Genome-Wide Scan for Linkage to Type 1 Diabetes in 2,496 Multiplex Families From the Type 1 Diabetes Genetics Consortium (original) (raw)
Abstract
OBJECTIVE
Type 1 diabetes arises from the actions of multiple genetic and environmental risk factors. Considerable success at identifying common genetic variants that contribute to type 1 diabetes risk has come from genetic association (primarily case-control) studies. However, such studies have limited power to detect genes containing multiple rare variants that contribute significantly to disease risk.
RESEARCH DESIGN AND METHODS
The Type 1 Diabetes Genetics Consortium (T1DGC) has assembled a collection of 2,496 multiplex type 1 diabetic families from nine geographical regions containing 2,658 affected sib-pairs (ASPs). We describe the results of a genome-wide scan for linkage to type 1 diabetes in the T1DGC family collection.
RESULTS
Significant evidence of linkage to type 1 diabetes was confirmed at the HLA region on chromosome 6p21.3 (logarithm of odds [LOD] = 213.2). There was further evidence of linkage to type 1 diabetes on 6q that could not be accounted for by the major linkage signal at the HLA class II loci on chromosome 6p21. Suggestive evidence of linkage (LOD ≥2.2) was observed near CTLA4 on chromosome 2q32.3 (LOD = 3.28) and near INS (LOD = 3.16) on chromosome 11p15.5. Some evidence for linkage was also detected at two regions on chromosome 19 (LOD = 2.84 and 2.54).
CONCLUSIONS
Five non–HLA chromosome regions showed some evidence of linkage to type 1 diabetes. A number of previously proposed type 1 diabetes susceptibility loci, based on smaller ASP numbers, showed limited or no evidence of linkage to disease. Low-frequency susceptibility variants or clusters of loci with common alleles could contribute to the linkage signals observed.
Type 1 diabetes develops when the insulin-secreting β-cells in the pancreas are depleted by an autoimmune process of unknown origin. Diagnosis occurs when sufficient β-cell mass is lost that blood glucose levels cannot be maintained in the physiologic range. Although insulin treatment for type 1 diabetes is life saving, development of effective preventive therapies could be enhanced by improved prediction and a better understanding of the underlying disease mechanism, particularly events occurring during the extended preclinical period. Such considerations have motivated studies directed toward identifying the genetic risk factors that contribute to the disorder that might provide novel insights into disease etiology and targets for preventive therapy.
A number of genome-wide scans for linkage to type 1 diabetes have been reported (1–7). All such studies consistently find evidence of linkage of type 1 diabetes to the HLA region on chromosome 6p21. An additional 15–20 regions with evidence of linkage to type 1 diabetes have been reported, but many of these regions have not been reproduced in independent studies. Several attempts have been made to address this problem through merging of datasets from different smaller studies (5,7), but differences in marker sets and allele coding have limited the power of these efforts.
To address issues of sample size and consistency and to clarify the evidence for linkage to type 1 diabetes, the Type 1 Diabetes Genetics Consortium (T1DGC) has conducted the largest affected sib-pair (ASP) linkage study in type 1 diabetes to date. This study includes many (n = 1,189) of the type 1 diabetes families used in previous smaller linkage studies, but the majority of families (n = 1,307) are newly recruited. More importantly, all samples, both retrospective and newly recruited, were independently genotyped expressly for this study at a single laboratory using a single platform and a common marker set. Analyses of these data provided evidence for linkage to type 1 diabetes in six regions on four different chromosomes.
RESEARCH DESIGN AND METHODS
The families included in the analyses were ascertained from nine different sources: the four T1DGC regional networks of Asia-Pacific (n = 228), Europe (n = 585), North America (n = 370), and the U.K. (n = 124); the Diabetes UK Warren 1 Repository (n = 429); the Human Biological Data Interchange repository (n = 424); the Joslin Diabetes Center (n = 112); the Steno Diabetes Center (n = 146); and Sardinia (n = 78). To be included in the study, a family had to have at least one ASP available for sampling; availability of one or both biological parents was preferred but not required. An individual was designated as affected if he or she had documented type 1 diabetes with onset at <35 years of age, had used insulin within 6 months of diagnosis, and had no concomitant disease or disorder associated with diabetes. A total of 2,496 families were genotyped. Most families, 2,287 (91.6%), had exactly two affected full siblings, 101 (4.0%) had three, and 7 (0.3%) had four or more. Among 2,657 ASPs in total, both parents were available for 69.6% of the ASPs, 18.9% had only a single parent available, and 11.6% had neither parent available. A few extended pedigrees also provided 13 affected half-sib pairs and 22 affected-affected avuncular pairs.
Genotyping.
Genotyping was carried out at the Center for Inherited Disease Research (CIDR) using the Illumina Human Linkage-12 Genotyping Beadchip, which contains 6,090 single nucleotide polymorphisms with an average genome-wide spacing of 0.58 cM.
Statistical analysis.
Before statistical genetic analyses, genotype data were evaluated for Mendelian errors using PedCheck (8). PREST (9) was used to estimate the likelihood of each specified relationship in pedigrees given the genotyping information. Based on these analyses, 43 families were removed from further analysis due to the presence of a duplicate family, a duplicate sample within the family, nonresolvable family errors, or missing genotype information. Merlin (10) was used to identify and resolve inconsistencies within families. Finally, 199 affected individuals who were not genotyped were removed from the analysis. The Kong and Cox linear nonparametric linkage (NPL) model (11) implemented in Merlin was used for the NPL linkage analysis. Marker-marker linkage disequilibrium was modeled in the linkage analysis using the maximum-likelihood clustered marker approach implemented in Merlin with a cutoff of _r_2 > 0.5.
RESULTS
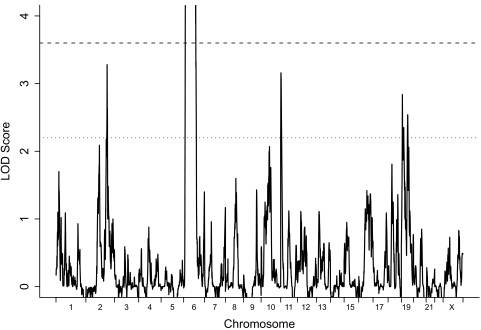
Figure 1 shows the results of the genome-wide analysis for linkage to type 1 diabetes in the entire T1DGC family collection. The average information content across all chromosomes was 0.924 and ranged from 0.904 on chromosome 20 to 0.939 on chromosomes six and X. Table 1 lists the regions where evidence for linkage exceeded the threshold for suggestive linkage (logarithm of odds [LOD] ≥2.2) in an ASP linkage study. The HLA region on chromosome 6p21.3 was the only region to display significant evidence of linkage to type 1 diabetes (LOD = 213.2). Four additional regions provided suggestive evidence of linkage. These included a region on chromosome 2q (LOD = 3.28), where there have been multiple past reports of linkage to type 1 diabetes, and the region surrounding the insulin gene at 11p15.5 (LOD = 3.16) that has been previously implicated in type 1 diabetes susceptibility by both linkage and association studies. The remaining two regions were on chromosome 19 (LOD = 2.84 and 2.54) and are separated from each other by 48.5 cM; evidence of linkage to chromosome 19 has been reported before but not to any level of confidence that justified specific attention. Other previously reported type 1 diabetes loci, which quite often were given type 1 diabetes designations, were not confirmed in the current study (Table 2).
FIG. 1.
Genome-wide evidence for linkage to type 1 diabetes. Evidence for linkage was evaluated using Merlin, and LOD scores were calculated using the Kong and Cox linear NPL model (10,11). LOD scores are plotted on the vertical axis versus chromosomal position on the horizontal axis. LOD scores at region 15.5–110.5 cM on chromosome 6 are larger than four and not shown. The dotted horizontal line indicates a LOD of 2.2. The dashed line indicates a LOD of 3.6.
TABLE 1.
Regions with suggestive or significant evidence of linkage to type 1 diabetes
| Chromosome | Position (cM) | LOD | P | LOD-1 support interval | Flanking markers* |
|---|---|---|---|---|---|
| 2 | 194.25 | 3.28 | 5.0 × 10−5 | 191.3–197.8 | rs1882395/rs1369842 |
| 6 | 52.0 | 213.2 | 8.0 × 10−216 | 51.0–52.5 | rs11908/rs412735 |
| 11 | 2.5 | 3.16 | 7.0 × 10−5 | 0–8.5 | rs741737/rs1609812 |
| 19 | 9.5 | 2.84 | 0.00015 | 7.5–26 | rs887270/rs1044250 |
| 19 | 58.0 | 2.54 | 0.0003 | 52–63 | rs1019937/rs1878926 |
TABLE 2.
Evidence for linkage in regions previously reported to harbor IDDM loci
| Locus | Chromosome | Defined by marker(s) | Marker tested | LOD | P |
|---|---|---|---|---|---|
| IDDM1 | 6p21.3 | HLA-DQ_β_1 | rs1019937 | 213.2 | 8.0 × 10−216 |
| IDDM2 | 11p15.5 | INS-VNTR | rs1004446 | 3.09 | 0.00008 |
| IDDM3 | 15q26 | D15S107 | rs906310 | 0.00 | 0.6 |
| IDDM4 | 11q13 | D11S1917 | rs923346 | 0.87 | 0.02 |
| IDDM5 | 6q25 | ESR | rs11902 | 0.00 | 0.5 |
| IDDM6 | 18q | D18S64 | rs1943418 | 0.68 | 0.04 |
| IDDM7 | 2q31 | D2S152 | rs696092 | 1.91 | 0.002 |
| IDDM8 | 6q27 | D6S264 | rs6910121 | 0.45 | 0.07 |
| IDDM9 | 3q21-25 | D3S1303 | rs1983380 | 0.40 | 0.09 |
| IDDM10 | 10cen | D10S193 | rs959629 | 1.07 | 0.013 |
| IDDM11 | 14q24.3 | D14S67 | rs1012921 | 0.13 | 0.2 |
| IDDM12 | 2q33 | CTLA4 | rs1947983 | 1.05 | 0.014 |
| IDDM13 | 2q34 | IGFBP2 | rs1851328 | 0.63 | 0.04 |
| IDDM15 | 6q | D6S283 | rs2207958 | 5.99* | 7.5 × 10−8 |
| IDDM16 | 14q32.3 | D14S293 | rs872945 | 0.05 | 0.3 |
| IDDM17 | 10q25 | D10S592 | rs151603 | 0.16 | 0.2 |
| IDDM18 | 5q33-34 | IL12B | rs270664 | 0.01 | 0.4 |
| 1q42 | D1S1617 | rs342817 | 0.00 | 0.5 | |
| 16q22-24 | D16S3098 | rs1037973 | 1.24 | 0.008 |
When considering evidence for linkage to type 1 diabetes on chromosome 6 that might possibly be due to loci other than the classic HLA genes, it was necessary to take into account the significant allele sharing at HLA, where LOD scores >3.6 were detected over an interval of ∼100 cM surrounding the locus. We searched for evidence of additional type 1 diabetes risk loci by calculating expected LOD (ELOD) scores based upon allele sharing at HLA along the length of chromosome 6 (supplementary Fig. 1, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1551/DC1). The observed LOD score exceeded the ELOD by 3.6 in three regions. One of these regions, from 81.5 to 101.5 cM (maximum LOD − ELOD = 10.8), overlapped with the location of the previously reported IDDM15 locus (6,12).
DISCUSSION
The T1DGC family collection includes the largest number of ASPs to be tested for linkage to type 1 diabetes in a genome-wide scan yet provides suggestive or significant evidence for only five loci linked to type 1 diabetes. In light of the numerous previous reports of loci linked to type 1 diabetes, we considered the relationship between the findings here and prior studies (Table 2).
The majority of studies, as in our current report, have observed significant evidence of linkage to type 1 diabetes at HLA, and there is well-confirmed evidence of disease association in the same region. Several additional loci on chromosome 6 (IDDM5, IDDM8, and IDDM15) have been proposed based on previous linkage studies (1,13,14). The current study provides little support for either IDDM5 or IDDM8 (Table 2). However, we did obtain a regional maximum LOD of 5.43, at 190.0 cM on chromosome 6q27, where IDDM8 was previously localized (13) when linkage disequilibrium was not modeled in the analysis. This region is characterized by a relatively strong intermarker linkage disequilibrium, which might explain this and previous reports of linkage in this region.
An additional region on chromosome 6q provided evidence for linkage to type 1 diabetes after adjusting for the effect of allele sharing at HLA. Previous studies have tested for linkage at IDDM15, which is located in this region, by calculating the expected LOD score at IDDM15 based upon the observed identical-by-descent (IBD) sharing at HLA and the recombination fraction between HLA and D6S283 (7,12). The excess of the observed IBD sharing compared with that which was expected was then used as a measure of the independent contribution of IDDM15. Applying this approach to the current dataset identified a broad (20 cM) region where the adjusted LOD exceeds 3.6. Uncertainties remain as to the precise location of IDDM15 because male and female recombination maps differ substantially in this region. Nevertheless, this region of increased LODs, after adjusting for the contribution of allele sharing at HLA, provides support for the existence of an additional linkage to type 1 diabetes in this region.
Chromosome 2q contains two confirmed type 1 diabetes risk loci near the IFIH1 and CTLA4 genes identified by association and three proposed loci (IDDM7, IDDM12, and IDDM13) for which suggestive evidence of linkage has been observed in some, but not all, studies (15–17). In the current report, there was no support for multiple distinct loci; suggestive evidence of linkage was observed in the 2q32.3 region, approximately midway between the previously proposed locations of IDDM7 and IDDM12. The INS locus at 11p15.5 is also well established as a risk locus for type 1 diabetes (18). Previous studies have provided significant evidence for both association and linkage to this site. The current study also finds suggestive evidence of linkage in this region.
Two regions on chromosome 19 displayed suggestive evidence of linkage to type 1 diabetes, which, given our sample sizes and marker map, is the most interesting data yet for this chromosome. So far, no associations with genome-wide significance have been reported in these regions.
Recently, genome-wide association (GWA) studies have identified at least 18 genomic regions with significant evidence of association to type 1 diabetes, but the majority of these loci, although common in the population, have relatively modest individual effects on risk (e.g., odds ratios [ORs] of 1.1–1.3) (19–22). Linkage studies are not optimal for detecting such loci where the risk alleles are common and have small individual effect sizes. Thus, it is not surprising that none of these newly identified loci from GWA studies fall within regions of even suggestive linkage in the current study. Several of our linkage peaks do include loci previously detected by association, but these tend to have large effects on risk such as HLA (OR 11.4) (23) or INS (2.38) (18).
The large sample size studied here, 2,658 ASPs from 2,496 families, provided us with unprecedented power to detect linkage to type 1 diabetes across the genome. While we detected one locus with significant evidence of linkage, and five more regions with suggestive evidence, our study provides little support for the majority of previously proposed type 1 diabetes loci. Our power to detect evidence of linkage to type 1 diabetes comes at a cost; the low frequency of families with multiple affected siblings required a world-wide recruitment effort. Therefore, we cannot exclude the possibility that loci with strong effects in particular populations might be obscured in the current analysis. For example, LOD = 3.21 was observed at 66.1 cM on chromosome 17 in the 370 T1DGC families recruited by the North American network, whereas no linkage was detected in the other eight populations (LOD scores ranging from −0.62 to 0.30).
In summary, the evidence for independent major gene effects in type 1 diabetes detectable by linkage (e.g., rare variants), which are shared across populations, appears to be limited to a few genes in the HLA complex: INS, CTLA4, IDDM15, and possibly two regions on chromosomes 19. The combination of these putative rare variant effects with common variants of smaller effect size, more readily detected by association-based approaches, accounts for the majority of risk for type 1 diabetes of which HLA class II alleles are by far the major contributor. Identification of candidate genes and variants in these and other regions will help to clarify the biochemical pathways and mechanisms that contribute to risk for this common autoimmune disease. The substantial collection of biospecimens and data assembled by the T1DGC, and available to diabetes researchers (www.t1dgc.org and www.t1dbase.org), should be an important resource for further characterizing those genomic regions identified by either linkage or association approaches as contributing to risk for type 1 diabetes.
Supplementary Material
Online-Only Appendix
Acknowledgments
This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. Further support was provided by a grant from the NIDDK (DK46635) to P.C. and a joint JDRF and Wellcome Trust grant to the Diabetes and Inflammation Laboratory at Cambridge, which also received support from the National Institute for Health Research Cambridge Biomedical Research Centre. Genotyping was performed by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University, contract no. N01-HG-65403.
No potential conflicts of interest relevant to this article were reported.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SC, Jenkins SC, Palmer SM, Balfour KM, Rowq BR, Farrall M, Barnett AH, Bain SC, Todd JA: A genome-wide search for human type 1 diabetes susceptibility genes.Nature 371: 130– 136, 1994 [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto L, Habita C, Beressi JP, Delepine M, Besse C, Cambon-Thomsen A, Deschamps I, Rotter JI, Djoulah S, James MR, Froguel P, Weissenbach J, Lathrop GM, Julier C: Genetic mapping of a susceptibility locus for insulin-dependent diabetes mellitus on chromosome 11q.Nature 371: 161– 164, 1994 [DOI] [PubMed] [Google Scholar]
- 3.Mein CA, Esposito L, Dunn MG, Johnson GC, Timms AE, Goy JV, Smith AN, Sebag-Montefiore L, Merriman ME, Wilson AJ, Pritchard LE, Cucca F, Barnett AH, Bain SC, Todd JA: A search for type 1 diabetes susceptibility genes in families from the United Kingdom.Nat Genet 19: 297– 300, 1998 [DOI] [PubMed] [Google Scholar]
- 4.Concannon P, Gogolin-Ewens KJ, Hinds D, Wapelhorst B, Morrison VA, Stirling B, Mitra M, Farmer J, Williams SR, Cox NJ, Bell GI, Risch N, Spielman RS: A second-generation screen of the human genome for susceptibility to insulin-dependent diabetes mellitus (IDDM).Nat Genet 19: 292– 296, 1998 [DOI] [PubMed] [Google Scholar]
- 5.Cox NJ, Wapelhorst B, Morrison VA, Johnson L, Pinchuk L, Spielman RS, Todd JA, Concannon P: Seven regions of the genome show evidence of linkage to type 1 diabetes in a consensus analysis of 767 multiplex families.Am J Hum Genet 69: 820– 830, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nerup J, Pociot F: A genomewide scan for type 1-diabetes susceptibility in Scandinavian families: identification of new loci with evidence of interactions.Am J Hum Genet 69: 1301– 1313, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Concannon P, Erlich HA, Julier C, Morahan G, Nerup J, Pociot F, Todd JA, Rich SS: Type 1 diabetes: evidence for susceptibility loci from four genome-wide linkage scans in 1,435 multiplex families.Diabetes 54: 2995– 3001, 2005 [DOI] [PubMed] [Google Scholar]
- 8.O'Connell JR, Weeks DE: PedCheck: a program for identification of genotype incompatibilities in linkage analysis.Am J Hum Genet 63: 259– 266, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Wilder K, McPeek MS: Enhanced pedigree error detection.Hum Hered 54: 99– 110, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Abecasis GR, Cherny SS, Cookson WO, Cardon LR: Merlin-rapid analysis of dense genetic maps using sparse gene flow trees.Nat Genet 30: 97– 101, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Kong A, Cox NJ: Allele-sharing models: LOD scores and accurate linkage tests.Am J Hum Genet 61: 1179– 1188, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delepine M, Pociot F, Habita C, Hashimoto L, Froguel P, Rotter J, Cambon-Thomsen A, Deschamps I, Djoulah S, Weissenbach J, Nerup J, Lathrop M, Julier C: Evidence of a non-MHC susceptibility locus in type I diabetes linked to HLA on chromosome 6.Am J Hum Genet 60: 174– 187, 1997 [PMC free article] [PubMed] [Google Scholar]
- 13.Luo DF, Bui MM, Muir A, Maclaren NK, Thomson G, She JX: Affected-sib-pair mapping of a novel susceptibility gene to insulin-dependent diabetes mellitus (IDDM8) on chromosome 6q25–q27.Am J Hum Genet 57: 911– 919, 1995 [PMC free article] [PubMed] [Google Scholar]
- 14.Luo DF, Buzzetti R, Rotter JI, Maclaren NK, Raffel LJ, Nistico L, Giovannini C, Pozzilli P, Thomson G, She JX: Confirmation of three susceptibility genes to insulin-dependent diabetes mellitus: IDDM4, IDDM5 and IDDM8.Hum Mol Genet 5: 693– 698, 1996 [DOI] [PubMed] [Google Scholar]
- 15.Copeman JB, Cucca F, Hearne CM, Cornall RJ, Reed PW, Ronningen KS, Undlien DE, Nistico L, Buzzetti R, Tosi R, Pociot F, Nerup J, Cornelis F, Barnett AH, Bain SC, Todd JA: Linkage disequilibrium mapping of a type 1 diabetes susceptibility gene (IDDM7) to chromosome 2q31–q33.Nat Genet 9: 80– 85, 1995 [DOI] [PubMed] [Google Scholar]
- 16.Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, Larrad MT, Rios MS, Chow CC, Cockram CS, Jacobs K, Mijovic C, Bain SC, Barnett AH, Vandewalle CL, Schuit F, Gorus FK, Belgian Diabetes Registry. Tosi R, Pozzilli P, Todd JA: The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes.Hum Mol Genet 5: 1075– 1080, 1996 [DOI] [PubMed] [Google Scholar]
- 17.Morahan G, Huang D, Tait BD, Colman PG, Harrison LC: Markers on distal chromosome 2q linked to insulin-dependent diabetes mellitus.Science 272: 1811– 1813, 1996 [DOI] [PubMed] [Google Scholar]
- 18.Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavesen MG, McCormack R, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Gillespie KM, Tuomilehto-Wolf E, Tuomilehto J, Bennett ST, Clayton DG, Cordell HJ, Todd JA: Remapping the insulin gene/IDDM2 locus in type 1 diabetes.Diabetes 53: 1884– 1889, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls.Nature 447: 661– 678, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C: A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene.Nature 448: 591– 594, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG: Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes.Nat Genet 39: 857– 864, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA: Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci.Nat Genet 40: 1399– 1401, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, Lavant E, Louey A, Moonsamy P: HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families.Diabetes 57: 1084– 1092, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online-Only Appendix