.B., Wang N.Y., Chakravarti A. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]