Exploring the mechanism of tryptophan 2,3-dioxygenase (original) (raw)
Abstract
The haem proteins TDO (tryptophan 2,3-dioxygenase) and IDO (indoleamine 2,3-dioxygenase) are specific and powerful oxidation catalysts that insert one molecule of dioxygen into L-tryptophan in the first and rate-limiting step in the kynurenine pathway. Recent crystallographic and biochemical analyses of TDO and IDO have greatly aided our understanding of the mechanisms employed by these enzymes in the binding and activation of dioxygen and tryptophan. In the present paper, we briefly discuss the function, structure and possible catalytic mechanism of these enzymes.
Keywords: haem; indoleamine 2,3-dioxygenase; kynurenine; oxygen; tryptophan 2,3-dioxygenase
Abbreviations: IDO, indoleamine 2,3-dioxygenase; hIDO, Homo sapiens IDO; TDO, tryptophan 2,3-dioxygenase; rmTDO, Ralstonia metallidurans TDO; xcTDO, Xanthomonas campestris TDO
Introduction
Haem-containing proteins have a wide variety of functions ranging from oxygen transport, storage and activation to simple single-electron transfers. TDO (tryptophan 2,3-dioxygenase) and IDO (indoleamine 2,3-dioxygenase) are members of a small family of haem enzymes that catalyse the aerobic metabolism of L-tryptophan to _N_-formylkynurenine. TDO was initially discovered in the 1930s and is found in both eukaryotes and prokaryotes [1]. TDO expression is normally restricted to the liver in mammals, although it has been identified in the brain and epididymis of some species [2,3]. IDO was first isolated from rabbit intestine in 1967 [4], and is found in many eukaryotes where it is expressed ubiquitously throughout the body, except in the liver. Some prokaryotic IDO-like proteins from Shewanella species have been identified [5], but to date these have no known function.
Sequence similarity between family members is low, with amino acid sequence alignment indicating an identity of approx. 10% between family members. Homology is only apparent from their three-dimensional structures and the conservation of certain amino acids in their active sites.
Why are TDO and IDO of interest?
A vital biochemical reaction
TDO and IDO activate molecular oxygen and catalyse its insertion into L-tryptophan to form _N_-formylkynurenine, in the first and rate-limiting step in the kynurenine pathway [6]. The kynurenine pathway processes over 90% of L-tryptophan utilized by humans. Although both enzymes catalyse the same reaction, TDO is highly substrate-specific, only dioxygenating L-tryptophan and some tryptophan derivatives substituted in the 5- and 6-positions. IDO displays wider substrate specificity, catalysing the dioxygenation of D-tryptophan, tryptamine and 5-hydroxytryptamine (serotonin) in addition to those substrates dioxygenated by TDO [7].
Tryptophan regulation
Both TDO and IDO play a central role in the regulation of tryptophan in the human body, controlling the kynurenine pathway and influencing serotogenic regulation [8]. IDO and TDO activity alters local tryptophan concentration and the build-up of pathway metabolites can lead to numerous physiological and pathophysiological conditions. These include cataract formation, AIDS-related dementia and ischaemic brain injury [6,8–10]. In addition, the local depletion of tryptophan by IDO or TDO is associated with an antimicrobial response. Some pathogens are sensitive to tryptophan degradation, and this may be an effective mechanism for controlling their ability to proliferate [11].
Immune regulation
Although both enzymes catalyse the same reaction, compartmentalization of IDO and TDO expression is thought to reflect their differing biological roles. IDO plays an active role in the human body's immune response, but there is little evidence to suggest that TDO plays a part in this process. Immune regulation by IDO is extremely complex, but two major roles have become evident [12,13]. First, IDO is a normal effector of peripheral tolerance in the immunoregulatory pathway of tryptophan metabolism. It helps to create an appropriate immune response, influencing the balance between tolerance and attack. Secondly, it is thought to play an essential role in acquired immune tolerance. IDO helps tumours to induce tolerance from the host's immune system by interacting with T-cells and depleting L-tryptophan levels [13], and it has consequently emerged as an attractive drug target in cancer and transplant treatments.
TDO and IDO structures?
Overall structure
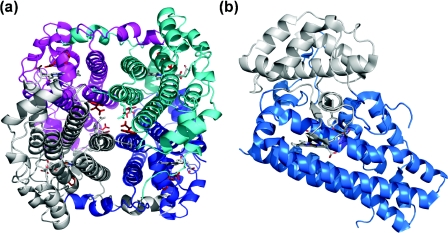
Recent crystallographic studies of xcTDO (Xanthomonas campestris TDO) [5], rmTDO (Ralstonia metallidurans TDO) [14] and hIDO (Homo sapiens IDO) [15] have aided our understanding of this family of haem enzymes. The crystal structures of xcTDO and rmTDO are essentially identical, sharing 47% sequence identity, and are intimately associated homotetrameric enzymes (Figure 1a). They are perhaps best described as a dimer of dimers because the N-terminal residues of each monomer form part of the substrate-binding site in an adjacent monomer. The N-terminal region lies above the distal face of the haem, providing part of the binding cavity. The proteins are completely helical, and a flexible loop (comprising residues 250–260 of xcTDO), involved in L-tryptophan binding, is observed just outside the haem pocket. This loop is only ordered in crystals grown in the presence of L-tryptophan, as substrate binding appears to induce its ordering.
Figure 1. Structures of (a) xcTDO and (b) hIDO.
(a) Structure of xcTDO. L-Tryptophan is shown in red, haem is grey, and separate monomers are shown in cyan, magenta, grey and blue. (b) Structure of hIDO. Haem is shown in grey, the smaller N-terminal domain is shown in grey, and the larger C-terminal domain is shown in marine.
IDO is a monomeric protein, folded into two distinct domains: a large (C-terminal) domain and a small (N-terminal) capping domain (Figure 1b). Contact between these domains is extensive, and they are joined by a long loop. The larger domain is essentially topologically identical with a monomer of TDO, and the structures are readily superposable [5,14,15]. The loop region connecting the two domains provides similar interactions in the active site to those provided by the N-terminal arm of TDO, forming part of the active-site cavity. A flexible loop exists just outside the haem pocket, like that of TDO, but is not fully resolved crystallographically, probably due to the lack of bound substrate in the active site.
Active-site environment
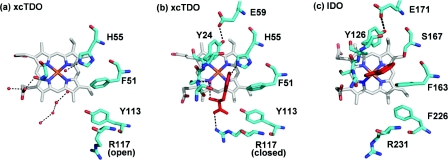
TDO and IDO are _b_-type haem proteins with histidine occupying the axial co-ordination position to the haem iron. The only structure available with substrate bound at the active site in the catalytically active ferrous state is xcTDO. The structure clearly shows the important substrate-binding interactions in the active site. The active-site pockets of xcTDO with and without L-tryptophan bound and that of hIDO are shown in Figure 2. Owing to high similarity between the active sites of TDO and IDO, it is reasonable to assume a similar substrate-binding mode in IDO. Interactions of the substrate with the active site can be split into two categories: interactions with the carboxy and ammonium groups of L-tryptophan, and those with the indole ring system.
Figure 2. Active-site residues of xcTDO and hIDO.
Active-site residues of (a) substrate-free xcTDO, (b) L-tryptophan-bound xcTDO, with L-tryptophan shown in red, and (c) hIDO, with 4-phenylimidazole (shown in red) bound.
In the structure of xcTDO, the carboxy group of L-tryptophan interacts with Arg117, Tyr113 and Thr254. Amino acid residues equivalent to Arg117 and Tyr113 are found in nearly all TDO and IDO proteins. This carboxy-binding motif appears to be essential for substrate binding; arginine reorients in the presence of substrate, co-ordinating the carboxy group of L-tryptophan with both ‘open’ and ‘closed’ conformations observed (Figures 2a and 2b). The closed conformation is only observed in TDO crystals grown in the presence of L-tryptophan. The substrate ammonium group is hydrogen-bonded to the side-chain hydroxy group of Thr254, the 7-propionate group of the haem, and a water molecule. Thr254 is involved in binding both the ammonium and carboxy moieties of L-tryptophan and is part of the flexible loop, just outside the haem pocket.
The substrate indole ring binds into a hydrophobic pocket, where it interacts with the side chain of Phe51 and several other hydrophobic residues, including Tyr24 and Tyr27 from the adjacent monomer. Both of these tyrosine residues are hydrogen-bonded to a glutamate residue, linking the N-terminal loop of the adjacent monomer to the active site, and this Tyr-Glu motif is conserved throughout TDO and IDO proteins.
Perhaps the most obvious difference between the active sites of TDO and IDO is the presence of His55 in xcTDO and its equivalent, Ser167 in hIDO (Figure 2). In TDO, the histidine side chain is hydrogen-bonded with the NH group of the indole ring of L-tryptophan, and a role for His55 would appear to be in regulating substrate binding. In comparison, Ser167 in IDO is incapable of forming this bond, and it appears unlikely that this residue plays any critical role in substrate binding or catalysis by IDO.
What is the catalytic mechanism?
Substrate binding
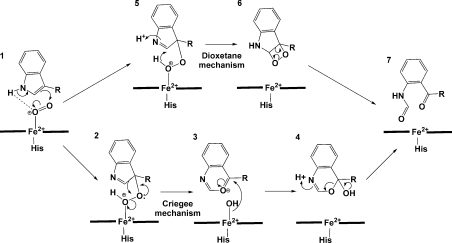
The proposed catalytic mechanism employed by this family of enzymes is shown in Figure 3. It is thought that the ferrous enzyme is required for catalytic activity and, upon substrate binding, there is an increase in the electrochemical midpoint potential for both TDO and IDO. This shift in the reduction potential reflects additional stabilization of the ferrous form.Such a thermodynamic stabilization of the reduced form would clearly favour reduction before substrate binding [5,16–18]. A recent study has suggested that cytochrome _b_5 is the electron transfer partner for IDO, providing ferrous active protein in vivo, consistent with these results.
Figure 3. Proposed catalytic mechanism of IDO and TDO.
Initial formation of the ternary complex (1) occurs by substrate binding to the ferrous protein, followed by dioxygen binding. The ternary complex activates O2 and catalysis proceeds via the formation of a hydroperoxide intermediate (2 and 5) that undergoes either a Criegee or a dioxetane rearrangement to form the product _N_-formylkynurenine (7).
For TDO, the initial formation of the ternary complex (Figure 3, 1) occurs by substrate binding followed by dioxygen binding to the ferrous protein. For IDO, the order of binding events is more complex, but the ternary complex must still be formed before the reaction can proceed. The ternary complex activates O2 and allows the otherwise spin-forbidden reaction to proceed.
Formation of the hydroperoxide intermediate
It has been proposed that formation of the hydroperoxide intermediate (Figure 3, 2 and 5) is catalysed by the loss of the indole proton. Two mechanisms are thought to be probable: base-catalysed deprotonation or proton abstraction by bound dioxygen [7]. If the substrate-binding mode in IDO is similar to that seen in xcTDO, proton abstraction by a proteinaceous base is only possible in TDO, with the active-site histidine (His55 in xcTDO) being the only basic residue in the active site of either enzyme (Figure 2). However, catalytic activity is maintained upon substitution of alanine for His55 in xcTDO, suggesting that base-catalysed deprotonation may not be essential for enzymatic activity [5]. Catalysis by the iron-bound dioxygen is generally proposed for IDO in the absence of a base, and seems reasonable from modelling simulations.
Rearrangement of the hydroperoxide intermediate
The rearrangement of the hydroperoxide intermediate to form the product could occur via either a Criegee or a dioxetane intermediate [7]. Little biochemical information currently exists to discriminate between these two differing pathways, and all discussion has been based on thermodynamic considerations and computer modelling simulations [7,14]. Thermodynamic considerations indicate that formation of the highly strained dioxetane intermediate and its high exothermicity of decomposition make this an unlikely route to be employed by an enzyme. In addition, no chemiluminescence has been detected on decomposition of the dioxetane. These data alone support the Criegee rearrangement for both IDO and TDO. However, further investigation is necessary to determine the nature of this rearrangement.
Conclusion
Recent crystallographic and biochemical analyses of TDO and IDO have aided our understanding of the mechanisms employed for binding and activating dioxygen by allowing identification of key active-site residues. Analysis of the active-site pockets of TDO and IDO shows that there are differences in the choice of residues employed in catalysis. However, how these changes have an impact on the catalytic mechanism and reaction intermediates employed by these enzymes is still unclear. Recent studies have led to possible future routes of investigation. These include: substitution of newly identified key active-site residues to identify their roles in catalysis; EPR and ENDOR (electron nuclear double resonance) studies of reaction intermediates, and computer modelling simulations.
Note added in proof (received 23 September 2008)
In a recent article by Chung et al. [19], density functional theory calculations on the catalytic mechanism of TDO and IDO have cast doubt on the relevance of the Criegee mechanism and raised the possibility of a radical addition pathway.
Acknowledgments
We thank the Wellcome Trust for financial support.
References
- 1.Kotake Y., Masayama I. The intermediary metabolism of tryptophan. XVIII. The mechanism of formation of kynurenine from tryptophan. Z. Physiol. Chem. 1936;243:237–244. [Google Scholar]
- 2.Ishiguro I., Naito J., Saito K., Nagamura Y. Skin L-tryptophan-2,3-dioxygenase and rat hair growth. FEBS Lett. 1993;329:178–182. doi: 10.1016/0014-5793(93)80217-i. [DOI] [PubMed] [Google Scholar]
- 3.Haber R., Bessette D., Hulihan-Giblin B., Durcan M.J. Identification of tryptophan 2,3-dioxygenase RNA in rodent brain. J. Neurochem. 1993;60:1159–1162. doi: 10.1111/j.1471-4159.1993.tb03269.x. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto S., Hayaishi O. Tryptophan pyrrolase of rabbit intestine: D- and L-tryptophan-cleaving enzyme or enzymes. J. Biol. Chem. 1967;242:5260–5266. [PubMed] [Google Scholar]
- 5.Forouhar F., Anderson J.L.R., Mowat C.G., Vorobiev S.M., Hussain A., Abashidze M., Bruckmann C., Thackray S.J., Seetharaman J., Tucker T., et al. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:473–478. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tankiewicz A., Pawlak D., Buczko W. Enzymes of the kynurenine pathway. Postepy Hig. Med. Dosw. 2001;55:715–731. [PubMed] [Google Scholar]
- 7.Sono M., Roach M.P., Coulter E.D., Dawson J.H. Heme-containing oxygenases. Chem. Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 8.Wirleitner B., Neurauter G., Schrocksnadel K., Frick B., Fuchs D. Interferon-γ-induced conversion of tryptophan: immunologic and neuropsychiatric aspects. Curr. Med. Chem. 2003;10:1581–1591. doi: 10.2174/0929867033457179. [DOI] [PubMed] [Google Scholar]
- 9.Wang R., Tang A.-G. Detection and clinical significance of kynurenine. Guowai Yixue Linchuang Shengwu Huaxue Yu Jianyanxue Fence. 2005;26:835–837. [Google Scholar]
- 10.Ohashi H., Saito K., Fujii H., Wada H., Furuta N., Takemura M., Maeda S., Seishima M. Changes in quinolinic acid production and its related enzymes following D-galactosamine and lipopolysaccharide-induced hepatic injury. Arch. Biochem. Biophys. 2004;428:154–159. doi: 10.1016/j.abb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 11.MacKenzie C.R., Heseler K., Muller A., Daubener W. Role of indoleamine 2,3-dioxygenase in antimicrobial defense and immunoregulation: tryptophan depletion versus production of toxic kynurenines. Curr. Drug Metab. 2007;8:237–244. doi: 10.2174/138920007780362518. [DOI] [PubMed] [Google Scholar]
- 12.Munn D.H. Indoleamine 2,3-dioxygenase, tumor-induced tolerance and counter-regulation. Curr. Opin. Immunol. 2006;18:220–225. doi: 10.1016/j.coi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Mellor A.L., Munn D., Chandler P., Keskin D., Johnson T., Marshall B., Jhaver K., Baban B. Tryptophan catabolism and T cell responses. Adv. Exp. Med. Biol. 2003;527:27–35. doi: 10.1007/978-1-4615-0135-0_3. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Kang S.A., Mukherjee T., Bale S., Crane B.R., Begley T.P., Ealick S.E. Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis. Biochemistry. 2007;46:145–155. doi: 10.1021/bi0620095. [DOI] [PubMed] [Google Scholar]
- 15.Sugimoto H., Oda S., Otsuki T., Hino T., Yoshida T., Shiro. Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basran J., Rafice S.A., Chauhan N., Efimov I., Cheesman M.R., Ghamsari L., Raven E.L. A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase. Biochemistry. 2008;47:4752–4760. doi: 10.1021/bi702393b. [DOI] [PubMed] [Google Scholar]
- 17.Papadopoulou N.D., Mewies M., McLean K.J., Seward H.E., Svistunenko D.A., Munro A.W., Raven E.L. Redox and spectroscopic properties of human indoleamine 2,3-dioxygenase and a His303Ala variant: implications for catalysis. Biochemistry. 2005;44:14318–14328. doi: 10.1021/bi0513958. [DOI] [PubMed] [Google Scholar]
- 18.Makino R., Sakaguchi K., Iizuka T., Ishimura Y. L-Tryptophan 2,3-dioxygenase; structure, function and interaction with substrate. Dev. Biochem. 1980;16:179–187. [Google Scholar]
- 19.Chung L.W., Li X., Sugimoto H., Spiro Y., Morokuma K. Density functional theory study on a missing piece in understanding of heme chemistry: the reaction mechanism for indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase. J. Am. Chem. Soc. 2008;130:12299–12309. doi: 10.1021/ja803107w. [DOI] [PubMed] [Google Scholar]