Induction of Pyruvate Dehydrogenase Kinase-3 by Hypoxia-inducible Factor-1 Promotes Metabolic Switch and Drug Resistance (original) (raw)
Abstract
The switch of cellular metabolism from mitochondrial respiration to glycolysis is the hallmark of cancer cells and associated with tumor malignancy. However, the mechanism of this metabolic switch remains largely unknown. Herein, we reported that hypoxia-inducible factor-1 (HIF-1) induced pyruvate dehydrogenase kinase-3 (PDK3) expression leading to inhibition of mitochondrial respiration. Promoter activity assay, small interference RNA knockdown assay, and chromatin immunoprecipitation assay demonstrated that hypoxia-induced PDK3 gene activity was regulated by HIF-1 at the transcriptional level. Forced expression of PDK3 in cancer cells resulted in increased lactic acid accumulation and drugs resistance, whereas knocking down PDK3 inhibited hypoxia-induced cytoplasmic glycolysis and cell survival. These data demonstrated that increased PDK3 expression due to elevated HIF-1α in cancer cells may play critical roles in metabolic switch during cancer progression and chemoresistance in cancer therapy.
Elevated glucose uptake and the switch of cellular metabolism from oxidative phosphorylation to aerobic glycolysis are hallmarks of cancer cells, a phenomenon known as the Warburg effect (1). It is considered that Warburg effect is a critical cellular metabolic adaptation to overexpression of hypoxia-inducible factor (HIF)2 in cancer cells, and elevated glycolysis is due to increased expression of genes encoding glucose transporters and glycolytic enzymes induced by HIF-1 (2–4). However, how cells switch metabolism from oxidative phosphorylation to aerobic glycolysis is still unclear.
Pyruvate dehydrogenase complex is responsible for catalyzing oxidative decarboxylation of pyruvate to produce acetylCoA and NADH to supply the procession of tricarboxylic acid cycle (also known as Krebs cycle) and mitochondrial respiration. Pyruvate dehydrogenase complex is a multienzyme complex consisting of three catalytic enzymes, E1, E2, and E3 (5). The E1 enzyme is also known as pyruvate dehydrogenase (PDH), which catalyzes the rate-limiting reaction of converting pyruvate to acetyl-CoA. The activity of PDH is primarily regulated by pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatase. PDK phosphorylates the α-subunit of PDH to suppress its enzymatic activity, whereas pyruvate dehydrogenase phosphatase dephosphorylates and thus activates PDH (5,6).
Four isotypes of PDKs (PDK1–4), encoded by distinct genes, have been identified in mammals. The expression levels of PDKs vary in a tissue-specific manner, suggesting that they may have different functions (7,8). In addition, the kinetic parameters and regulation of PDKs are also different among different isogenes. The expression of PDK1 can be up-regulated by hypoxia (9,10), whereas the expression of PDK2 is elevated in liver, kidney, and mammary gland during starvation (11). High fat diet and diabetes can enhance PDK4 expression (12,13). However, the regulation of PDK3 gene expression was never reported before.
The activity of PDKs is largely determined by the binding capacity of PDKs to the lipoyl domain of E2. The binding affinity of PDK3 to E2 is the greatest (relative order: PDK3 > PDK1 = PDK2 > PDK4) (14); therefore, it is not surprising that the enzyme activity of PDK3 is the highest among all PDKs (25-fold higher than the activity of the least active PDK2) (7). Furthermore, high concentration of pyruvate inhibits the activity of PDK1, -2, and -4, but not PDK3 (7,15). This unique feature implicates the potential importance of PDK3 in the metabolic switch of cancer cells and makes it the most prominent new candidate as a target for cancer therapy.
When the size of solid tumor is greater than 1 mm3, cells will face hypoxic stress due to slow growth of blood vessels (16). One of cell's responses to hypoxia is through the HIF-regulated gene expression to modulate several biological processes such as angiogenesis, proliferation, migration, apoptosis, and metabolism (17). HIF is a heterodimeric transcription factor consisting of the α (HIF-1α, HIF-2α or HIF-3α) and β subunits (18). HIF-1β, also known as aryl hydrocarbon receptor nuclear translocator, is constitutively expressed, whereas the protein of HIF-α is inducible under hypoxia (18). Therefore, the α subunits of HIF are more important in regulating gene expression under hypoxia.
Previous reports showed that the HIF-α subunits are expressed in a tissue-specific manner. HIF-1α is expressed ubiquitously in human tissues, whereas HIF-2α is expressed in restricted tissues such as lung, endothelium, and carotid body (19–21). However, recent data indicate that HIF-2α is expressed in numerous other cell types, including kidney fibroblasts, hepatocytes, intestinal epithelial cells, pancreatic interstitial cells, cardiomyocytes, and type II pneumocytes (22,23). To date, accumulating evidence shows that HIF-1α and HIF-2α are involved in tumor progression, whereas the expression and function of HIF-3α remain uncharacterized (17,24,25).
Given that PDK3 is the only PDK for which the enzymatic activity is not inhibited by a high concentration of pyruvate, it may play a critical role in causing metabolic switch of cancer cells during progression. Therefore, we aimed to investigate the possible role of HIFs in PDK3 expression and the consequent effect of PDK3 expression in cancer cells. Results from this study provide important information for unraveling the mechanism of cancer malignancy and for considering new anticancer strategy by selecting the novel PDK3 as a molecular target.
EXPERIMENTAL PROCEDURES
_Cell Culture and Hypoxia Treatment_—Cell lines, including HeLa, IMR32, and colo320DM, were maintained in specific culture media recommended by the American Type Culture Collection and were supplemented with 10% fetal bovine serum. The human endometrial stromal cells were purified and cultured in Dulbecco's modified Eagle's medium/F-12 medium with 10% fetal bovine serum, as previously described (26,27). Between 16 and 18 h before undergoing hypoxia, cells were plated at a density of 5 × 105 cells per 30-mm glass dish. The medium was changed at 1 h before treatment to assure an adequate amount of nutrient and growth factor. Hypoxic treatment was carried out in an incubator with 1% O2, 5% CO2, and 94% N2, or with dimethyloxaloylglycine (DMOG) or desferrioxamine (DFO).
_RNA Isolation and Real-time RT-PCR_—Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the standard procedure. RNA concentrations and quality were determined using UV absorption at 260 nm and 280 nm. Total RNA (500 ng) was subjected to reverse transcription at 42 °C for 60 min followed by real-time PCR amplification using a thermal cycler (ABI 7900, Applied Biosystems, Foster City, CA) as previously described (28). The primer sequences used in this study are listed inTable 1 except the 18S rRNA, which was reported previously (29).
TABLE 1.
List of primes used in this study
| Gene | Primer sequencesa | Amplicon length | _T_m |
|---|---|---|---|
| °C | |||
| PDK1 | F: 5′-GAAGATCTAGATCTAAACCTGCCTCCCATTC-3′ | 1419 | 58 |
| R: 5′-CCCAAGCTTAAGCTTCCCGCTAGAGAAGCCACA-3′ | |||
| PDK2 | F: 5′-GAAGATCTAGATCTTGAAAGGCTGTGAGCAAGG-3′ | 1413 | 58 |
| R: 5′-CCCAAGCTTAAGCTTCGCGACTTTGGTTGTGGT-3′ | |||
| PDK3 | F: 5′-GAAGATCTAGATCTCGTCCACTGAAGGCACAAAAGC-3′ | 1454 | 58 |
| R: 5′-CCCAAGCTTAAGCTTGGCGCACAGACCCGCCTA-3′ | |||
| PDK4 | F: 5′-GGGGTACCGGTACCTTGCAGTGAGCCGAGATG-3′ | 1590 | 58 |
| R: 5′-CTAGCTAGCTAGCACTGTGGCTGGCTTGAGG-3′ | |||
| PDK3 | F: 5′-CCCAAGCTTGTCTAGGCGGGTCTGTGC-3′ | 1279 | 58 |
| cDNA | R: 5′-CGGGGTACCTTGTAATGGAAATCAAGGTGGT-3′ | ||
| PDK1 | F: 5′CGGATCAGAAACCGACACA3′ | 106 | 60 |
| R: 5′ACTGAACATTCTGGCTGGTGA3′ | |||
| PDK2 | F: 5′-AAGGACACCTACGGCGATG-3′ | 83 | 60 |
| R: 5′-ATGGAGATGCGGCTGAGG-3′ | |||
| PDK3 | F: 5′-TTAATAAGTCCGCATGGCGC-3′ | 83 | 60 |
| (RT-PCR) | R: 5′-TGAAGCATCCCTGGGTTCAC-3′ | ||
| PDK3 | F: 5′-CCGGACAAAACACAAACGTC-3′ | 260 | 55 |
| (ChIP-PCR) | Nested F: 5′-ATAGGGCACGTTTGCGAAG-3′ | 185 | 55 |
| R: 5′-CAGCAGCAGCTCCAGGAC-3′ | |||
| PDK3 distal | F: 5′-TCTGTTCCAGAATCCCAACC-3′ | 110 | 55 |
| (ChIP-PCR) | R: 5′-GGGCATTCAAAACAAGGAAA-3′ |
_Western Blot_—Equal amounts of proteins were resolved by SDS-PAGE and transferred to a polyvinyl difluoride membrane, which was then blocked with 5% skim milk and incubated with specific antibodies. After washing and incubation with horseradish peroxidase-conjugated second antibodies, bound antibody was detected using the enhanced chemiluminescence system (PerkinElmer Life Sciences). The blots were then stripped with stripping buffer (100 mm 2-mercaptoethanol, 2% SDS, and 62.5 mm Tris-HCl, pH 6.7) and reprobed with different antibodies. Antibodies used in this study are: anti-HIF-1α (Novus Biologicals, NB100–449), anti-HIF-2α (Novus Biologicals, NB100–122), anti-HIF-1β (BD Biosciences, 611078), anti-PDK1 (Stressgen, KAP-PK112E), anti-PDK2 (Santa Cruz Biotechnology, sc-14486), anti-PDK3 (Novus Biologicals, H00005165-M01), and anti-PDK4 (Santa Cruz Biotechnology, sc-14495).
Plasmids, Transfection, and Promoter Activity Assays_—The sequences of human PDK1–4 promoters were retrieved from the Ensembl data base. Human PDK1–4 promoters were amplified using PCR with pairs of primers (Table 1). After digestion by restriction enzyme, human_PDK1–4 promoters were cloned to the pGL3 basic vector (Promega, Madison, WI). Alternatively, two 18-base oligonucleotides (forward: 5′-ctagCGCGTACGTGCAGCAACC-3′, reverse: 5′-tcgaGGTTGCTGCACGTACGCG-3′) corresponding to human PDK3 HRE matrix (–236/–219) were synthesized and cloned into SV40-driven pGL3 plasmid after annealing. Mutation of the putative HRE sequence was achieved by replacing the bases RCGTG with RAAAG.
HeLa cells were placed on 24-well plates for luciferase assays. Commercial plasmids containing a cytomegalovirus-driven β-galactosidase reporter system (Promega) were co-transfected into the cells using Lipofectamine 2000 (Invitrogen). After transfection, the cells were incubated in 10% fetal bovine serum Dulbecco's modified Eagle's medium/F-12 medium for 6 h. The medium was then replaced, and cells were subjected to DFO or hypoxic treatments for 24 h. Luciferase assays were done using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Briefly, 50 μl of luciferase substrates was added to 20 μl of lysate, and luciferase activity was measured using a 20/20 luminometer (Turner Designs, Sunnyvale, CA). The level of β-galactosidase was determined by _A_420. Each luciferase assay experiment was performed in duplicate and repeated as indicated in the figure legends.
_Chromatin Immunoprecipitation-PCR Assay_—The protocol used was as described before (30,31) with modifications. In brief, proteins (HIF-1α) and DNA in normoxia-, hypoxia-, or DFO-treated cells (1 × 106 cells) were cross-linked by incubation for 10 min at room temperature with a final concentration of 1% formaldehyde. After aspiration of the formaldehyde, the cells were washed twice with ice-cold phosphate-buffered saline containing protease inhibitors and scraped into a conical tube. Genomic DNA was sheared to lengths of 0.3–0.5 kb by sonicating the cell lysate. Two percentage of the diluted lysate was kept for input control. The chromatin solution was pre-cleared with mixtures containing bovine serum albumin, salmon sperm DNA, and protein A-Sepharose. Anti-HIF-1α antibody or rabbit IgG (for negative control) was added to the supernatant fraction and subjected to chromatin immunoprecipitation assay following standard procedures. After separated from protein, genomic DNA was amplified by real-time PCR using specific primers (Table 1).
siRNA_—Short interference RNAs (siRNAs) against human HIF-1α, HIF-2α, PDK1, PDK3, and GC content-matched scramble control (Table 2) were purchased from Invitrogen. The siRNA was used to transfect cells with a final concentration of 40 nm. After transfection, cells were incubated in normoxia or hypoxia for 24 h and subjected to real-time RT-PCR quantification or Western blot analysis as described above. In a separated experiment, siRNA-transfected cells were subjected to anti-cancer drug treatment as described below. Specificity of siRNA was determined by quantification of an interferon-γ-sensitive 2′,5′-oligoadenylate synthetase gene_OAS1 (32).
TABLE 2.
List of siRNAs used in this study
| Name | Catalog | Sequence |
|---|---|---|
| siPDK1_1 | HSS107771 | AAGUACUGAACAUUCUGGCUGGUGA |
| siPDK1_2 | HSS107772 | AUAUAAUACAAAUCACACAGACGCC |
| siPDK3_1 | HSS107778 | UGGAUAAGUCUUCUUUACCCAAAGU |
| siPDK3_2 | HSS107776 | UUCUCACAUGCAUUAUCUCUCCCGA |
| siHIF-1α_1 | 12938-122 | CCAUGAGGAAAUGAGAGAAAUGCUU |
| siHIF-1α_2 | 12938-122 | CCACAGUGCAUUGUAUGUGUGAAUU |
| siHIF-2α_1 | HSS103261 | GGCCAGGUGAAAGUCUACAACAACU |
| siHIF-2α_2 | HSS103263 | AGAAAGAUCAUGUCGCCAUCUUGGG |
_Induction of Lactic Acid Production and Cell Survival_—Cells were cultured under normoxia or hypoxia conditions for 24 h. Media were collected for quantification of lactic acid production using a commercially available EIA kit (BioVision, Mountain View, CA) according to the procedure recommended by the manufacturer. Cells were then washed by phosphate-buffered saline for three times, trypsinized, and then counted under inverted microscope by trypan blue exclusion method. In some cases, clinically used anti-cancer drugs, cisplatin and paclitaxel, were administered to culture medium prior to normoxia or hypoxia treatment.
_TUNEL Assay_—The apoptotic cells were detected by using a commercial detection kit (TdT-FragEL™ DNA Fragmentation Detection Kit, QIA33, Calbiochem) according to the manufacturer's procedure. Cells were fixed by 4% formaldehyde at room temperature for 10 min and rehydrated by 1× Tris-buffered saline (20 mm Tris, pH 7.6, 140 mm NaCl) at room temperature for another 15 min. Cells were then incubated with 0.2% Triton X-100 at room temperature for 10 min for permeabilization followed by incubation with 3% H2O2 at room temperature for 5 min. After equilibration by 1× terminal deoxynucleotidyl transferase (TdT) equilibration buffer at room temperature for 30 min, cells were incubated with TdT labeling reaction mixture to label the exposed 3′-OH ends of DNA fragments with biotin-labeled and unlabeled deoxynucleotides catalyzed by TdT at 37 °C for 1.5 h. The labeling reaction was terminated by incubating the cells with stop solution at room temperature for 5 min. After blocking the cells by the blocking buffer, cells were incubated with streptavidin-horseradish peroxidase-conjugated solution at room temperature for 30 min to detect the biotinylated nucleotides. The labeled sample was detected by diaminobenzidine reaction to generate an insoluble colored substrate at the site of DNA fragmentation. For each experiment, at least 1000 cells were counted to determine the positive rate of apoptotic nuclei.
_Statistical Analysis_—The data are expressed as means ± S.E. of the mean (S.E.). Differences between groups were analyzed using one-way analysis of variance in commercial statistical software (GraphPad Prism 4.02, GraphPad Software, San Diego, CA). Tukey's procedure was used to test differences between groups found significant using the_F_-test. In some occasions, Student's t test was used to compare differences between particular groups. Statistical significance was set at p < 0.05.
RESULTS
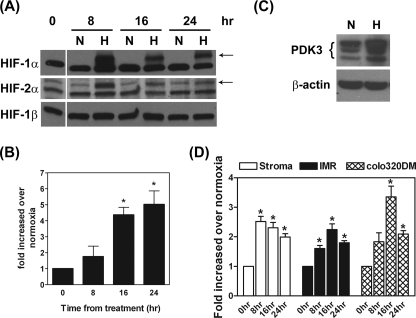
_Hypoxia Induces PDK3 Expression in Normal and Cancer Cells_—To evaluate the expression of PDK3 regulated by HIF, HeLa cells were cultured in 1% O2 (hypoxia) or 21% O2 (normoxia) for various times, and expression of PDK3 mRNA was quantified by real-time RT-PCR. Hypoxia treatment significantly up-regulated HIF-1α and HIF-2α in HeLa cells, whereas the constitutively expressed HIF-1β was not changed (Fig. 1_A_). Expression of PDK3 mRNA was up-regulated at 16 and 24 h after hypoxia treatment (Fig. 1_B_). Concordantly, the level of PDK3 protein was also elevated (Fig. 1_C_).
FIGURE 1.
Hypoxia induces PDK3 expression in normal and cancer cells. A, HeLa cells were cultured under normoxia (N) or hypoxia (H) conditions for 8, 16, and 24 h. Cells were lysed in radioimmune precipitation assay buffer, and expression of HIF-1α, HIF-2α, and HIF-1β was detected via Western blot using specific antibodies.Arrows indicate specific hypoxia-induced HIF-1α and HIF-2α proteins, respectively. The lower bands are nonspecific signals. B, HeLa cells were cultured under normoxia or hypoxia conditions for 8, 16, or 24 h, and expression of PDK3 mRNA was quantified by real-time RT-PCR (n = 6). Asterisks indicate significant differences from the 0-h control. C, representative image shows the expression of PDK3 protein in HeLa cells under normoxia (N) and hypoxia (H) conditions for 24 h. This experiment was repeated for four times and the result was similar. D, human endometrial stromal cells (primary culture, n = 6 using different batches of cells), IMR32 cells (n = 3), and colo320DM cells (n = 3) were cultured under normoxia or hypoxia conditions for 8, 16, and 24 h, and expression of PDK3 mRNA was quantified by real-time RT-PCR. Asterisks indicate significant differences compared with the 0-h control.
To further investigate the role of HIF in PDK3 expression, DFO, an iron chelater that causes HIF-1α accumulation, and the prolylhydroxylase inhibitor, DMOG, were used to block HIF-1α and/or HIF-2α degradation, and levels of PDK3 were detected. Treatment with DFO and DMOG dose-dependently induced PDK3 mRNA expression (supplemental Fig. S1_A_). Concomitantly, DFO and DMOG both induced PDK3 protein expression, accompanied by increased HIF-1α and HIF-2α accumulation in the cell (supplemental Fig. S1_B_). These results demonstrated that hypoxia-induced PDK1 and PDK3 expression was mediated by HIF-1α and/or HIF-2α.
Next, we tested whether elevated expression of PDK3 mediated by hypoxia was a common event among different cell types. The neuroblastoma cancer cell line IMR32, colorectal cancer cell line colo320DM, and primary cultured human endometrial stromal cells were incubated under normoxia or hypoxia conditions as described above. Similar to the results seen in HeLa cells, hypoxia induced PDK3 gene expression in these three types of cells (Fig. 1_D_). This result demonstrated that up-regulation of PDK3 under hypoxia condition is a common event in normal and cancer cells.
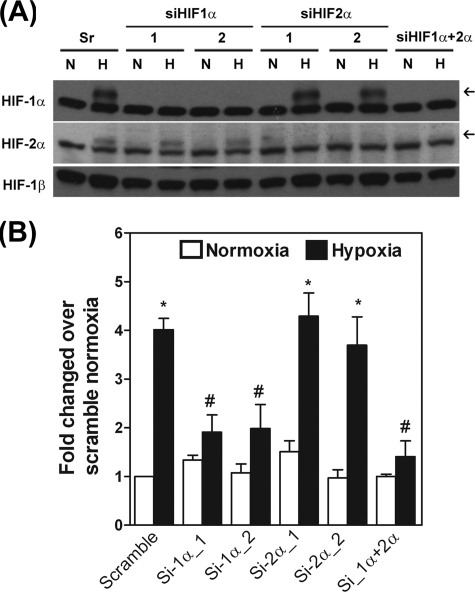
Expression of PDK3 Is Induced by Hypoxia through HIF-1_α, but Not HIF-2_α—Because both HIF-1α and HIF-2α were up-regulated under hypoxia condition, we aimed to determine whether expression of PDK3 by hypoxia was regulated by HIF-1α, HIF-2α, or both. Two individual sets of siRNA against human HIF-1α and HIF-2α, respectively, were used to knock down the endogenous HIF-1α and/or HIF-2α to address this question. After assuring the efficiency and specificity of siRNAs (Fig. 2_A_ and data not shown), HIF-1α and HIF-2α knockdown HeLa cells were cultured under hypoxia condition, and the expression of PDK3 mRNA was quantified by real-time RT-PCR. Expression of PDK3 was increased in cells transfected with scramble siRNA under hypoxia. Knocking down HIF-1α markedly inhibited hypoxia-induced PDK3 expression (Fig. 2_B_). On the other hand, the hypoxia-induced PDK3 expression was not affected by knocking down HIF-2α, whereas double knockdown of HIF-1α and HIF-2α exerted a similar effect as that by single knockdown of HIF-1α (Fig. 2_B_). These results demonstrated that hypoxia increases the expression of PDK3 via HIF-1α but not by HIF-2α.
FIGURE 2.
Up-regulation of PDK3 is mediated by HIF-1α. A, HeLa cells were transfected with siRNAs against HIF-1α, HIF-2α, HIF-1α plus HIF-2α, or GC content-matched scramble control. After transfection for 6 h, cells were cultured under normoxia or hypoxia conditions for 24 h, and levels of HIF-1α, HIF-2α, and HIF-1β were detected by Western blot using specific antibodies. The experiment was repeated three times with similar results. B, HeLa cells were transfected with siRNAs against HIF-1α, HIF-2α, HIF-1α plus HIF-2α, or GC content-matched scramble siRNA. After transfection for 6 h, cells were cultured under normoxia or hypoxia conditions for 24 h, and expression of mRNA for PDK3 was quantified by real-time RT-PCR (n = 3). Asterisks indicate significant differences from normoxia group.#, indicates significant difference from the hypoxia-treated scramble group.
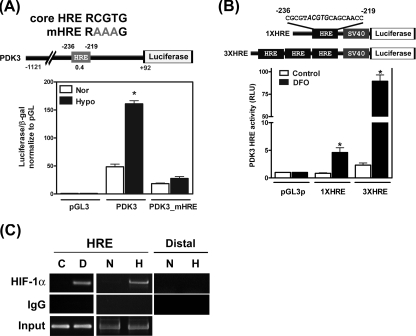
Hypoxia-induced PDK3 Expression Is Regulated at the Transcriptional Level_—To investigate effects of HIF on the expression of PDK3 and detail molecular mechanisms of such regulation, we employed a bioinformatic platform (The Binding Element Searching Tool,thebest.binfo.ncku.edu.tw/thebest) to predict the presence of possible binding elements of HIF (HRE) in the promoter regions of human PDK3 gene. There is one HRE located at –219/–236 of human PDK3 promoter (Fig. 3_A_). To verify whether the bioinformatic-predicted HRE was functional, promoter of PDK3 was cloned into reporter plasmid and luciferase activity was determined. The promoter activity of PDK3 was increased in hypoxia (1% O2)-treated HeLa cells (Fig. 3_A_). The bioinformatic prediction also revealed that there are two HREs in the human PDK1 gene, one HRE with a very low score in human PDK2 gene, and no HRE in human PDK4 (supplemental Fig. S2_A). In agreement with the bioinformatic prediction, promoter activity of PDK1 was increased under hypoxia treatment, whereas the promoter activities of PDK2 and PDK4 were not affected by hypoxia treatment (supplemental Fig. 2, B and_C_). To further confirm that hypoxia-induced promoter activities in PDK3 and PDK1 were mediated by predicted HREs, the sequences of HREs were mutated to abolish HIF-1 binding capacity. Mutation of the HRE sequence in PDK3 completely abolished promoter activity under hypoxia treatment (Fig. 3_A_). Similarly, mutation of HRE2 completely inhibited hypoxia-induced PDK1 promoter activity, whereas mutation of HRE1 in PDK1 only partially reduced PDK1 promoter activity (supplemental Fig. S2_B_). Again, treatment with DFO significantly increased the promoter activities of PDK3 and PDK1, whereas PDK2 promoter activity was not affected by DFO treatment (supplemental Fig. S2_D_). The result was consistent with the predicted score of each HRE in the PDK promoter. The HRE with higher scores (HRE in PDK3 and HRE2 in PDK1) was more responsive, whereas the two HREs with very low scores (HRE1 in PDK1 and HRE in PDK2) exerted minimal or no responsiveness to hypoxia.
FIGURE 3.
Induction of PDK3 by HIF-1α is controlled at the transcriptional level. A, HeLa cells were transiently transfected with control plasmid (pGL3), PDK3, or core HRE mutated PDK3 (PDK3-mHRE) reporter constructs and then cultured under normoxia (21% O2) or hypoxia (1% O2) for 24 h. The reporter plasmid containing β-galactosidase (β_-gal_) was co-transfected for internal control. Data show mean ± S.E. (S.E.) of four independent experiments performed in duplicate.Asterisks indicate significant differences between hypoxia and normoxia groups by unpaired t test. The promoter construct of PDK3 with predicted HRE (gray boxes), score of HRE (numbers under HRE boxes), and the sequences of core HRE and mutated core HRE (mHRE) are shown on top of the figure. B, HeLa cells were co-transfected with β-galactosidase and SV40-driven pGL3 reporter plasmids containing no (pGL3p), one copy (1XHRE), or three copies (3XHRE) of predicted PDK3 HRE sequences and subjected to vehicle (control) or DFO treatment for 24 h. Promoter activities were determined by measuring luciferase and β-galactosidase activities. Data show mean ± S.E. from three independent experiments. Asterisks indicate significant differences between DFO and control groups. C, HeLa cells were cultured under normoxia (N) or hypoxia (H) condition or treated with DFO (D) for 12 h, and binding of HIF-1α to the PDK3 HRE was detected by chromatin immunoprecipitation-PCR assay. Normal rabbit IgG was used as negative control. Un-precipitated genomic DNA (2%) was used for input control. A second set of primer that locates 3.5 kb upstream of predicted HRE site (designed as distal) was also used to amplify the HIF-1α antibody pulled down DNA as control. The experiment was repeated three times with similar results.
Next, we tested whether the predicted HRE was sufficient to drive the PDK3 promoter under hypoxia. Reporter constructs containing one or three predicted HRE matrices of PDK3 (termed 1XHRE and 3XHRE, respectively) were transiently transfected into HeLa cells, and luciferase activity was determined after DFO treatment. Treatment of 1XHRE-transfected HeLa cells with 10 mm DFO significantly enhanced the luciferase activity (Fig. 3_B_). Moreover, the induction of luciferase activity in cells transfected with 3XHRE was further enhanced (47-fold greater than that in cells transfected with 1XHRE) (Fig. 3_B_). Because these reporter constructs contain only the predicted HIF-binding matrix and cells were treated with DFO, the result clearly demonstrated that up-regulation of PDK3 was indeed controlled by binding of HIF transcription factor to the predicted HRE and that the predicted HRE was sufficient for HIF-1-dependent PDK3 up-regulation.
The binding of HIF-1α to the promoter of PDK3 was further confirmed by chromatin immunoprecipitation assay. Using anti-HIF-1α antibody and PCR primers designed around the HRE site, we found HIF-1α specifically bound to the promoters of PDK3 in HeLa cells treated with DFO or hypoxia (Fig. 3_C_). In contrast, primers designed at a distal region (3.5 kb upstream of the transcription start site) failed to amplify PDK3 amplicon in HIF-1α immunoprecipitated DNA (Fig. 3_C_). These results provided the first evidence to demonstrate the direct binding of HIF-1α to the PDK3 HRE within the living cells.
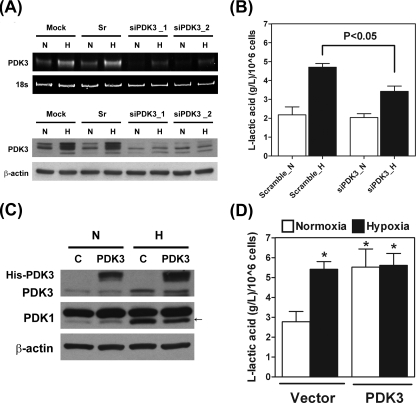
_Up-regulation of PDK3 Induced Lactic Acid Production_—The phosphorylation of PDH by PDK will switch the glucose metabolism from mitochondrial respiration to cytoplasmic glycolysis and result in accumulation of lactic acid. We thus investigated the production of lactic acid to elucidate the functional role of PDK3. Two sets of siRNA against PDK3 were used to knock down the expression of PDK3. Administration of both siPDK3_1 and siPDK3_2 effectively reduced the cellular levels of PDK3 mRNA and protein, whereas the GC content-matched scramble siRNA had no effect (Fig. 4_A_). Concomitantly, production of lactic acid induced by hypoxia was significantly inhibited in siPDK3-treated cells (Fig. 4_B_). To further confirm the role of PDK3 in lactic acid production, full-length cDNA encoding human PDK3 was transfected into HeLa cell. Forced expression of PDK3 increased lactic acid production even under normoxia condition (Fig. 4, C and_D_). These data clearly demonstrated that PDK3 was necessary and sufficient for the induction of metabolic switch.
FIGURE 4.
Up-regulation of PDK3 expression results in increased lactic acid production. A, HeLa cells were transfected with siRNA against PDK3 (siPDK3_1 and siPDK3_2), GC content matched scramble control (Sr), or transfection reagent only (Mock) and cultured under normoxia or hypoxia condition for another 24 h. Levels of PDK3 mRNA and protein were determined by RT-PCR and Western blot, respectively. These experiments were repeated three times with similar results. B, HeLa cells were transfected with siRNA against PDK3 (siPDK3_1 was used) or GC content-matched scramble control (Sr) and cultured for 24 h. Production of lactic acid was determined by using an EIA kit. Data show mean ± S.E. of three independent experiments. C, HeLa cells were transiently transfected with empty vector (C) or histidine-tagged full-length human PDK3 cDNA and cultured under normoxia (N) or hypoxia (H) condition for 24 h. Expression of endogenous and his-tagged PDK3 protein was detected by Western blot. D, HeLa cells were transiently transfected with empty vector (Vector) or full-length human PDK3 cDNA (PDK3) and cultured under normoxia or hypoxia condition for 24 h. Production of lactic acid was measured by using an EIA kit. Data show mean ± S.E. of four independent experiments.Asterisks indicate significant difference from the normoxia-vector group.
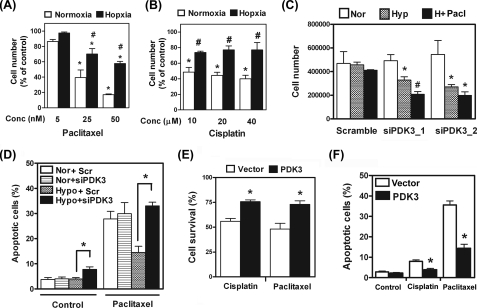
_Induction of PDK3 under Hypoxia Inhibited Drug-induced Cell Death_—It is known that cancer cells with increased lactic acid production due to HIF-induced metabolic switch are more resistant to chemotherapy. We thus aimed to test whether up-regulation of PDK3 by HIF-1α would play a role in increased drug resistance. Two clinically used anti-cancer drugs, paclitaxel and cisplatin, were selected for drug resistance testing. Administration of paclitaxel dose-dependently killed cancer cells under normoxia (Fig. 5_A_). However, when cultured under hypoxia condition, the percentage of cells killed by paclitaxel was significantly reduced (Fig. 5_A_). Similar results were also observed when cisplatin was used (Fig. 5_B_).
FIGURE 5.
Up-regulation of PDK3 by hypoxia increases drug resistance. A, HeLa cells were treated with different doses of paclitaxel and cultured under normoxia or hypoxia conditions for 24 h. Numbers of live cells were counted and expressed as percentage of control. Data show mean ± S.E. of four independent experiments. Asterisks indicate significant differences from untreated group. #, indicates significant difference between normoxia and hypoxia groups. B, HeLa cells were treated with different doses of cisplatin and cultured under normoxia or hypoxia condition for 24 h. Numbers of live cells were counted and expressed as percentage of control. Data show mean ± S.E. of four independent experiments. Asterisks indicate significant differences from untreated group. #, indicates significant difference between normoxia and hypoxia groups. C, HeLa cells were transfected with siRNA against PDK3 (siPDK3_1 and siPDK3_2) or GC content-matched scramble siRNA and cultured under normoxia (Nor), hypoxia (Hyp), or hypoxia plus 25 nm paclitaxel (H + Pacl) for 24 h. Cells were then washed three times with phosphate-buffered saline, and attached live cells were counted under a microscope in the presence of trypan blue dye. Data show mean ± S.E. of four independent experiments.Asterisks indicate significant difference from normoxia groups;#, indicates different from both normoxia and hypoxia groups.D, HeLa cells were transfected with siRNA against PDK3 (siPDK_1) or GC content-matched scramble siRNA and cultured under normoxia or hypoxia and treated with or without 25 nm paclitaxel for 24 h. Apoptotic cells were analyzed by TUNEL assay as described under “Experimental Procedures.” Data show mean ± S.E. of three independent experiments using different batches of cells. E, HeLa cells were transiently transfected with empty vector (Vector) or full-length human PDK3 cDNA (PDK3) and cultured under normoxia condition with vehicle, or cisplatin (20 μm), or paclitaxel (25 nm) for 24 h. Numbers of live cells were counted and expressed as percentage of control (without drug treatment). Data show mean ± S.E. of three independent experiments. Asterisk indicates significant difference between PDK3-overexpressed and vector-only groups. F, HeLa cells were treated as described in E except that TUNEL assay was used to determine the number of apoptotic cells. Asterisk indicates significant difference between PDK3-overexpressed and vector-only groups.
To demonstrate that increased drug resistance under hypoxia was caused by up-regulation of PDK3, we knocked down PDK3 in HeLa cells and re-evaluated drug-induced cell killing. Results demonstrated that knocking down PDK3 alone did not cause cell death under normoxia condition, whereas the survival rate of PDK3-knockdown cells was markedly reduced under hypoxia (Fig. 5_C_). Furthermore, hypoxia-mediated increased resistance to paclitaxel was abolished in PDK3 knockdown cells (Fig. 5_C_). Results from TUNEL assay demonstrated that knocking down PDK3 increased the number of apoptotic cells (Fig. 5_D_ and supplemental Fig. S3) suggesting that the reduced cell number was due to increased cell death. On the other hand, forced overexpression of PDK3 significantly enhanced cell survival against paclitaxel or cisplatin treatment under normoxia condition (Fig. 5,E and F). Taken together, these results demonstrated that up-regulation of PDK3 indeed played a pivotal role in hypoxia-induced drug resistance.
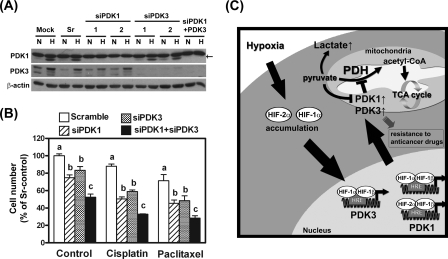
_Up-regulation of PDK1 and PDK3 by HIF Additively Increased Drug Resistance_—Because both PDK1 and PDK3 were up-regulated under hypoxia condition, we were interested to see the roles of these two proteins on HIF1α-induced drug resistance. Transient transfection of HeLa cell with siRNA against PDK1 ablated PDK1 expression (Fig. 6_A_). Similar to what was observed in PDK3 knockdown cells, ablation of PDK1 resulted in increased cell death under hypoxia (Fig. 6_B_). Double knockdown of both PDK1 and PDK3 caused more cell death under hypoxia (Fig. 6_B_) suggesting an additive effect on cell survival. When these cells were treated with cisplatin or paclitaxel, PDK1-deficient cells exert similar percentage of cell death compared with PDK3-knockdown cells (Fig. 6_B_). Simultaneously, knockdown PDK1 and PDK3 resulted in further increasing susceptibility to drug killing (Fig. 6_B_). Together, these data provide compelling evidence to demonstrate that PDK1 and PDK3 played an additive effect in HIF1α-induced drug resistance.
FIGURE 6.
The additive effect of PDK1 and PDK3 in HIF-induced drug resistance. A, HeLa cells were transiently transfected with siRNAs against PDK1 (si1–1 and si1–2), PDK3 (si3–1 and si3–2), both PDK1 and PDK3 (si1 + 3), or scramble control and subjected to normoxia or hypoxia treatment for 24 h. Expression of PDK1, PDK3, and β-actin was detected by Western blot. The arrow indicates specific PDK1 signal. B, HeLa cells were transiently transfected with siRNAs as indicated and subjected to vehicle (control), cisplatin (20 μm), or paclitaxel (25 nm) treatment under hypoxia condition for 24 h. Numbers of live cells were counted and expressed as percentage of scramble-control (sr-control). Data show mean ± S.E. of three independent experiments. Different letters indicate significant difference among the siRNA-transfected groups within each drug treatment group. C, schematic drawing summarizes effect of hypoxia or HIF in PDK1 and PDK3 expression and the downstream effects on metabolic switch and drug resistance.
DISCUSSION
Increased glucose uptake and aerobic glycolysis are hallmarks of energy biogenesis of cancer cells. Switching the cellular metabolism from mitochondrial respiration to cytoplasmic glycolysis not only represents a means for adaptation to hypoxia but also a pivotal transition for cell survival and propagation. However, how this metabolic switch is controlled remains enigmatic. Phosphorylation and thus inactivation of PDH by PDK blocks the conversion of pyruvate to acetyl-CoA and the initiation of mitochondrial respiration. By using bioinformatic and molecular biological approaches, we demonstrated that expression of PDK3 is induced by HIF-1α and proved that up-regulation of PDK3 is critical in controlling metabolic switch and drug resistance in cancer cells (Fig. 6_C_).
Almost 80 years ago, Warburg observed that cancer cells consume more glucose than normal cells, and the way cancer cells generate ATP is through conversion of pyruvate to lactic acid even under normal oxygen supply, a process known as aerobic glycolysis or the Warburg effect (1). Not much about the mechanism of Warburg effect was known until recently when up-regulation of glycolytic enzymes by HIF was identified (9,10,33). In the glucose metabolic pathway, PDK inhibits the conversion of pyruvate to acetyl-CoA thus blocking the entry to the Krebs cycle by phosphorylation and inactivation of PDH. Such action of PDKs inhibits mitochondrial respiration and shifts the cellular biogenesis to cytoplasmic glycolysis. By screening the entire PDK family, we demonstrated that hypoxia can induce PDK1 and PDK3 but not PDK2 or PDK4 expression. The lack of induction of PDK2 by hypoxia was different from previous report in A549 (non-small-cell lung cancer) cells (34). The discrepancy is not known but may be due to difference in cell types used. Next, we pinpointed the exact binding site of HIF-1α on PDK3 by bioinformatic prediction and site-directed mutagenesis approaches. More importantly, we characterized that HIF-1α was essential for PDK3 up-regulation while HIF-2α did not contribute to up-regulation of PDK3. Our current data provide evidence to support distinct functions of HIF-1α and HIF-2α and advance our understanding in hypoxia-regulated PDK3 expression.
Cellular biogenesis is altered under hypoxia condition or HIF-1α overexpression; however, how HIF-1α switches mitochondrial respiration to cytoplasmic glycolysis is not completely understood. In this study, we demonstrated that elevation of PDK3 under hypoxia-induced HIF-1α accumulation results in increased lactic acid production indicating mitochondrial respiration is inhibited and cytoplasmic glycolysis is enhanced. We further demonstrated that forced expression of PDK3 is sufficient to induce lactic acid production under normoxia condition, whereas knocked down PDK3 expression inhibited hypoxia-induced lactic acid production. These data provide evidence to explain an important part of the underlying mechanism of metabolic switch under hypoxia condition.
A growing body of evidence indicates that overexpression of HIF-1α is linked to poor prognosis in human caner and is associated with treatment failure and increase mortality (35). Recent study further demonstrated that mitochondrial respiration-deficient cells have a survival advantage (36). In non-deficient cells, however, up-regulation of PDK1 and PDK3 thus inhibiting mitochondrial respiration may also exert similar effects. Indeed, in this study we demonstrated that hypoxic cells are more resistant to drug-induced cell death. However, the survival advantage under hypoxia was abolished when PDK3 was ablated by siRNA. In contrast, overexpression of PDK3 alone was sufficient to increase drug resistance even under normoxia condition. These data demonstrated that PDK3-mediated metabolic switch plays important roles in hypoxia-induced increased resistance to anti-cancer drugs and provided a molecular mechanism to explain, at least in part, the nature of drug resistance in hypoxic tumor. Recently, it was reported that PDK1 is also induced by hypoxia (9,10). Indeed, our data confirmed this notion and further extended it by pinpointing the exact HIF-1α binding site in PDK1 promoter. Interestingly, ablation of PDK1 by siRNA attenuated hypoxia-induced drug resistance similar to that in PDK3 knockdown cells. Double knockdown of PDK1 and PDK3 additively reduced cell survival under hypoxia. Effects of cisplatin and paclitaxel on cell killing were profoundly enhanced in double knockdown cells compared with scramble siRNA-treated or single PDK1/PDK3 knockdown cells, suggesting the redundant function of these two enzymes. The results also point out the importance of thorough evaluation of members of the gene family to gain a comprehensive picture of their biological functions.
The importance of PDK3 in physiologic or pathologic condition was overlooked in the past partly due to a previous report indicating that PDK3 is exclusively expressed in heart and skeletal muscle by Northern hybridization (8). However, using RT-PCR and/or Western blot, we found that PDK3 is widely expressed in many cell types (Fig. 1).3 Here, we demonstrated that hypoxia induced PDK3 in several normal and cancer cells suggesting it is a common event rather than a unique phenomenon of HeLa cell. The significance of up-regulation of PDK3 by hypoxia is 2-fold. First, the kinase activity and binding affinity of PDK3 are the highest among PDKs (7,14). Induction of PDK3 by hypoxia would efficiently block the conversion of pyruvate to acetyl-CoA and reduce oxygen consumption. Thus, PDK3 serves as a gatekeeper to ensure efficient blockage of PDH activity. Second, it is known that high concentration of pyruvate inhibits the activity of PDK1, -2, and -4, but not PDK3 (7,15). In HIF-1α-overexpressed cancer cells, inactivation of PDH by HIF-induced up-regulation of PDKs would result in the accumulation of pyruvate, and the accumulated pyruvate would inactivate the enzymatic activities of PDK1, -2, and -4, but not PDK3. Therefore, induction of PDK3 guarantees the continuously shutdown of mitochondrial respiration. This is especially important in many HIF-1α-overexpressed cancer cells where pyruvate was also elevated. Taken together, up-regulation of PDK1 and PDK3 by HIF-1α, which is elevated in cancer cells due to hypoxia or oncogene-induced overexpression, may provide a reasonable explanation for the phenomenon of Warburg effect.
In conclusion, by using bioinformatic and molecular biological approaches, we have demonstrated that hypoxia induces PDK1 and PDK3 expression in cancer cells. Up-regulation of PDK1 and PDK3 under hypoxia induces metabolic switch and drug resistance. Given its unique feature in enzyme kinetics and biological function, PDK3 may represent the critical molecule that controls the metabolic switch and drug resistance in cancer cells, especially those with elevated HIF-1α levels. Our current findings provide novel insights about pathological processes of cancer cells and suggest that PDK3 may be a new and pivotal target for tumor therapy.
Supplementary Material
[Supplemental Data]
Acknowledgments
We thank The Bioinformatics Center at the National Cheng Kung University for help in bioinformatic analysis of HIF binding sites.
*
This work was supported by the National Research Program of Genome Medicine (Grant NSC94-3112-B-006-010) and the Landmark Project (Grant A-0123) of National Cheng Kung University, Taiwan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
S⃞
The on-line version of this article (available athttp://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
2
The abbreviations used are: HIF, hypoxia-inducible factor; E1, pyruvate dehydrogenase; E2, dihydrolipoyl acetyltransferase; E3, dihydrolipoyl dehydrogenase; PDH, pyruvate dehydrogenase; PDK, pyruvate dehydrogenase kinase; DMOG, dimethyloxaloylglycine; DFO, desferrioxamine; RT, reverse transcription; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; HRE, binding element of HIF; siRNA, short interference RNA.
3
C.-W. Lu, S.-C. Lin, K.-F. Chen, Y.-Y. Lai, and S.-J. Tsai, unpublished data.
References
- 1.Warburg, O., Wind, F., and Neglers, E. (1930) The Metabolism of Tumors Constable and Co., London, pp. 254–270
- 2.Iyer, N. V., Kotch, L. E., Agani, F., Leung, S. W., Laughner, E., Wenger, R. H., Gassmann, M., Gearhart, J. D., Lawler, A. M., Yu, A. Y., and Semenza, G. L. (1998) Genes Dev. 12 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seagroves, T. N., Ryan, H. E., Lu, H., Wouters, B. G., Knapp, M., Thibault, P., Laderoute, K., and Johnson, R. S. (2001) Mol. Cell. Biol. 21 3436–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greijer, A. E., van der Groep, P., Kemming, D., Shvarts, A., Semenza, G. L., Meijer, G. A., van de Wiel, M. A., Belien, J. A., van Diest, P. J., and van der Wall, E. (2005) J. Pathol. 206 291–304 [DOI] [PubMed] [Google Scholar]
- 5.Patel, M. S., and Korotchkina, L. G. (2001) Exp. Mol. Med. 33 191–197 [DOI] [PubMed] [Google Scholar]
- 6.Roche, T. E., and Hiromasa, Y. (2007) Cell Mol. Life Sci. 64 830–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowker-Kinley, M. M., Davis, W. I., Wu, P., Harris, R. A., and Popov, K. M. (1998) Biochem. J. 329 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudi, R., Bowker-Kinley, M. M., Kedishvili, N. Y., Zhao, Y., and Popov, K. M. (1995) J. Biol. Chem. 270 28989–28994 [DOI] [PubMed] [Google Scholar]
- 9.Kim, J. W., Tchernyshyov, I., Semenza, G. L., and Dang, C. V. (2006) Cell Metab. 3 177–185 [DOI] [PubMed] [Google Scholar]
- 10.Papandreou, I., Cairns, R. A., Fontana, L., Lim, A. L., and Denko, N. C. (2006) Cell Metab. 3 187–197 [DOI] [PubMed] [Google Scholar]
- 11.Wu, P., Blair, P. V., Sato, J., Jaskiewicz, J., Popov, K. M., and Harris, R. A. (2000) Arch. Biochem. Biophys. 381 1–7 [DOI] [PubMed] [Google Scholar]
- 12.Holness, M. J., Kraus, A., Harris, R. A., and Sugden, M. C. (2000) Diabetes 49 775–781 [DOI] [PubMed] [Google Scholar]
- 13.Wu, P., Inskeep, K., Bowker-Kinley, M. M., Popov, K. M., and Harris, R. A. (1999) Diabetes 48 1593–1599 [DOI] [PubMed] [Google Scholar]
- 14.Tuganova, A., Boulatnikov, I., and Popov, K. M. (2002) Biochem. J. 366 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker, J. C., Yan, X., Peng, T., Kasten, S., and Roche, T. E. (2000) J. Biol. Chem. 275 15773–15781 [DOI] [PubMed] [Google Scholar]
- 16.Brown, J. M., and Giaccia, A. J. (1998) Cancer Res. 58 1408–1416 [PubMed] [Google Scholar]
- 17.Ke, Q., and Costa, M. (2006) Mol. Pharmacol. 70 1469–1480 [DOI] [PubMed] [Google Scholar]
- 18.Semenza, G. (2002) Biochem. Pharmacol. 64 993–998 [DOI] [PubMed] [Google Scholar]
- 19.Ema, M., Taya, S., Yokotani, N., Sogawa, K., Matsuda, Y., and Fujii-Kuriyama, Y. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian, H., Hammer, R. E., Matsumoto, A. M., Russell, D. W., and McKnight, S. L. (1998) Genes Dev. 12 3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian, H., McKnight, S. L., and Russell, D. W. (1997) Genes Dev. 11 72–82 [DOI] [PubMed] [Google Scholar]
- 22.Rosenberger, C., Mandriota, S., Jurgensen, J. S., Wiesener, M. S., Horstrup, J. H., Frei, U., Ratcliffe, P. J., Maxwell, P. H., Bachmann, S., and Eckardt, K. U. (2002) J. Am. Soc. Nephrol. 13 1721–1732 [DOI] [PubMed] [Google Scholar]
- 23.Wiesener, M. S., Jurgensen, J. S., Rosenberger, C., Scholze, C. K., Horstrup, J. H., Warnecke, C., Mandriota, S., Bechmann, I., Frei, U. A., Pugh, C. W., Ratcliffe, P. J., Bachmann, S., Maxwell, P. H., and Eckardt, K. U. (2003) FASEB J. 17 271–273 [DOI] [PubMed] [Google Scholar]
- 24.Holmquist-Mengelbier, L., Fredlund, E., Lofstedt, T., Noguera, R., Navarro, S., Nilsson, H., Pietras, A., Vallon-Christersson, J., Borg, A., Gradin, K., Poellinger, L., and Pahlman, S. (2006) Cancer Cell 10 413–423 [DOI] [PubMed] [Google Scholar]
- 25.Haase, V. H. (2006) Am. J. Physiol. 291 F271–F281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai, S. J., Wu, M. H., Chen, H. M., Chuang, P. C., and Wing, L. Y. (2002) Endocrinology 143 2715–2721 [DOI] [PubMed] [Google Scholar]
- 27.Tsai, S. J., Wu, M. H., Lin, C. C., Sun, H. S., and Chen, H. M. (2001) J. Clin. Endocrinol. Metab. 86 5765–5773 [DOI] [PubMed] [Google Scholar]
- 28.Chuang, P. C., Sun, H. S., Chen, T. M., and Tsai, S. J. (2006) Mol. Cell. Biol. 26 8281–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, H. S., Hsiao, K. Y., Hsu, C. C., Wu, M. H., and Tsai, S. J. (2003) Endocrinology 144 3934–3942 [DOI] [PubMed] [Google Scholar]
- 30.Chen, K. F., Lai, Y. Y., Sun, H. S., and Tsai, S. J. (2005) Nucleic Acids Res. 33 5190–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, M. H., Chen, K. F., Lin, S. C., Lgu, C. W., and Tsai, S. J. (2007) Am. J. Pathol. 170 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridge, A. J., Pebernard, S., Ducraux, A., Nicoulaz, A. L., and Iggo, R. (2003) Nature Genet. 34 263–264 [DOI] [PubMed] [Google Scholar]
- 33.Fantin, V. R., St-Pierre, J., and Leder, P. (2006) Cancer Cell 9 425–434 [DOI] [PubMed] [Google Scholar]
- 34.Bonnet, S., Archer, S. L., Allalunis-Turner, J., Haromy, A., Beaulieu, C., Thompson, R., Lee, C. T., Lopaschuk, G. D., Puttagunta, L., Bonnet, S., Harry, G., Hashimoto, K., Porter, C. J., Andrade, M. A., Thebaud, B., and Michelakis, E. D. (2007) Cancer Cell 11 37–51 [DOI] [PubMed] [Google Scholar]
- 35.Zhong, H., De Marzo, A. M., Laughner, E., Lim, M., Hilton, D. A., Zagzag, D., Buechler, P., Isaacs, W. B., Semenza, G. L., and Simons, J. W. (1999) Cancer Res. 59 5830–5835 [PubMed] [Google Scholar]
- 36.Pelicano, H., Xu, R. H., Du, M., Feng, L., Sasaki, R., Carew, J. S., Hu, Y., Ramdas, L., Hu, L., Keating, M. J., Zhang, W., Plunkett, W., and Huang, P. (2006) J. Cell Biol. 175 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]