Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts (original) (raw)
Abstract
Metazoan genes are encrypted with at least two superimposed codes: the genetic code to specify the primary structure of proteins and the splicing code to expand their proteomic output via alternative splicing. Here, we define the specificity of a central regulator of pre-mRNA splicing, the conserved, essential splicing factor SFRS1. Cross-linking immunoprecipitation and high-throughput sequencing (CLIP-seq) identified 23,632 binding sites for SFRS1 in the transcriptome of cultured human embryonic kidney cells. SFRS1 was found to engage many different classes of functionally distinct transcripts including mRNA, miRNA, snoRNAs, ncRNAs, and conserved intergenic transcripts of unknown function. The majority of these diverse transcripts share a purine-rich consensus motif corresponding to the canonical SFRS1 binding site. The consensus site was not only enriched in exons cross-linked to SFRS1 in vivo, but was also enriched in close proximity to splice sites. mRNAs encoding RNA processing factors were significantly overrepresented, suggesting that SFRS1 may broadly influence the post-transcriptional control of gene expression in vivo. Finally, a search for the SFRS1 consensus motif within the Human Gene Mutation Database identified 181 mutations in 82 different genes that disrupt predicted SFRS1 binding sites. This comprehensive analysis substantially expands the known roles of human SR proteins in the regulation of a diverse array of RNA transcripts.
Metazoan genomes are encoded with multiple overlapping layers of information required for the precise control of gene expression. The splicing code has co-evolved with the genetic code and regulates the post-transcriptional expression of protein-coding genes (for review, see Wang and Cooper 2007). In the nucleus, splicing is required to remove intervening sequences (introns) from precursor messenger RNAs (pre-mRNAs) and to correctly join protein-encoding regions (exons) together. Inclusion of an exon into the mature mRNA is regulated by _cis_-acting RNA elements known as exonic or intronic splicing enhancers and silencers (ESE, ISE, and ESS, ISS, respectively) that function to recruit _trans_-acting RNA-binding proteins. In the cytoplasm, these same RNA elements are decoded by tRNAs and the ribosome in order to template protein synthesis. Alternative splicing allows a single gene to express many different protein isoforms by including all, some, or none of a specific exon sequence in the mRNA (for review, see Maniatis and Tasic 2002). Current estimates suggest that at least 70% of protein-coding genes undergo alternative splicing (Wang and Cooper 2007). However, understanding how these events are regulated and coordinated represents a major challenge.
Classification of functional _cis_-acting RNA elements on a global scale is required to begin the arduous task of defining the specific outputs from every human gene (for review, see Wang and Burge 2008). Combinations of elegant computational studies and biochemical assays are now beginning to address this important problem in gene regulation. A variety of bioinformatics strategies have identified hundreds of putative _cis_-acting sequences that are enriched with regulated exons or introns. These studies have included sequences that function as independent, nonredundant regulatory units and those that work together and have therefore co-evolved (Fairbrother et al. 2002; Zhang and Chasin 2004; Zhang et al. 2005; Wang et al. 2006; Xiao et al. 2007; Friedman et al. 2008). One caveat is that the cognate RNA-binding proteins often escape identification. In contrast, biochemical studies such as the selected evolution of ligands by exponential enrichment (SELEX) can reveal the nucleotide sequences recognized by specific RNA-binding proteins (Tuerk and Gold 1990; Tacke and Manley 1995; Liu et al. 1998; Cavaloc et al. 1999; Smith et al. 2006). However, these sequences are often degenerate and lack sufficient specificity to reveal the global organization of protein–RNA interactions. One of the most powerful methodologies used to examine interactions of RNA-binding proteins with their cognate targets is the RNA immunoprecipitation-microarray experiment (RIP-chip). RIP-chip is similar to chromatin IP-microarray analysis (ChIP-chip) with the exception that it is RNA–protein rather than DNA–protein interactions that are assayed (Tenenbaum et al. 2002; Sanchez-Diaz and Penalva 2006). RIP-chip is a robust method that has been applied to both yeast and metazoan systems and can reveal relationships between transcripts and regulatory RNA-binding proteins. Interpretation of RIP-chip experiments, however, requires caution, owing to several technical considerations. In the absence of cross-linking reagents, RNA-binding proteins are free to dissociate from their endogenous RNA targets and reassociate with higher affinity binding sites, thereby giving rise to a risk of false discovery (Mili and Steitz 2004). Formaldehyde cross-linking of intact cells can preserve in situ protein–RNA interactions but can also induce protein–protein cross-links, thereby increasing the likelihood that RNA targets associated with other RNA-binding proteins may be inadvertently copurified. Despite these issues, numerous studies using RIP-chip have suggested that RNA-binding proteins exhibit distinct binding specificities (Brown et al. 2001; Hieronymus and Silver 2003; Gerber et al. 2004; Gama-Carvalho et al. 2006; Olson et al. 2007). These observations have led to the formulation of the hypothesis that the coordinated post-transcriptional control of functionally related transcripts is organized by specific RNA-binding proteins (Keene 2007).
The RIP-related cross-linking immunoprecipitation (CLIP) method sidesteps several of the pitfalls described above (Ule et al. 2003, 2005a). The primary advantages of this method over the other standard RNA IP methods include the following: (1) photo cross-linking of intact cells preserves the in situ RNA-binding specificity, (2) partial RNase digestion liberates the protein-binding site from the full-length transcript, (3) stringent purification conditions decrease contamination, thereby enhancing the specificity of the assay, and (4) cloning and sequencing of purified RNA fragments directly identifies both the genomic locus from which the RNA transcript was derived and the region recognized by individual RNA-binding proteins. CLIP analysis has been successfully performed in a study of the murine RNA-binding protein NOVA, a neural gene-specific alternative splicing factor involved in the human neurological disorder, paraneoplastic opsoclonus myoclonus ataxia (POMA) (Ule and Darnell 2006). CLIP analysis of NOVA identified a network of pre-mRNAs encoding proteins involved in postsynaptic functions that are regulated by NOVA at the level of alternative splicing. These data allowed the creation of a “genomic map” capable of predicting alternative splicing events based upon the NOVA binding position (Ule et al. 2006).
Splicing factor arginine/serine-rich 1 (SFRS1, also known as SF2, ASF, ASF/SF2, and SF2/ASF) is a highly conserved, essential pre-mRNA splicing factor with dual functions in constitutive and alternative splicing (Ge et al. 1991; Krainer et al. 1991). SFRS1 is a member of a large protein family known as the serine and arginine-rich proteins (SR proteins). SR proteins have a modular domain structure comprising one or two amino-terminal RNA recognition motifs (RRMs) and a carboxyl-terminal domain composed almost exclusively of alternating serine and arginine repeats. SR proteins function at early stages of spliceosome assembly and play an important role in specifying splice site selection (for review, see Lin and Fu 2007). In cultured cells, SFRS1 shuttles between the nucleus and the cytoplasm and also participates in post-splicing RNA processing reactions, including mRNA export, stability, nonsense-mediated decay, and translation (Caceres et al. 1998; Huang and Steitz 2001; Lemaire et al. 2002; Sanford et al. 2004; Zhang and Krainer 2004). Although many of the biochemical roles of SR proteins in pre-mRNA splicing can be carried out by other family members, SFRS1 is absolutely required during both murine and nematode embryogenesis and for the maintenance of genome stability in mammalian cell culture models (Wang et al. 1996; Longman et al. 2000; Li and Manley 2005; Xu et al. 2005). SFRS1 is a proto-oncogene, located at 17q21.3-q22, which is amplified in many types of human tumor (Karni et al. 2007). The comprehensive identification of SFRS1 mRNA targets promises to improve our understanding of the diverse biological roles of SFRS1.
Despite numerous in vitro and in vivo studies, the RNA-binding specificity of SFRS1 is not fully understood. Here, we combine CLIP with high-throughput sequencing (CLIP-seq). CLIP-seq proffers a comprehensive and cost-effective system for the identification of biologically relevant _cis_-acting RNA elements recognized by specific RNA-binding proteins in the context of their cellular environment. Our data provide an unprecedented evaluation of the RNA-binding specificity of the essential splicing factor SFRS1. CLIP-seq identified a purine-rich consensus motif, which is present in a majority of exonic, intronic, ncRNA, and intergenic transcripts coprecipitated by SFRS1. Analysis of exonic RNA fragments bound by SFRS1 revealed an enrichment of binding sites between 21 and 40 nt from the 5′ or 3′ splice sites. This sequence and its positional specificity were then used to predict SFRS1 binding sites that have been disrupted by mutations causing human inherited disease. This analysis identified 181 mutations (in 82 different genes) associated with genetic disease that abolished putative binding sites for SFRS1. These results suggest that defective protein–RNA interactions may play a rather broader role in human inherited disease than has previously been anticipated.
Results
Cross-linking immunoprecipitation of SFRS1
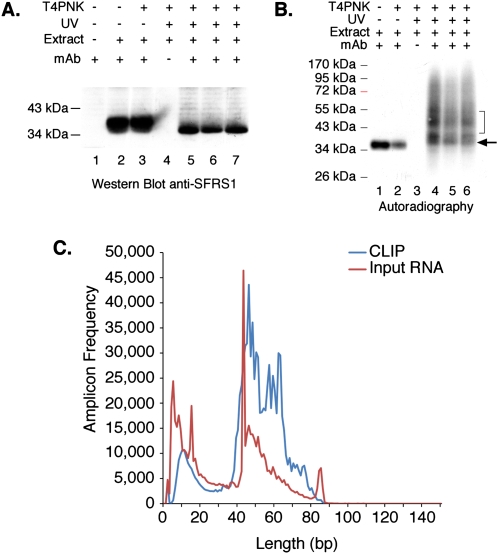
We used CLIP to identify _cis_-acting RNA elements recognized by splicing factor SFRS1. Three independent cultures of human embryonic kidney cells (HEK293T) exposed to UV and one control culture without UV irradiation were used to prepare whole-cell extracts as previously described (Sanford et al. 2005). Following DNase and RNase treatment of the extracts, SFRS1 was precipitated using mAb 96 (Hanamura et al. 1998). Immunoprecipitation of SFRS1 was confirmed by Western blot analysis. Precipitation of SFRS1 is dependent upon antibody and independent of UV irradiation (Fig. 1A; cf. lanes 2,3,5–7, and lane 4). Radiolabeled SFRS1–RNA complexes were visualized by autoradiography in parallel with the Western blot analysis. Precipitation of the 32P-labeled complexes by the SFRS1 antibody was dependent upon UV cross-linking (Fig. 1B, cf. lanes 1,2 and 4–6). Surprisingly, SFRS1 can be radiolabeled in the absence of UV irradiation. However, incorporation of 32P is not dependent upon T4PNK, indicating that this signal is unrelated to SFRS1–RNA complexes (Fig. 1B, lanes 1,2). The bracket in Figure 1B designates the region purified from nitrocellulose membranes and corresponding to a 10–15 kDa increase in the apparent molecular weight of free SFRS1 (Fig. 1B, compare bracket to arrowhead marking the expected migration of SFRS1). RNA extracted from the nitrocellulose membrane was then amplified by reverse transcription polymerase chain reaction (RT–PCR). For comparison, nonselected input RNA was purified and amplified from RNase-treated HEK293T whole-cell extracts. We directly sequenced CLIP-derived and input amplicons using the 454 Life Sciences (Roche) Genome Sequencer FLX system (Margulies et al. 2005). As expected from the mobility of the SFRS1–RNA complexes, the majority of purified RNA fragments were between 45 and 65 nt in length (Fig. 1C).
Figure 1.
CLIP of SFRS1 from HEK293T cells and amplicon sequencing. (A) Western blot analysis of SFRS1 immunopreciptation (IP) from control and UV-cross-linked cells. CLIP was performed in three independent cultures of HEK293T cells (lanes 5–7). SFRS1 was detected with the same antibody used for IP. The blot was visualized with chemiluminescence (Pierce Super Signal). This panel demonstrates the specificity of the IP; no signal was detected in control IP samples (lane 4), but SFRS1 was efficiently precipitated from both UV-irradiated and control cell extracts. (B) Autoradiograph of 32P-labeled SFRS1–RNA complexes immobilized on nitrocellulose membrane. Following extensive washes of the immunoprecipitated complex, the beads were incubated with T4PNK (NEB) and [γ32P]ATP in order to phosphorylate the 5′ end of RNA fragments. In the absence of UV irradiation, SFRS1 can become phosphorylated. However, this does not require T4PNK (cf. lanes 1 and 2). These data suggest that a subset of SFRS1 is copurified with an SR protein kinase even under these stringent conditions. Importantly, in the absence of UV irradiation, no slower migrating protein–RNA complexes are observed (cf. lanes 1,2 and 4–6). The arrow indicates the position of “free” SFRS1 and the bracket defines the region of nitrocellulose excised from the blot. (C) Histogram comparing the amplicon length distribution with numbers of reads. Amplicons prepared from both CLIP (blue line) and nonselected input RNA samples (red line) were sequenced using the 454 FLX platform and short read reagents. As expected from B, the majority of amplicons (after removal of both linker sequences) are between 40 and 60 bp in length.
High-throughput sequencing
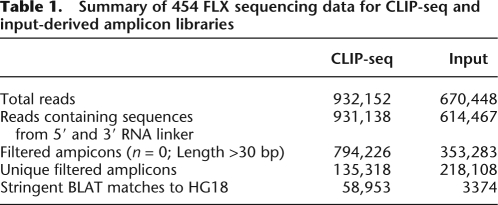
SFRS1-bound RNAs were analyzed in four independent CLIP-seq experiments. In total, 932,152 reads were obtained from SFRS1-bound RNA. In addition, 670,448 reads from nonselected input RNA were generated from three independent experiments for comparison (Table 1). A total of 99.8% and 91.6% of sequences derived from CLIP and input amplicon libraries contained information from both 5′ and 3′ RNA linkers, indicating that the vast majority of RNA fragments were sequenced completely. The sequences corresponding to RNA fragments were then filtered, based upon a minimum allowable size of 30 bp and no ambiguous base calls. After this filtering step, all redundant amplicon sequences were removed, leaving a pool of 135,318 and 218,108 CLIP- or input-derived RNA fragments, respectively. The unique RNA fragments were aligned to the human genome using the BLAST-like Alignment Tool (BLAT) (Kent 2002). RNA fragments derived from CLIP or input RNA samples mapped to 58,953 and 3374 loci, respectively, indicating that the CLIP sequences were far more diverse than those derived from the input samples. Overlapping clusters of unique amplicons were used to define the contiguous sequence blocks (henceforth referred to as “blocks”). Blocks derived from nonselected total RNA served as a reference sample. We assume that these unselected blocks correspond to the most abundant transcripts and may be a potential source of contamination in the CLIP-seq data set. Therefore, blocks common to both the input and CLIP-seq were excluded from further analysis, despite the possibility that some very abundant RNAs might actually bind to SFRS1. By applying this stringent filter, the abundant class may have been lost. However, this conservative approach ensured that only extensively enriched sequences would have been captured. Only 5% of the CLIP-derived blocks were present in the reference set.
Table 1.
Summary of 454 FLX sequencing data for CLIP-seq and input-derived amplicon libraries
SFRS1 consensus motif
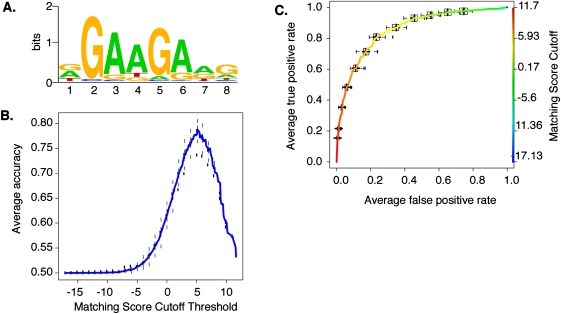
Identification of the consensus site for SFRS1 is important in order to understand mechanisms of splice-site selection. We used the motif-finding algorithm Multiple EM for Motif Elicitation (MEME) (Bailey and Elkan 1995) to identify RNA-binding sequences shared by sequenced fragments that cross-link to SFRS1 in vivo. There were 681 unique sequence blocks present in at least three of the four CLIP-seq experiments and absent from the input sequences. We randomly split this set in half, using 340 to train the MEME algorithm, and holding the remaining 341 blocks in reserve as part of a gold standard data set. After picking a single representative amplicon at random from each sequence block, MEME identified a purine-rich octamer containing a GAAGAA core (Fig. 2A). This motif is similar to several SFRS1 recognition sites previously identified by binding SELEX experiments and by mutational analysis of splicing enhancers in the fibronectin extra-domain A and cardiac troponin T alternative cassette exons (Caputi et al. 1994; Ramchatesingh et al. 1995; Tacke and Manley 1995). Computational methods searching for ESEs have also identified and validated the GAAGAA sequence as a functional splicing enhancer (Fairbrother et al. 2002). We then calculated a positional weight matrix (PWM) from the SFRS1 consensus motif. The predictive power of the PWM was evaluated using two different statistical plots: the accuracy curve, which calculates the accuracy of the binding-site prediction as a function of matching score cutoff thresholds, and the receiver operating characteristic curves (ROC), which evaluate sensitivity and specificity of the binding-site model (Fig. 2B,C). For each plot, we used the PWM to scan a gold standard data set, consisting of amplicons from the remaining 341 blocks as a positive component and an equal number of sequences picked at random from intergenic deserts as a negative component. These measurements established the maximum accuracy, sensitivity, and specificity of PWM as 78%, 81%, and 77%, respectively. We therefore consider that the consensus binding-site model presented in Figure 2 has a high probability of correctly identifying SFRS1 binding sites in silico.
Figure 2.
Modeling the in situ SFRS1 consensus-binding motif. (A) The MEME algorithm was used to identify a consensus motif from 300 amplicons selected at random from a total of 641 blocks common to three out of four CLIP-seq experiments. This calculation was repeated 20 times. The motif with the highest sensitivity and specificity (see C) is depicted here. The likelihood of finding this motif at random is <1 × 10−107. (B) The averaged Accuracy plot for each positional weight matrix calculated from the MEME results. The prediction accuracy of the SFRS1 site was plotted as a function of matching score cutoff threshold. Maximum accuracy (78%) was achieved at a cutoff score of 5.2. This calculation was repeated 40 times using the gold standard data set. Error bars correspond to the standard deviation from the mean accuracy. (C) The averaged receiver operator characteristic curve of the SFRS1 consensus site model. This plot evaluates the sensitivity (true positive rate of discovery) and specificity (false positive discovery rate) as functions of matching score cutoff threshold. The ROC evaluates the ability of the PWM to discriminate between positive (CLIP-seq derived) and negative components (55-bp fragments selected from intergenic deserts) of the gold standard data set. This calculation was repeated 40 times. Error bars correspond to standard deviation from the mean sensitivity and specificity.
Classification of SFRS1 binding sites
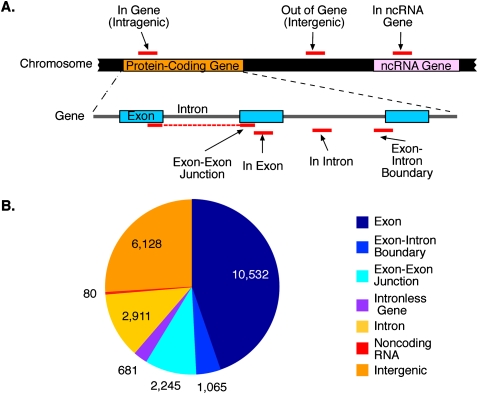
The UCSC Known Gene and Rfam databases were used to annotate each sequence block. Figure 3A depicts the strategy used for annotation. A total of 23,699 blocks were identified in the SFRS1 CLIP-seq experiment (Fig. 3B). The majority (73%) of these blocks mapped to loci annotated as protein-coding genes. Of the 17,365 blocks present within protein-coding genes, 83% were associated with exonic sequences. We subclassified these exon-associated blocks into those that were contained exclusively within a single exon (10,532 blocks; 60%), those that spanned an exon–exon junction (2245; 13%), those that spanned an exon–intron boundary (1065; 6%), and those contained within an intronless gene (681; 4%). The remaining 17% (2911) of blocks mapping to protein-coding genes were present within introns. Three files containing all of the genomic coordinates of blocks targeted by SFRS1, blocks present in the input sample, and the positions of SFRS1 consensus sites can be found in the Supplemental material.
Figure 3.
Classification of _cis_-acting RNA elements bound by SFRS1. (A) Annotation strategy for classifying sequence blocks identified by CLIP-seq. Following alignment of amplicon sequences to the human genome, blocks of overlapping sequences were defined and subsequently annotated using the UCSC Known Gene and Rfam databases. Blocks were classified at the gene level (“In Gene,” “Out of Gene,” and “In ncRNA”) and at the transcript level (“In Exon,” “In Intron,” “Exon–Intron Boundary,” and “Exon–Exon Junction”). None of the blocks presented here overlapped amplicon sequences obtained from input RNA samples. (B) Quantification of block annotations. The majority of blocks were classified as “In Gene” and were predominantly associated with exon sequences. Slightly more than 25% of blocks were defined as intergenic, but binding sites for SFRS1 within these transcripts were found to be highly conserved over evolutionary time (See Supplemental Fig. 2).
SFRS1 binding sites associated with alternative splicing
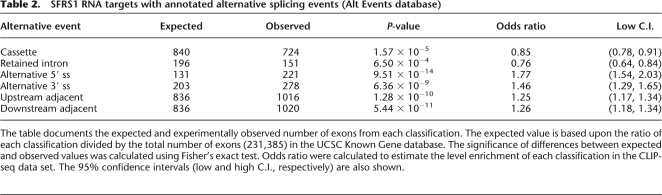
SFRS1 is a well-characterized splicing factor with roles in the regulation of alternative splicing (for review, see Lin and Fu 2007). We extracted a nonredundant version of the AltEvents database from the UCSC Genome Browser and used it to classify SFRS1 target exons based upon their relationship to constitutive or alternative splicing (Table 2). Some 88.8% (12,304) of the exonic binding sites of SFRS1 were localized to constitutive exons, whereas only 11.2% (1538) had evidence of alternative splicing in this database. Alternative cassette exons were the single-most abundant classification, followed by exons containing alternative 3′ or 5′ splice sites. Retained introns were the least common. The observed levels of these exons within the SFRS1 CLIP-seq data set differed significantly from the expected levels of each classification based upon their ratios to all exons annotated by the UCSC Known Gene database (Table 2). Both cassette exons and retained introns were underrepresented in the pool of SFRS1 targets (Fisher's exact test P < 1.6 × 10−5 and 6.5 × 10−4, respectively). In contrast, exons with alternative 5′ and 3′ splice sites were enriched in the CLIP-seq data relative to the genome (Fisher's exact test P < 9.5 × 10−14 and 6.4 × 10−9, respectively). Recent work from Biamonti and coworkers (Ghigna et al. 2005) suggests that binding sites for SFRS1 in constitutive exons may regulate the inclusion or exclusion of adjacent cassette exons. We found that SFRS1 bound in equal proportions to exons immediately upstream or downstream from cassette exons (5′ or 3′ adjacent exons, respectively). We observed a significant enrichment in the CLIP-seq data set relative to the human genome for binding sites located within 5′ and 3′ adjacent exons (Fisher's exact test P < 1.3 × 10−10 and 5.4 × 10−11, for upstream and downstream exons, respectively).
Table 2.
SFRS1 RNA targets with annotated alternative splicing events (Alt Events database)
Intronless genes
Not all intragenic blocks fell within intron-containing genes; 681 binding sites in 332 intronless mRNAs were identified in this experiment. A total of 100 blocks mapped to 30 different histone genes. Several other interesting intronless genes identified as SFRS1 targets include those encoding transcription factors, such as JUN, JUND, SOX4, SOX12, FOXC1, and those encoding post-translational regulators, such as UBC, SUMO1, and SUMO2. These findings are consistent with roles for shuttling SR proteins in the nuclear export of histone 2a mRNA (Huang and Steitz 2001). However, our experiment suggests that SFRS1 regulates the post-transcriptional expression of many functionally diverse intronless genes. One subtle difference between intronless and intron-containing targets is a modest enrichment of SFRS1 binding sites within the UTRs of intronless genes relative to spliced transcripts (Supplemental Fig. 1). Some 43% of blocks mapping to intronless genes were present in UTRs as compared with 25% of blocks within spliced transcripts, respectively (Fisher's exact test P < 0.004; Supplemental Fig. 1). However, the biological significance of these data is unclear.
Intergenic and noncoding RNA targets
A significant proportion (26%) of all blocks mapped to regions not annotated as protein-coding genes. According to the Rfam database (Griffiths-Jones et al. 2003), only 80 of the 6218 intergenic blocks have been annotated as noncoding RNA genes. Among the ncRNAs represented in the SFRS1 CLIP-seq data are 23 snoRNAs, three microRNA precursors, and the 5.8S rRNA, XIST and MALAT1 RNAs. It is possible that the remainder may correspond to as yet undiscovered ncRNAs. Phylogenetic conservation was used to evaluate the significance of SFRS1 binding sites in RNA transcribed from intergenic loci. The conservation scores, or phastCons scores, were downloaded from the UCSC Genome Browser and reflect the overall conservation among 17 vertebrate species (Felsenstein and Churchill 1996; Siepel et al. 2005). We determined the mean conservation score for each nucleotide within 1200 bp of the center of each intergenic SFRS1 binding site. These data were then compared with intergenic regions randomly selected from the human genome (Supplemental Fig. 2). The majority of SFRS1-bound RNA transcripts derived from intergenic regions were found to be highly conserved across multiple vertebrate lineages, suggesting a high degree of negative selective pressure at these sites.
SFRS1 binds to a subset of functionally related mRNAs
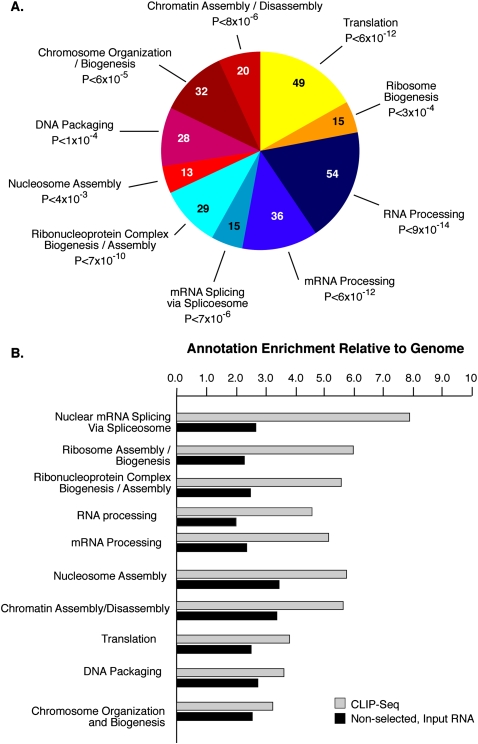
Keene (2007) has proposed that RNA-binding proteins may coordinately regulate the post-transcriptional expression of functionally related genes. To determine whether particular classes of genes may be influenced by SFRS1, Gene Ontology (GO) annotations were assigned to SFRS1 mRNA targets present in three out of four CLIP-seq experiments using EASE (Expression Analysis Systematic Explorer) (Hosack et al. 2003). Annotation enrichment was ascertained by computing the EASE score for each GO classification. The top 10 (based on Holm-corrected EASE scores) (Hosack et al. 2003) most enriched classes of mRNA bound by SFRS1 are shown in Figure 4A. Based on biological process annotations, involving several redundant layers, it is clear that the most enriched SFRS1 RNA targets encode proteins involved in gene expression in general and RNA processing specifically. In order to avoid potential bias due to highly abundant transcripts, we also defined the GO of the nonselected, RNase-digested input RNA. The enrichment of each annotation term in both the CLIP-seq and nonselected input RNA relative to the human genome was calculated and ranked using EASE Scores. Relative to both the genome and nonselected input RNA, SFRS1 target mRNAs were found to be highly enriched for genes involved in biological processes related to pre-mRNA splicing, RNA processing, and ribosome biogenesis (Fig. 4B). In contrast, mRNAs encoding proteins functioning in processes such as chromatin and nucleosome assembly did not differ significantly between SFRS1 RNA targets and nonselected RNase-treated input RNA samples, despite being enriched relative to the genome as a whole. As expected, the nonselected RNase-treated input RNA samples contained a much wider array of GO-terms than the CLIP-seq targets (data not shown).
Figure 4.
SFRS1 targets are enriched in mRNAs encoding RNA-binding proteins. (A) The top 10 classes of Gene Ontology terms enriched in the CLIP data set relative to the expected ratios in the DAVID database (Hosack et al. 2003). The top 10 classes of targets are all related to gene expression. The numbers of genes observed in each category are indicated in the pie chart. Holm-corrected EASE scores are given for each category (Hosack et al. 2003). (B) Comparison of the top 10 CLIP-enriched Gene Ontology terms with nonselected input mRNA samples. The annotation enrichment relative to the genome is plotted for both CLIP (gray bars) and input (black bars) derived mRNAs. mRNAs encoding splicing factors are most highly enriched in CLIP.
Validation of SFRS1–RNA interactions
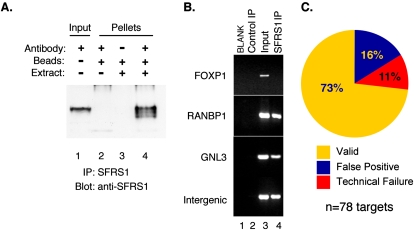
CLIP-seq analysis of SFRS1 identified thousands of potential RNA targets. As with any high-throughput method, it is necessary to gauge the accuracy of the CLIP-seq targets using a secondary assay. In order to validate the SFRS1–RNA interactions, we used the RNA–immunoprecipitation (RIP) assay to determine whether 78 RNA transcripts selected at random from the CLIP-seq data set interact with SFRS1 under native conditions. SFRS1 was found to be efficiently and specifically immunoprecipitated from whole-cell extracts prepared from HEK293T cells (Fig. 5A, cf. lanes 3 and 4). The increased mobility of SFRS1 observed in lane 4 is due to partial dephosphorylation of SFRS1 and can be blocked by inclusion of phosphatase inhibitors during the IP incubation (data not shown). Coprecipitated RNA was then isolated from input, control IP, and anti-SFRS1 IP samples and analyzed by RT–PCR (Fig. 5B; Supplemental Fig. 3). Of the 78 randomly selected target transcripts, 58 were detectable in both the input and anti-SFRS1 IP samples but not in RNA isolated from the control IP. These validated targets include many intergenic transcripts, demonstrating that these unannotated RNA transcripts are associated with SFRS1 in vivo. Nine interactions were classified as false positive on the basis that the transcript was either detected in both the control and SFRS1 IP (see Supplemental Fig. 3, ZNF66) or present only in the input sample, but absent from the IP. It is possible that some nonvalidated interactions between SFRS1 and target RNAs were undetectable under non-cross-linking conditions. Eleven RNA targets were classified as technical failures, since no detectable signal was observed in any RNA sample. In total, ∼73% of randomly selected SFRS1 target transcripts could be validated by the RIP assays (Fig. 5C). These data were in good agreement with the statistical evaluation of the SFRS1 consensus motif that correctly recognized ∼78% of target RNA fragments.
Figure 5.
Validation of SFRS1–RNA interactions by RNA–IP RT–PCR. (A) Western blot analysis of proteins precipitated by the anti-SFRS1 monoclonal antibody. SFRS1 was detected in both the input extract (lane 1) and the material immunoprecipitated with anti-SFRS1 (lane 4) but not the control beads (lanes 2,3). The blot was visualized as described in Figure 1. (B) Examples of RT–PCR analysis of endogenous SFRS1–mRNA complexes. RNA extracted from the control IP, input extract, and SFRS1 IP, was reverse transcribed using oligo dT and Superscript III (Invitrogen). A total of 78 different primer sets were used to amplify specific transcripts from cDNA. (C) Summary of RT–PCR validation of 78 randomly selected sequence blocks identified by CLIP-seq. Validated interactions correspond to detectable PCR product in both the input and SFRS1 IP samples. False positive transcripts correspond to PCR products present in the input and the control IP. Technical failures yielded no PCR products from any cDNA sample, indicating that the transcript could not be directly validated under these conditions.
The SFRS1 consensus motif is enriched near the boundaries of exons identified by CLIP-seq
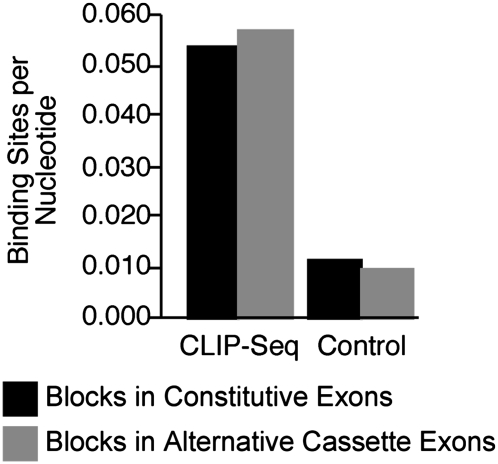
Our data suggest that the robust consensus motif presented in Figure 2 represents the canonical binding site for SFRS1. However, only a small proportion of blocks were used to generate the model. We next asked whether the consensus motif was enriched in blocks derived from exon sequences bound by SFRS1 relative to randomly selected exonic sequences. The binding-site density (number of binding sites exceeding the matching score cutoff threshold per nucleotide) for 8693 blocks present in constitutive exons and 426 blocks within alternative cassette exons was calculated. The average binding-site density was compared with an equal number of 55-nt sequence blocks selected at random from nontargeted exons. Figure 6 indicates that the consensus site is highly enriched within exonic sequence blocks captured by CLIP-seq as compared with 55-nt blocks selected at random from exons across the genome (P < 2.2 × 10−22, Wilcoxon test). Additionally, these calculations indicate that each sequence block contains, on average, ∼1.7 consensus binding sites. The number of binding sites for SFRS1 within exonic sequence blocks ranges from as few as 0 matches to the consensus motif to as many as 16 sites. We also observed a modest enrichment in consensus-site density in alternative cassette exons versus constitutive exons cross-linked to SFRS1, suggesting that, on average, there are slightly more binding sites for SFRS1 in alternative exons (P < 0.005, Wilcoxon test).
Figure 6.
The SFRS1 consensus motif was enriched in blocks identified by CLIP relative to randomly selected blocks from exon sequences. The average number of SFRS1 consensus sites per nucleotide was determined and plotted for sequence blocks in 8693 constitutive (black bars) and 426 alternative cassette exons (gray bars) identified by the CLIP-seq experiment. For the control group, 55 bp regions were selected at random from equal numbers of constitutive or alternative cassette exons not contained in the pool of SFRS1 CLIP-seq data. The Wilcoxon test confirmed the mean binding sites per nucleotide were significantly different for CLIP-seq and control exons (P < 10−22). Binding sites for SFRS1 in alternative cassette exons were found to be modestly enriched relative to constitutive exons (P < 0.005).
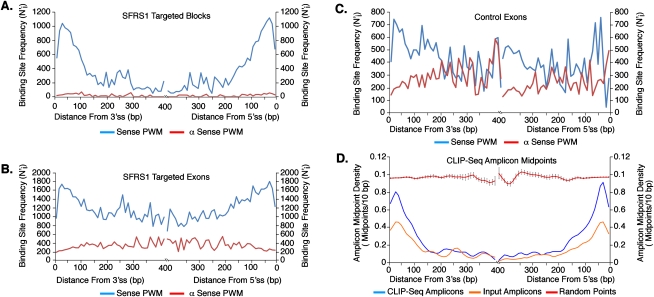
The spatial relationship between binding-site positions and splice sites can provide important mechanistic insights into molecular functions of RNA-binding proteins (Ule et al. 2006). Biochemical studies suggest that SR proteins function at early stages of spliceosome assembly and promote recognition of splice sites (Fu 1993; Kohtz et al. 1994; Graveley et al. 2001; Shen and Green 2004). Indeed, the proximity of SR protein-binding sites to splice sites is positively correlated with the in vitro splicing efficiency of reporter pre-mRNAs (Graveley and Maniatis 1998). To determine whether the distribution of SFRS1 binding sites is restricted to specific positions within exons, we scanned experimentally observed blocks, full-length exons targeted by CLIP-seq, and randomly selected exons from the genome with PWMs corresponding to the SFRS1 consensus site and the reverse complement of the motif. The distance (in base pairs, bp) from each consensus site was measured to the nearest 5′ or 3′ splice site. The frequency of binding sites at specific nucleotide positions was determined and adjusted for the uneven length distribution of human exons (Majewski and Ott 2002). The highest frequency of SFRS1 consensus sites were located in blocks near exon–intron boundaries, specifically between 20 and 41 bp from 5′ and 3′ splice sites (Fig. 7A, blue lines). In contrast, the antisense PWM failed to detect a significant number of matching sites within the CLIP-seq blocks (Fig. 7A, red lines). We performed two additional calculations to ensure that the observed positions of SFRS1 consensus sites were not biased by our analytical methods. First, we compared the distribution of sites identified by the sense and antisense PWMs in full-length exons identified by CLIP-seq and randomly selected exons from the human genome (Fig. 7B, blue and red lines, respectively). Again, SFRS1 consensus sites identified by the sense PWM exhibited a clear bias toward the exon boundaries. In contrast, sites identified by the antisense PWM are evenly distributed across CLIP-seq exons. Both PWMs identified matching sequences in randomly selected exons. However, only a weak bias toward exon boundaries was observed in the case of the sense PWM, whereas the antisense motif showed no apparent bias (Fig. 7C, blue and red lines, respectively). These data demonstrate that although consensus SFRS1 binding sites are present throughout exon sequences, experimentally observed blocks, presumably containing engaged binding sites for SFRS1, are enriched at fixed positions relative to splice sites. The distribution of SFRS1 consensus sites in alternative cassette exons was found to be virtually identical to those in constitutive exons (data not shown). Finally, we directly examined the positional distribution of the amplicons themselves, relative to splice sites. For each amplicon sequence that was derived from an exon or exon–intron boundary, we measured the distance in base pairs (bp) from the midpoint of the amplicon sequence to the nearest 5′ or 3′ splice site. As with previous calculations, we determined the frequency of amplicon midpoints in 10-bp bins extending away from the splice sites. Each amplicon midpoint was counted only once with respect to either a 5′ or 3′ splice site, thereby ensuring that the genomic coordinates of each midpoint measurement contributed only to the nearest splice site. The amplicon midpoint frequencies were directly compared with an equal number of randomly selected points from exon sequences extracted from the human genome and from amplicon sequences derived from input RNA samples. The average frequencies for each bin were calculated over 40 replicate samplings. Figure 7D demonstrates that the midpoints of CLIP-seq amplicons were restricted to specific positions relative to 5′ and 3′ splice sites (Fig. 7D, blue lines). Amplicons picked from the input samples also showed a slight positional bias relative to 5′ and 3′ splice sites (Fig. 7D, orange lines). However, CLIP-seq amplicon midpoints were clearly enriched relative to the input samples. In contrast, randomly selected control “midpoints” displayed no positional bias (Fig. 7D, red lines). This analysis is independent of the positional weight matrix and therefore directly confirms that SFRS1 binding sites are enriched at specific distances (∼20–41 nt) relative to splice sites. Given that the median lengths of internal constitutive and cassette exons identified by CLIP-seq were found to be 142 and 158 nt, respectively, which are somewhat longer than their counterparts in the UCSC Known Gene database (125 and 108 nt, respectively), the data presented above reflect a clear positional bias of SFRS1 binding sites.
Figure 7.
SFRS1 binding sites are enriched at fixed positions relative to splice sites. The adjusted frequency of SFRS1 consensus sites within 10-bp bins (N′) at a specific position relative to splice sites (i) was calculated by multiplying the number of consensus sites observed by the total number of exons divided by exon ≥2i in length. In A–C, the blue and red lines represent the sense or antisense PWMs, respectively. (A) Positions of SFRS1 binding sites within sequence blocks identified by CLIP-seq. (B) Positions of SFRS1 binding sites across full-length exons targeted by SFRS1. (C) Positions of SFRS1 binding sites across randomly selected exons from the human genome. (D) The distance from splice sites to the midpoints of CLIP-seq amplicons was calculated as described above. CLIP-seq and Input Amplicon midpoints (blue and orange lines, respectively) were compared with randomly selected “points” picked from exons selected at random from the genome (red lines). This comparison demonstrated that the amplicons identified by CLIP were enriched relative to the input samples at the boundaries of exons. Likewise, randomly selected “points” differ dramatically with respect to the experimentally observed amplicon midpoints.
Many human disease mutations disrupt SFRS1 consensus sites
Single-nucleotide substitutions or point mutations often alter the genetic code by producing aberrant protein products. However, although nonsense mutations introduce premature termination codons into the open reading frames of disease genes, it is often much more difficult to rationalize the pathogenic basis of missense and synonymous mutations. One reason is that point mutations can manifest their detrimental effects through RNA processing. It is now well established that defects in pre-mRNA splicing and the regulation of alternative splicing can induce heritable disease in humans (for review, see Wang and Cooper 2007). Studies of the BRCA1, SMN, CFTR, GH1, and ATM genes (among others) have demonstrated that all classes of point mutations, including nonsense mutations, can disrupt exonic splicing regulatory elements and induce aberrant alternative splicing (Teraoka et al. 1999; Liu et al. 2001; Cartegni and Krainer 2002; Moseley et al. 2002; Kashima and Manley 2003; Pagani et al. 2003). Based on these results and others, a considerable effort to identify splicing-relevant mutations using PWM generated by both binding and functional SELEX is now underway (Smith et al. 2006). However, as stated above, different approaches for identifying the binding specificity of SFRS1 yield results that do not always concur. These differences serve to confound our understanding of the pathology of human inherited disease.
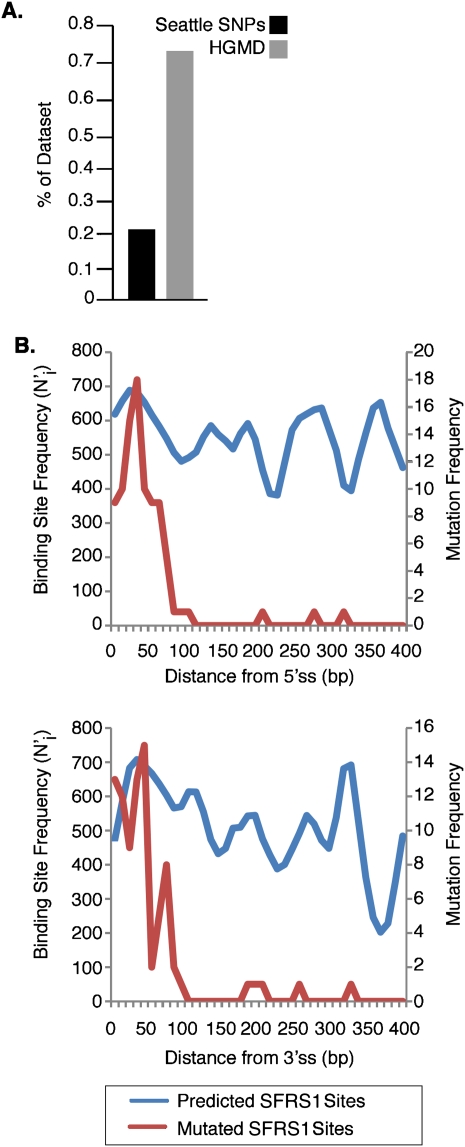
To investigate the potential impact of human disease-causing mutations on RNA processing involving SFRS1, exons from the Human Gene Mutation Database (HGMD; http://www.hgmd.org) were scanned with the PWM generated by CLIP-seq. This data set comprised 21,700 single-nucleotide substitutions giving rise to either missense, synonymous, or nonsense mutations either causing or associated with human inherited disease (Stenson et al. 2003). We scored mutations that abolished predicted SFRS1 binding sites relative to the wild-type allele, based on the thresholds established in Figure 2. As a control, exons from the SeattleSNPs database (http://pga.gs.washington.edu), containing 1436 validated human polymorphisms, were also scanned with the PWM. The high allele frequencies of these polymorphisms are broadly indicative of their functional neutrality. In total, we identified 181 disease-causing single-nucleotide substitutions (0.83%) in 82 different genes that ablate potential binding sites for SFRS1. Missense mutations accounted for the largest percentage (57%) of lost SFRS1 binding sites, whereas nonsense mutations made up the remaining 43% of mutations. In contrast, the SeattleSNPs database contained only three different polymorphic sites (0.21%) that were predicted to give rise to the loss of an SFRS1 binding site. We therefore found that substitutions resulting in the loss of SFRS1 binding sites were enriched approximately fourfold in the HGMD mutation data set relative to the control data set (Fig. 8A) (Fisher's exact test, P < 10−5). These data are consistent with previous studies showing that purifying selection reduces single-nucleotide substitutions in exonic positions harboring splicing regulatory sequences (Majewski and Ott 2002; Fairbrother et al. 2004; Parmley et al. 2006).
Figure 8.
Disruption of SFRS1 binding sites can cause human inherited disease. (A) Single-nucleotide substitutions causing loss of predicted SFRS1 binding sites in the Human Gene Mutation Database (http://www.hgmd.org) and the SeattleSNPs database (http://pga.gs.washington.edu) were identified by scanning reference and mutated exon sequences with the SFRS1 PWM. The proportion of entries in each database giving rise to a loss of SFRS1 sites was then plotted, and a statistically significant difference between the HGMD and SeattleSNPs data sets observed (P < 10−5; Fisher's exact test). (B) Disease mutations resulting in the loss of SFRS1 binding sites were found to be largely confined to exon boundaries. The top and bottom panels plot mutated sites relative to the 5′ and 3′ splice sites, respectively. The blue line in each plot represents the distribution of SFRS1 binding sites throughout the HGMD exons. The red lines correspond to the distribution of sites ablated by disease-causing mutations. These data demonstrate that although binding sites for SFRS1 can be predicted across HGMD exons, disease mutations tend only to disrupt those SFRS1 binding sites that are in close proximity to splice sites.
We next posed the question of where the mutations causing the loss of SFRS1 binding sites were located relative to splice sites (Fig. 8B). First, we determined the positions of all potential SFRS1 binding sites within exons represented in the HGMD. Potential SFRS1 binding sites were present throughout the disease gene exons and, as expected, these predicted binding sites showed very little positional bias (Fig. 8B, blue lines). In contrast, the majority of mutated SFRS1 binding sites were enriched in positions within 50 bp of the nearest 5′ or 3′ splice site (Fig. 8B, red lines). These data suggest that human disease mutations that disrupt potential SFRS1 binding sites are located in positions that are wholly compatible with their being physiological binding sites for SFRS1. In support of this conclusion, several of the mutations identified in this computational screen are already known to induce aberrant alternative splicing of the endogenous pre-mRNA in patients. These include three nonsense mutations in MLH1 (K461X) (Liu et al. 2001), ATM (E1978X) (Teraoka et al. 1999), and GH1 (E84X) (Moseley et al. 2002), as well as two missense mutations in GH1 (E85G) (Moseley et al. 2002) and NPHP1 (G342R) (Betz et al. 2000).
Discussion
High-throughput DNA sequencing is rapidly changing the landscape of genomic research (Wold and Myers 2008). Our study is perhaps the first to utilize high-throughput sequencing to analyze protein–RNA interactions. We used the 454 FLX system (Roche) for amplicon sequencing based upon several considerations including read length and the well-validated platform. The read length of the 454 FLX system, using a short read kit, allows for 120–150 bp reads and is ideal for completely sequencing the RNA fragment and linkers produced by CLIP, ensuring that all sequence information is preserved. The longer read length provided by the 454 platform facilitated mapping of sequences to the genome and allowed for detection of many exon–intron and exon–exon junctions in the amplicon library. The primary consideration for future CLIP-seq experiments is clearly an increased sequencing throughput. Issues arising from the natural abundance of different RNA transcripts and the preferential PCR amplification during library preparation have the potential to introduce many redundant reads. The work presented here cannot be viewed as comprehensive, because significant new sequences were discovered in each of the four CLIP-seq experiments analyzed. However, the majority of the novel sequences share at least one statistically significant match to the consensus SFRS1 binding site. Therefore, for the specific application of CLIP-seq, data generated by the 454 platform are akin to a large-scale sampling or snapshot. Systems such as the SOLiD platform from ABI, which promise significantly increased throughput, have the potential to deliver truly comprehensive CLIP-seq analyses.
The work presented above represents a significant step toward elucidating the roles of SFRS1 in post-transcriptional gene expression. Perhaps more importantly, these experiments demonstrate the potential of CLIP-seq to illuminate the recognition code of RNA-binding proteins and their in situ binding sites. A comprehensive evaluation of protein–RNA interactions is critical for understanding how RNA-binding proteins positively or negatively regulate post-transcriptional processes such as alternative splicing. As future CLIP-seq experiments increase the catalog of known protein–RNA interactions, efforts to integrate binding-site data with functional genomic approaches have the potential to reveal the global organization of post-transcriptional regulatory networks in mammalian cells (Ule et al. 2006; Wang and Burge 2008).
A significant advantage of CLIP-seq is the large amount of raw data generated by the high-throughput sequencing of amplicons. These data facilitate the elucidation of consensus sites using motif-finding algorithms such as MEME. The motif presented in Figure 2 was the only statistically significant sequence identified by MEME. The robust nature of the binding-site model allowed for high-resolution mapping of SFRS1 binding sites within the amplicon data. This is important because future in silico analyses should focus on these positionally restricted windows for identification of SFRS1-regulated exons. Such an approach is exemplified by the search for SFRS1 binding sites abolished by inherited mutations causing human genetic disease. We identified 181 exonic mutations in 82 different disease genes that abolish putative SFRS1 binding sites. Nearly 87% of these mutations were located within 50 bp of the nearest splice site, a region already demonstrated by CLIP-seq to be enriched in SFRS1 binding sites. It is quite possible that mutations falling outside of the preferred zone of SFRS1 binding will have little impact on RNA processing. However, at least five of the mutations identified here have already been correlated with changes in alternative splicing. Given that none of the mutations we identified were apparent in previously published reports identifying large numbers of splicing-relevant disease mutations, the pathological impact of exonic mutations upon splicing could turn out to be quite significant. Our findings argue that defective RNA processing, typically considered unusual in cases of nonsynonymous disease mutations, could actually be the rule rather than the exception.
The CLIP method, developed in the Darnell laboratory at Rockefeller University, was first used to identify RNA targets of the splicing regulator NOVA. NOVA and SFRS1 are very different types of splicing factors and these differences are clearly reflected in their in situ RNA-binding specificities elucidated by CLIP. NOVA and SFRS1 engage RNA through structurally distinct RNA-binding domains. The K-homology RNA-binding domain of NOVA recognizes a pyrimidine-rich YCAY motif that is nearly threefold more abundant in RNA fragments bound by NOVA relative to nontargeted sequences. In contrast, our study shows that SFRS1 binds a purine-rich octamer with a GAAGAA core. This motif is highly enriched in exons bound by SFRS1 relative to randomly selected exon sequences (Fig. 6). The binding sites for both proteins within pre-mRNA are restricted to specific positions. Intronic and 3′ UTR binding sites are most prevalent in the NOVA targets, whereas internal exonic binding sites are strongly preferred by SFRS1 (Fig. 7). The positional binding specificities of both proteins can provide insight into their mechanisms of action. By comparing alternative splicing patterns of target transcripts in NOVA knockout and wild-type mice, Ule and coworkers were able to deduce how different positions of NOVA binding sites influenced splice site selection (Ule et al. 2006). Although we have not yet established the functionality of each binding site for SFRS1 identified by CLIP-seq, based upon the well established roles of SFRS1 in pre-mRNA splicing, we speculate that exonic binding sites are likely to function as splicing enhancers. However, because the majority of blocks identified by CLIP-seq are classified as exonic, it is not possible to determine whether these binding events occur during spliceosome assembly or instead at a later stage of mRNA processing. In contrast, blocks spanning exon–intron boundaries clearly represent interactions with pre-mRNA, whereas those spanning exon junctions are derived from spliced mRNA. Fewer than 5% of all blocks (1065) mapped to exon–intron boundaries and only 2245 mapped to exon junctions. Given the proximity of these blocks to splice sites, it is possible that the same RNA elements recognized by SFRS1 influence pre-mRNA splicing and subsequent cytoplasmic steps of post-transcriptional gene expression. Recent work from our laboratory identified binding sites in several mRNAs that are engaged by SFRS1 in both nuclear and cytoplasmic/polysomal mRNA fractions of the cell (Sanford et al. 2008). Clearly, further functional studies are required to elucidate the functions of SFRS1 binding sites identified by CLIP-seq.
Another interesting finding from our study is that binding sites for SFRS1 are enriched in exons that are adjacent to alternative cassette exons (Table 2). A previous study demonstrated that SFRS1 binding sites in a constitutive exon regulated the skipping of an upstream alternative cassette exon in the receptor tyrosine kinase RON gene (Ghigna et al. 2005). We propose that SFRS1 may play a prominent role in regulating this mode of competitive exon skipping by activating downstream splice sites. Finally, there are also significant differences between the functions of proteins encoded by mRNAs targeted by NOVA and SFRS1. NOVA mRNA targets tend to encode proteins involved in pre- and postsynaptic function as well as neuronal inhibition (Ule et al. 2003, 2005b). In contrast, mRNAs encoding other RNA-binding proteins are overrepresented in the collection of SFRS1 targets. These include a statistically significant enrichment of other splicing factors (Fig. 4). We are confident that the enrichment of RNA-binding protein messages is biologically significant for several reasons. First, comparisons of SFRS1 targets with mRNAs present in the nonselected input RNA samples demonstrate that transcript abundance alone does not account for the enrichment of RBP mRNAs in the CLIP-seq data set. Secondly, recent experiments describe auto- and _trans_-regulatory post-transcriptional networks involved in homeostatic control of RNA-binding protein expression (Lareau et al. 2007; Ni et al. 2007; Barberan-Soler and Zahler 2008; Saltzman et al. 2008). Two hallmarks of this mechanism include alternative splicing-coupled nonsense-mediated decay (AS-NMD) and ultraconserved _cis_-acting regulatory elements within coding exons of RBP mRNAs (Bejerano et al. 2004). In many cases, the ultraconserved regions of RBP genes overlap alternative exons with the potential to induce NMD (Bejerano et al. 2004; Lareau et al. 2007; Ni et al. 2007). In total, we identified eight out of 111 known ultraconserved regions within exonic sequences. Included in these ultraconserved binding sites are several other genes encoding RNA-binding proteins such as SFRS1 itself, SFRS6, HNRNPM, PCBP2, and CLK4 encoding SR protein kinase (Supplemental Fig. 4). We suggest that SFRS1 may be involved in controlling RBP homeostasis.
Methods
Cell culture
Human embryonic kidney (HEK293T) cells were cultured in DMEM (Sigma), supplemented with 10% fetal calf serum, and incubated at 37°C in the presence of 5% CO2. For each CLIP experiment, cells were grown to 75% confluence in 15-cm plates.
Cross-linking immunoprecipitation (CLIP)
CLIP analysis of SFRS1 was performed as described (Ule et al. 2003) with the following modifications relating to extract preparation and RNase treatment. Whole-cell lysates were prepared from UV-treated or control cells as previously described (Sanford et al. 2005). The soluble extract was treated with 30 U of RQ DNase 1 for 20 min at 37°C. The reactions were terminated by the addition of 20 mM EDTA. Subsequently, ribosomal subunits were cleared by centrifugation of the extract at 100,000_g_ using an Optima Max ultracentrifuge (Beckman Coulter) in a TLA120.2 rotor for 20 min. Cleared extracts were then treated with a dilute cocktail of RNase A/T1 (Ambion) at a final dilution of 1:10,000 for 20 min at 37°C. A total of 200 U of RNaseOut (Invitrogen) was then added to the extract. Proteins were then partially denatured by addition of an equal volume of buffer A (2× PBS, 0.2% SDS, 1% NP-40). An aliquot of each UV-treated extract was used to prepare input RNA fragments. The remainder of the extract was used for immunoprecipitation with anti-SFRS1 monoclonal antibody. Extracts were treated with proteinase K (Ambion) at a final concentration of 2 mg/mL, phenol extracted twice, and ethanol precipitated. The trimmed input RNA was then ligated to the 3′ RNA linker, followed by the 5′ RNA linker, and used as templates for RT–PCR as previously described (Ule et al. 2005a). Gel-purified amplicons from the primary RT–PCR were reamplified for 15 cycles using HPLC-purified primers that were complementary to the RNA linkers, but also contained the 454 capture sequences (Margulies et al. 2005) as described in the original CLIP protocol (Ule et al. 2005a). Amplicons were gel purified from 2% NuSieve Agarose gels using the QIAX II Gel Extraction kit (QIAGEN).
High-throughput sequencing of amplicons
Prior to sequencing, the quality and quantity of gel-purified amplicons was assessed using a DNA LabChip1000 on an Agilent 2100 BioAnalyzer. High-throughput sequencing was performed using the Genome Sequencer FLX system (Roche Diagnostics) following standard protocols (Margulies et al. 2005). Titration runs were performed for all samples.
Primers
All DNA oligonucleotides (sequences available on request) were synthesized by IDT, Inc.
454 capture primers:
- P5454A (HPLC-purified): 5′-GCCTCCCTCGCGCCATCAGAGGGAGGACGATGCGG-3′
- P3454B (HPLC-purified): 5′-GCCTTGCCAGCCCGCTCAGCCGCTGGAAGTGACTGACAC-3′
Mapping of high-throughput sequencing data to the human genome
Several QC steps were implemented prior to mapping amplicon sequences to the human genome. We removed any amplicon sequences that did not include a recognizable match to 5′ and 3′ RNA linkers used for amplifying the RNA library. Once amplicons with both linker sequences had been identified, sequences <30 bp were removed, as were sequences containing ambiguous base calls. Finally, to avoid complications from preferential amplification during PCR, redundant identical amplicon sequences were filtered out from each experiment and only representative amplicons were retained. In order to study the binding of SFRS1 on a genome-wide scale, the filtered amplicons were aligned using BLAT (Kent 2002) against human genome assembly hg18, March 2006 (NCBI build 36, accessed Oct. 18, 2007) (Karolchik et al. 2008). BLAT sequences containing more than 80% repetitive sequence were removed, and only one mismatch or one gap was allowed so that single nucleotide polymorphisms (SNPs) and splicing junctions could be accommodated. The annotation strategy focused upon loci containing overlapping unique amplicons. We refer to these regions as sequence blocks (blocks). Blocks from each CLIP-seq experiment were annotated using the UCSC Known Gene database (http://genome.ucsc.edu/; accessed on Oct. 18, 2007) (Karolchik et al. 2008) and the Rfam database. The annotation data was then subclassified in order to determine the number of blocks targeting specific genomic structures (exon, intron, exon–intron boundary, intergenic, etc.) and to determine the number of unique genomic structures identified in each experiment. To identify alternatively spliced exons bound by SFRS1, all binding sites were mapped against the Alternative Event Database (derived from AltEvent track in the UCSC Genome Browser [Karolchik et al. 2008]). The binding sites that were not located in alternatively spliced exons were by default designated as constitutive exons (the set of unique exons in the UCSC Known Gene database excluding alternative spliced exons related to AltEvent track).
Modeling and statistical evaluation of the SFRS1 consensus-binding motif
In order to establish precisely where SFRS1 binds, the Multiple Em for Motif Elicitation (MEME) algorithm (version 3.5.7; http://meme.sdsc.edu/meme/intro.html) (Bailey et al. 2006) was used to determine the consensus motif of amplicons in CLIP-hit blocks that did not overlap input blocks. We focused on the 681 blocks detected by three out of four CLIP samples. A single amplicon sequence was randomly selected from each block. A total of 300 of the randomly selected sequences were used to perform MEME analysis and 300 sequences were used as the positive component of “gold standard” sequences to evaluate the predictive power of the derived consensus motif. This procedure was repeated 20 times. The ROC curve was selected that had the maximum area under the curve (AUC), and its corresponding PWM (positional weight matrix) was taken as the final prediction of the SFRS1 consensus motif (Fig. 2). The PWM can be found in Supplemental Table 1. During each ROC analysis, 40 groups of background sequences were selected to compare with the gold standard sequences. The background sequences were identical in length to the gold standard data set but were selected at random from intergenic desert regions (defined for practical purposes as having no genes within 100,000 bp upstream or downstream) from the chromosomes contributing to each gold standard sequence; any blocks from the CLIP experiments were deleted. After scanning each gold standard (true positive) and background sequence (false positive) using PWM derived from MEME, we computed the binding scores for each octamer, based upon which TP (true positive), TN (true negative), FP (false positive), and FN (false negative) rates were calculated at different score cutoff thresholds. Averaging 40 groups of these data, we plotted a final averaged ROC curve, precision-recall curve and accuracy curve using the ROCR package (Sing et al. 2005). As mentioned above, we selected the ROC curve that gave the maximum AUC as our final result; its predictive power is illustrated in Figure 2B. We adopted the cutoff value 5.2, corresponding to the maximum accuracy of prediction, as the threshold to ascertain whether or not a given octamer was likely to be a bona fide binding site. To evaluate the relationship of the SFRS1 PWM to the majority of CLIP-seq data, the average number of binding sites per nucleotide was calculated for blocks from alternative cassette or constitutive exons identified by CLIP-seq or an equal number of blocks selected at random from protein-coding genes. Wilcoxon's signed-rank test was used to evaluate the statistical significance of the data.
Gene Ontology analysis
We identified all genes targeted by three out of four CLIP samples, and then excluded those genes that were targeted by any INPUT sample. The gene list containing these genes was then input into EASE (Hosack et al. 2003) (Expression Analysis Systematic Explorer, version 2.0; http://david.abcc.ncifcrf.gov/ease) to compute the overrepresented functional categories in “Biological Process,” “Cellular Component,” and “Molecular Function” systems (Hosack et al. 2003). EASE scores (modified Fisher's exact test probabilities by penalizing the count of positive agreement by one) were computed for all categories in each system. Holm's correction method was applied to the data in order to identify the most significantly overrepresented gene categories, because the genes contained in each GO category were not mutually exclusive. We also performed a comparison between the ratios of genes hit by CLIP and the background gene lists (CLIP–hit ratio and expected ratio). These data provided evidence for annotation enrichment relative to the entire genome. Because transcript abundance may also influence the protein–RNA interactions identified by CLIP-seq, we also used analyzed mRNAs identified in the nonselected input RNA samples. Comparison of the annotation enrichment between the CLIP-seq and input RNA samples was important as a means to identify specific targets that could have originated from highly transcribed genes.
SFRS1 binding-site frequency relative to splice sites
Blocks identified by CLIP-seq or randomly selected exon sequences from the human genome were scanned using the SFRS1 consensus PWM to identify statistically significant binding sites (those with binding scores above the matching score threshold of 5.2). As an additional control, we generated a PWM corresponding to the reverse complementary sequence of the SFRS1 binding site (see Supplemental Table 1). The frequency of binding sites (N) at each nucleotide position (i) relative to a splice site was determined by recording the genomic coordinates for each exonic binding site at the fifth position of the consensus motif. Binding-site quantity was adjusted for the uneven distribution of exon lengths by dividing the binding-site frequency at a given position (Ni) by the number of exons of at least 2i in length (Majewski and Ott 2002). We favored this approach over normalization of exon length because we felt that the absolute position, rather than the proportional position, of each _cis_-acting element was more likely to be biologically meaningful. The adjusted frequency (Ni’) of binding sites was plotted in 10-nt bins relative to the 5′ or 3′ splice site. Amplicon midpoints were mapped by selecting sequences at random from blocks identified by CLIP-seq. Because SFRS1 CLIP-seq identified many more exons that the input amplicons, we compared the density of amplicon midpoints in each 10-bp bin. Midpoint density is calculated by dividing the number of midpoints at each position (i) by the number of exons at least 2i in length. The distribution of midpoints from targeted and input amplicons was compared with randomly selected positions within exon sequences. This negative control calculation was repeated 40 times.
Identification of SFRS1 binding sites predicted to be disrupted by human disease mutations
Two variation data sets were used to identify the possible role of SFRS1 binding-site mutation in the contexts of human inherited disease and interindividual variation: (A) germ-line pathogenic substitutions from the Human Gene Mutation Database; (B) single-nucleotide polymorphisms from the SeattleSNPs resequencing project. All nucleotide variants were mapped to the human genome using the BLAST-like alignment tool (BLAT) (Kent 2002). If genomic variants mapped to multiple exons (overlapping), then both exons were considered in the analysis. A total of 40,397 coding-region pathogenic nucleotide substitutions were retrieved from the Human Gene Mutation Database (HGMD; http://www.hgmd.org) (Stenson et al. 2003). However, we only examined those 21,700 single-nucleotide substitutions from 440 genes (53.7% of all substitutions) that were located within internal coding exons. The 1436 SNPs derived from the SeattleSNPs resequencing project (http://pga.mbt.washington.edu/), selected for their high allele frequency, were assumed to be functionally neutral.
Mapping mutations predicted to disrupt SFRS1 binding sites
For each nucleotide substitution, a data set comprising the wild-type and corresponding mutant exons was compiled. Using these data and a sliding window of 8 bp to evaluate the SFRS1 position weight matrix (PWM) using a threshold of 5.2, SFRS1 target sites were determined within the wild-type and mutant exons. Mutations reducing the matching score to levels below the score cut-off threshold of 5.2 were classified as loss of binding and vice versa for gain of binding. The net loss or gain for the set of 8-mers was then used to determine whether the result was an overall loss or gain of SFRS1 at any given position. The frequency and positional distribution of mutations predicted to give rise to a loss of SFRS1 binding sites was determined as described above.
Validation of SFRS1–RNA interactions
Anti-SFRS1 monoclonal antibody (60 uL cell culture supernatant/IP) was bound to 60 μL (packed bead volume) recombinant Protein A Sepharose CL-4B beads (Invitrogen) in 1.0 mL 0.1 M sodium phosphate buffer (pH 8.1). For negative control precipitations, 40 μL beads were equilibrated with 0.1 M sodium phosphate buffer (pH 8.1). Antibody-bound or control beads were then washed 3× in 1.0 mL lysis buffer (20 mM Tris·HCl at pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.5% NP40, 0.5% Triton X100, one mini complete protease inhibitor tablet [Roche Diagnostics]). A total of 20 μL antibody-bound beads were held in reserve for a Western-blotting negative control sample (beads, antibody, no extract). Whole cells were prepared by extracting HEK293T cells in lysis buffer as described above. Soluble extract (1/50th) was retained as an input reference sample for Western blotting and RT–PCR analysis. Equal amounts of soluble extract were incubated with 40 μL anti-SFRS1 beads or control beads at 4°C for 1 h on a rotating mixer. Beads were then washed 4× with lysis buffer. One-third of the beads were used for Western blot analysis of precipitated protein; the remaining beads were used for RNA extraction with Tri Reagent LS (Sigma) following the manufacturer's protocol. Equal amounts of RNA (∼500–600 ng) were treated with 1 U of RQ DNase (Promega Corp.) for 20 min at 37°C. RQ DNase was inactivated by the addition of EGTA and incubation at 65°C for 10 min. Equal amounts of input RNA, anti-SFRS1 IP, or control IP (typically 400 ng) were used for cDNA synthesis with oligo dT cellulose and SuperScript III reverse transcriptase (Invitrogen) following the manufacturer's specifications. A total of 25 ng of cDNA (or ddH2O for no template controls) was used as a template for all PCR assays. PCR reactions were performed in 96-well format, 25 μL/well (1× R-Taq mix [MidWest Scientific] and 200 nM primers) in an Eppendorf EP Mastercycler (Germany) using the following cycle conditions: 95°C 5 min; 35 cycles of: 95°C 30 sec, 59°C 30 sec, 72°C 60 sec; 72°C 5 min. PCR products were analyzed on 2% agarose gels and visualized by staining with ethidium bromide.
Acknowledgments
We thank A. Krainer for generously providing anti-SF2 (SFRS1) monoclonal antibody. We thank M. Ares and A. Zahler for comments on the manuscript and J. Bruzik, J. Caceres, and D. Unni for helpful discussions. Finally, we thank the reviewers for their efforts in providing a thorough critique of our manuscript. Amplicon sequencing was performed at the Indiana University Center for Genomics and Bioinformatics (Bloomington, Indiana). This work was supported by an American Heart Association Scientist Development Grant to J.R.S. (0830206N), grants from the U.S. National Institutes of Health to J.R.S (1R01GM085121), H.J.E, and S.D.M (K22LM009135 and R01LM009722), and financial support from BIOBASE GmbH to D.N.C. and M.M.
Footnotes
References
- Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S., Zahler A.M. Alternative splicing regulation during C. elegans development: Splicing factors as regulated targets. PLoS Genet. 2008;4:e1000001. doi: 10.1371/journal.pgen.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Betz R., Rensing C., Otto E., Mincheva A., Zehnder D., Lichter P., Hildebrandt F. Children with ocular motor apraxia type Cogan carry deletions in the gene (NPHP1) for juvenile nephronophthisis. J. Pediatr. 2000;136:828–831. [PubMed] [Google Scholar]
- Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Caceres J.F., Screaton G.R., Krainer A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Casari G., Guenzi S., Tagliabue R., Sidoli A., Melo C.A., Baralle F.E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R. Disruption of an SFRS1-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Cavaloc Y., Bourgeois C.F., Kister L., Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Fairbrother W.G., Holste D., Burge C.B., Sharp P.A. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:e268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., Churchill G.A. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- Friedman B.A., Stadler M.B., Shomron N., Ding Y., Burge C.B. Ab initio identification of functionally interacting pairs of cis-regulatory elements. Genome Res. 2008;18:1643–1651. doi: 10.1101/gr.080085.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M., Barbosa-Morais N.L., Brodsky A.S., Silver P.A., Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Zuo P., Manley J.L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P.M., Green M.R., Riva S., Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Graveley B.R., Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- Graveley B.R., Hertel K.J., Maniatis T. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA. 2001;7:806–818. doi: 10.1017/s1355838201010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A., Caceres J.F., Mayeda A., Franza B.R., Jr, Krainer A.R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H., Silver P.A. Genome-wide analysis of RNA–protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Jr, Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Steitz J.A. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- Keene J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J.D., Jamison S.F., Will C.L., Zuo P., Luhrmann R., Garcia-Blanco M.A., Manley J.L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer A.R., Mayeda A., Kozak D., Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lemaire R., Prasad J., Kashima T., Gustafson J., Manley J.L., Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes & Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lin S., Fu X.D. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Liu H.X., Zhang M., Krainer A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.X., Cartegni L., Zhang M.Q., Krainer A.R. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- Longman D., Johnstone I.L., Caceres J.F. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S., Steitz J.A. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley C.T., Mullis P.E., Prince M.A., Phillips J.A., III An exon splice enhancer mutation causes autosomal dominant GH deficiency. J. Clin. Endocrinol. Metab. 2002;87:847–852. doi: 10.1210/jcem.87.2.8236. [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S., Blanchette M., Park J., Savva Y., Yeo G.W., Yeakley J.M., Rio D.C., Graveley B.R. A regulator of Dscam mutually exclusive splicing fidelity. Nat. Struct. Mol. Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F., Buratti E., Stuani C., Baralle F.E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- Parmley J.L., Chamary J.V., Hurst L.D. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- Ramchatesingh J., Zahler A.M., Neugebauer K.M., Roth M.B., Cooper T.A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A.L., Kim Y.K., Pan Q., Fagnani M.M., Maquat L.E., Blencowe B.J. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz P., Penalva L.O. Post-transcription meets post-genomic: The saga of RNA binding proteins in a new era. RNA Biol. 2006;3:101–109. doi: 10.4161/rna.3.3.3373. [DOI] [PubMed] [Google Scholar]
- Sanford J.R., Gray N.K., Beckmann K., Caceres J.F. A novel role for shuttling SR proteins in mRNA translation. Genes & Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Ellis J.D., Cazalla D., Caceres J.F. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Coutinho P., Hackett J.A., Wang X., Ranahan W., Caceres J.F. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Green M.R. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SFRS1-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Tacke R., Manley J.L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum S.A., Lager P.J., Carson C.C., Keene J.D. Ribonomics: Identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: Underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ule J., Darnell R.B. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule J., Jensen K., Mele A., Darnell R.B. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005a;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005b;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Wang G.S., Cooper T.A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang Z., Burge C.B. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Takagaki Y., Manley J.L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Wang Z., Xiao X., Van Nostrand E., Burge C.B. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold B., Myers R.M. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- Xiao X., Wang Z., Jang M., Burge C.B. Coevolutionary networks of splicing cis-regulatory elements. Proc. Natl. Acad. Sci. 2007;104:18583–18588. doi: 10.1073/pnas.0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R., Jr, Ye Z., Liu F., et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Zhang X.H., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Krainer A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Zhang X.H., Leslie C.S., Chasin L.A. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]