A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate (original) (raw)
Abstract
The resolution of the chicken consensus linkage map has been dramatically improved in this study by genotyping 12,945 single nucleotide polymorphisms (SNPs) on three existing mapping populations in chicken: the Wageningen (WU), East Lansing (EL), and Uppsala (UPP) mapping populations. As many as 8599 SNPs could be included, bringing the total number of markers in the current consensus linkage map to 9268. The total length of the sex average map is 3228 cM, considerably smaller than previous estimates using the WU and EL populations, reflecting the higher quality of the new map. The current map consists of 34 linkage groups and covers at least 29 of the 38 autosomes. Sex-specific analysis and comparisons of the maps based on the three individual populations showed prominent heterogeneity in recombination rates between populations, but no significant heterogeneity between sexes. The recombination rates in the F1 Red Jungle fowl/White Leghorn males and females were significantly lower compared with those in the WU broiler population, consistent with a higher recombination rate in purebred domestic animals under strong artificial selection. The recombination rate varied considerably among chromosomes as well as along individual chromosomes. An analysis of the sequence composition at recombination hot and cold spots revealed a strong positive correlation between GC-rich sequences and high recombination rates. The GC-rich cohesin binding sites in particular stood out from other GC-rich sequences with a 3.4-fold higher density at recombination hot spots versus cold spots, suggesting a functional relationship between recombination frequency and cohesin binding.
Genome resources for chicken have accumulated over the past decade, culminating in the publication of a draft sequence of the chicken genome (Hillier et al. 2004). Despite the availability of a high-quality genome sequence and detailed physical maps (Wallis et al. 2004), high-resolution linkage maps continue to be critical for the identification of genomic regions affecting phenotypic traits. In chicken, the consensus linkage map published in 2000 (Groenen et al. 2000) not only has remained the reference map in genetic studies, but also served together with the physical BAC contig maps (Wallis et al. 2004) and additional linkage maps (Kerje et al. 2003; Jacobsson et al. 2004) as the major reference for anchoring whole-genome shotgun sequence contigs to specific chromosomes and chromosome positions.
Low pass sequencing of three domestic chickens (commercial broiler, experimental layer, and Chinese Silkie), achieving 0.25× coverage per individual in addition to the Red Jungle fowl female sequenced at 6.6× coverage, generated a resource of 2.8+ million single nucleotide polymorphisms (SNPs) (International Chicken Polymorphism Map Consortium 2004). Although these SNPs represent an enormous resource for genetic studies, variation in recombination frequencies in the genome makes it difficult to predict what the actual genetic distance is for closely spaced markers. A high-resolution linkage map will facilitate fine mapping of quantitative trait loci (QTLs) mapped at lower resolution in classical linkage analysis using microsatellite markers. Increasing the marker density of the linkage map further enables the analyses of genomic sequences associated with high recombination rates. Accurate high-density linkage maps have been shown to be critical for linkage studies (Daw et al. 2000; Fingerlin et al. 2006) and improved high-density linkage maps have recently become available in human (Matise et al. 2007) and mouse (Shifman et al. 2006). These high-resolution maps indicated a strong correlation between recombination hot spots and high density of the 7-nucleotide oligomer CCTCCCT both in human (Myers et al. 2005) and mouse (Shifman et al. 2006).
The fact that the chicken genome is composed of different subsets of chromosomes, generally referred to as macro- and microchromosomes exhibiting structural differences, adds another dimension to these associations. It is well established that microchromosomes exhibit higher recombination rates compared with those in macrochromosomes (Rodionov 1996). Upon completion of the draft chicken genome sequence, it became clear that chromosome size shows correlations with recombination rates, gene density, gene size, CpG island density, and overall GC content.
The map presented here incorporates 8599 SNPs in addition to 669 selected markers (primarily microsatellites) that were previously included in the consensus linkage map. Inclusion of the microsatellites ensures accurate transfer of map positions of previous QTL analysis to the current improved consensus linkage map. This new improved map allowed us to address basic questions concerning the chicken meiotic map such as: (1) Do the same sequence features that affect recombination in mammals play a similar role in birds? (2) Are recombination hot spots characterized by specific structural features or conserved sequences? (3) Are any such structural features causative of increased recombination or merely the result of the higher recombination at those locations? (4) Are there differences in recombination between sexes and populations?
Results
Markers and genotyping quality
In total, 12,945 SNPs were selected and screened in three large-scale SNP studies using three existing mapping populations; the Wageningen (WU) and East Lansing (EL) mapping populations used to obtain the chicken consensus linkage map (Groenen et al. 2000) and a Red Jungle fowl/White Leghorn intercross (Kerje et al. 2003). Comparable high success rates for genotyping calls were obtained in all three experiments that ranged from 88% to 92%. Only SNPs where the three groups of genotypes AA, AB, and BB grouped within clear distinct clusters were considered to be reliable. About 87% of the SNPs with reliable call rates were informative in at least one of the three mapping populations. Eventually, approximately 10,000 SNPs were selected for map construction. A few percent of these markers showed non-Mendelian inheritance, possibly caused by the presence of null alleles. Where the non-Mendelian inheritance was limited to a single family, the genotypes were set to zero for that particular family, whereas the marker was removed entirely from the analysis when the problem was observed in multiple families. The number of informative meioses per marker varied from 10 to 242, with an average of 85.
The comparisons of the male vs. female map appeared to be a good aid in identifying markers with obvious genotyping errors. Regions on the map showing extreme differences in male vs. female recombination were always analyzed in detail using flips and by removing individual markers and groups of markers from the map and observing the effect that this had on the map. Individual markers that resulted in map inflation often contained an excess of double recombinants. At the marker density used, such double recombinants are a clear indication for genotyping errors, and therefore were excluded from the final analysis.
Map construction
The EL and WU mapping populations have been used extensively for linkage mapping (Groenen and Crooijmans 2003), and the total number of markers previously mapped on the consensus map using these two populations is 2261 (Schmid et al. 2005). The majority of these markers are microsatellites and amplified fragment length polymorphisms (AFLPs). Because many of these microsatellites have been used extensively in QTL mapping experiments, it was decided to include these markers in the current analysis to be able to align previous linkage data with the new map. In the present study, only two of the original 10 families from the WU population (Groenen et al. 1998) were used, and only markers that were informative in these two families could be included in the analysis. Furthermore, it was decided to exclude the AFLP markers from the analysis because of the lower reliability of the genotypes of this type of marker in the existing data set, and because in the last decade these markers have not been used in any published study in chicken. The only exceptions were those AFLP markers that had not been assigned to any of the existing linkage groups, because we reasoned that these might aid in the development of new linkage groups for some of the missing microchromosomes.
We initially attempted to update the linkage map starting with the existing consensus linkage map. However, that approach was abandoned and map construction was started with the most informative SNP markers, followed by adding microsatellite and other markers at a later stage. The main reason to do so was the observation that the number of genotyping errors was significantly higher in the older marker data as compared with the SNP genotypes obtained by the Illumina GoldenGate assay (Fan et al. 2006). Eventually, 669 markers from the previous consensus map could be added to the current map. A total of 8599 SNPs were included, bringing the total number of markers on the current linkage map to 9268 (Supplemental Table 1). The high marker density in the current map enabled us to spot potential genotyping errors in the microsatellite data. These were identified as double recombinants using the CRI-MAP (Green et al. 1990) “chrompic” option and removed from the data.
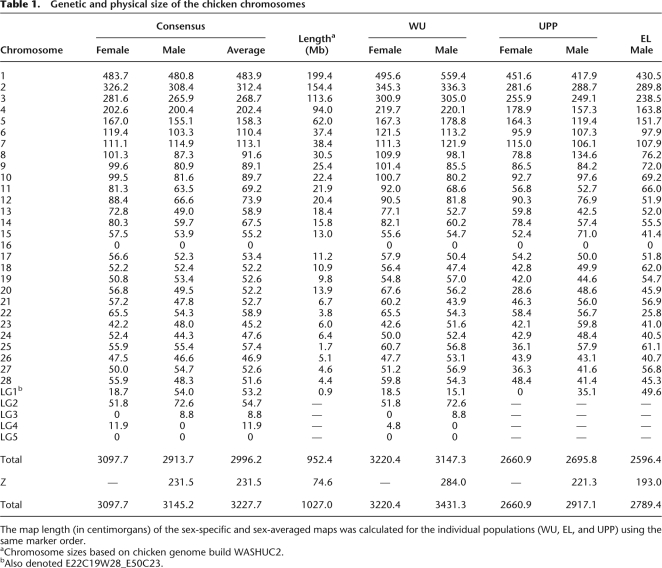
The total length of the current sex average map is 3228 cM, which is considerably shorter than the 4200 cM in the previous linkage map for chicken (Schmid et al. 2005). Excluding the four new linkage groups, the difference between the old and new map is even more striking (3152 vs. 4200 cM). It is now clear that the old map was inflated due to genotyping errors. We also revealed highly significant (P < 0.001) differences between populations as regards the total map length (Table 1); the result was obtained by running the “fixed” option of CRI-MAP on the data from each population separately using the same marker order (Supplemental Table S3). The observed differences between the populations appears to be a rather general trend across the genome rather than being caused by specific regions on the different chromosomes (Supplemental Fig. S1). The WU male map was 16% and 22% longer than the Uppsala (UPP) and EL male maps, respectively, excluding LG2–LG7 that were not included in all maps. Similarly, the WU female map was 18% longer than the UPP female map. These differences are primarily caused by longer linkage maps for the macrochromosomes, including chromosome Z.
Table 1.
Genetic and physical size of the chicken chromosomes
Genome assembly and missing chromosomes
Our new map consists of 34 linkage groups and covers at least 29 of the 38 autosomes. If the four new linkage groups (LG2–LG5) each represent an individual chromosome, the number of autosomes covered by the current map is 33, which means that still at least five of the microchromosomes are not represented by a linkage group. The new linkage groups, as well as the linkage group representing chromosome 16, only contain a small number of loci. Furthermore, LG2 is comprised only of AFLP markers, and therefore no sequences can be assigned to this particular linkage group/chromosome. Finally, 26 markers that could not be placed on any of the 34 linkage groups showed linkage to one other unassigned marker likely representing 13 regions of the genome not yet covered by the linkage map (Supplemental Table S2).
The linkage and sequence maps are generally in good agreement (Supplemental Table S1). There are close to 100 marker pairs (1%) that were in a slightly different order on the two maps, but the majority of these represent two adjacent markers and do not affect the overall alignments of the maps. There are two regions where the two maps deviate considerably. The first region is the end of chromosome 1 between 463 and 483 cM, where the linkage data indicate that several of the sequence contigs in the sequence map should be inverted or the order changed. The new order results in a decrease in size of the map of 3.7 cM, and the difference in log10 likelihood for the two maps was 12.5 in favor of the map presented in this study. The second region is the entire chromosome 25, where many of the sequence contigs clearly are in the wrong order and location. When the option fixed was used with the markers on chromosome 25 in the order as they appear on the sequence map, the resulting linkage map doubled in size to 114.9 cM. The difference in log10 likelihood for the two maps was 58, in favor of the map presented in this study.
Male vs. female recombination rates
In many species, the frequency of recombination during meiosis is lower in the heterogametic sex (Haldane 1922). Thus, in mammalian species, the female linkage map is longer than the male one. For instance, in humans, the average map length is 60% longer in females (Matise et al. 2007). Differences in male vs. female recombination rate in chicken were examined by performing a sex-specific analysis. As shown in Table 1, there is no significant difference in total length between the male and female maps. A sex-specific analysis was not possible for the EL population, because being a backcross population, only a male-specific map was calculated based on the segregating F1 male. The difference between the sex-specific maps based on the WU population was only 3%, while no overall difference was observed for the UPP population. Thus, the heterogeneity in recombination rates between populations is much more prominent than the heterogeneity between sexes (Table 1).
Recombination hot spots
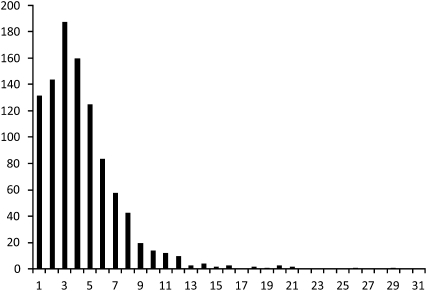
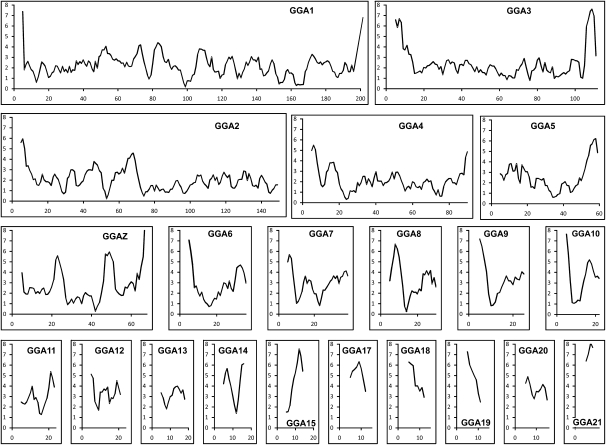
Recombination frequencies were calculated for nonoverlapping bins of ∼1 Mb with marker positions defining the ends of each of the intervals (Fig. 1). Excluding the four new linkage groups (LG2–LG5) that could not be aligned with the genome assembly, the current physical size covered by the remaining 9242 markers is 1020 Mb. With a size of 3176 cM for the sex-average linkage map and an average bin size of 1 Mb, this brings the average genetic size of a 1-Mb bin to 3.11 cM. Interestingly, the range observed within the 1014 bins of 1 Mb is extremely large, ranging from 0 to close to 20 cM (Fig. 1); three regions with a genetic size of up to 40 cM are observed on chromosomes 22 and 25, but these were excluded from the analysis because they most likely represent errors in the most recent sequence assembly (see below). A similar result was observed using 2-Mb sized intervals with an average size of 5.89 cM per 1.96 Mb, and the estimates per interval were in the range of from 0 to 38.6 cM. Interestingly, although the majority of recombination hot spots are located on microchromosomes, a small number is located on macrochromosomes. Generally, these recombination hot spots tend to be located at the distal part of the chromosomes (Fig. 2).
Figure 1.
Distribution of recombination rates for 1-Mb bins in the chicken genome. The _x_-axis shows the recombination rate as centimorgan/megabase. The _y_-axis shows the number of bins.
Figure 2.
Recombination rates of chicken chromosomes. Recombination rates were calculated for 1-Mb–sized bins and plotted as a sliding window showing the average recombination frequency for a window of 5 Mb. The scale on the _x_-axis is in megabases and the scale on the _y_-axis represents the recombination rate as centimorgan/megabase. Figures for chromosomes 22–28 are not included because of the small size of these chromosomes, resulting in uninformative plots.
Correlation of recombination with sequence parameters
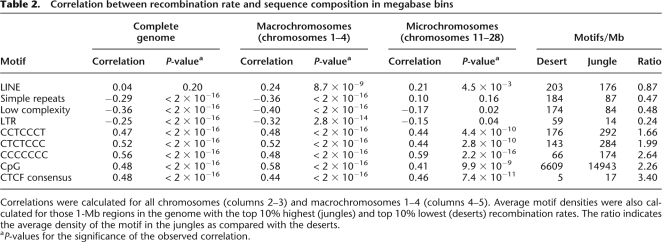
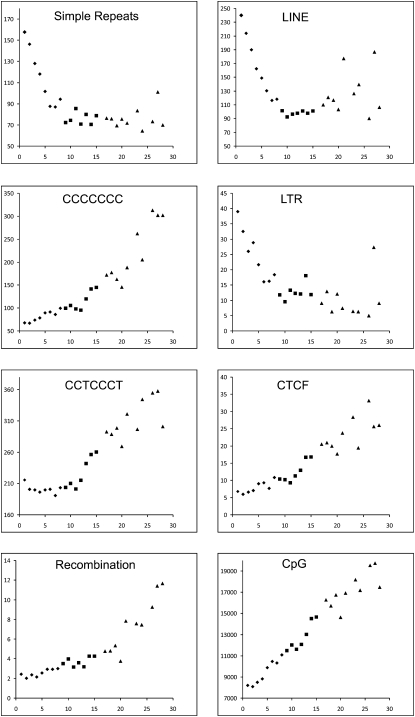
Generally, recombination rates are strongly correlated with GC content, an observation that is consistent with a bias toward GC in mismatch repair used during the gene conversion process (Spencer et al. 2006). Several studies in human and mouse have shown a highly significant correlation between recombination and specific sequence features, in particular the sequence CCTCCCT (Myers et al. 2005; Shifman et al. 2006). To examine whether this correlation also exists in chicken, we analyzed the occurrence of this and related sequences within megabase-sized intervals of the chicken genome (Table 2). Recently, it was shown that cohesin-binding sites closely resemble the binding site for the CCCTC-binding factor (CTCF) (Wendt et al. 2008). Because both cohesin and CTCF bind similar GC-rich sequences, the consensus sequence for cohesin (CCNCCNGGNGG) was also tested. Finally, the correlation was also calculated between the recombination frequency and repetitive element content (LINE, LTR, simple repeats, and low-complexity sequences). Consistent with previous studies in mammals, the results clearly showed a strong positive correlation between recombination rate and GC-rich sequences and a negative correlation with AT-rich sequences. Earlier studies on chicken already indicated a strong correlation between recombination frequency and chromosome size (Hillier et al. 2004), and the same correlation can be seen between the different sequence parameters and chromosome size (Fig. 3). To test whether the observed correlation between the sequence parameters and the recombination frequency was only caused by the differences in density of these elements on the macrochromosomes vs. the microchromosomes, correlations were also calculated separately for the four largest macrochromosomes (Table 2). With the exception of the LINE elements, the same correlations were observed within the macrochromosomes.
Table 2.
Correlation between recombination rate and sequence composition in megabase bins
Figure 3.
Distribution of average recombination rates and sequence motif densities across chromosomes in chicken. The density plots for chromosomes 1–15 and 17–28 are shown. The scale on the _x_-axis represents the chromosome number and the _y_-axis represents the average number of elements per megabase or recombination rate in centimorgan/megabase. (♦) Macrochromosomes; (■) intermediate chromosomes; (▲) microchromosomes.
In order to get further insight into what degree the sequence composition affects recombination rates, we compared the sequence composition of those 1-Mb intervals with the top 10% highest (“recombination jungles”) and the top 10% lowest (“recombination deserts”) recombination rates (Table 2); this approach has previously been used in mouse (Shifman et al. 2006). The largest difference is seen for LTRs, which have a fourfold higher incidence in recombination deserts as compared with recombination jungles. Most striking is the 3.4 times higher number of CTCF-binding sites in recombination jungles.
Discussion
The current updated version of the chicken consensus linkage map is a considerable improvement compared with the previous versions (Groenen et al. 2000; Schmid et al. 2005). There are two major reasons for this improvement. The first is the fivefold higher marker density of the current map. This higher marker density allowed for the detection of double recombinants affecting a single marker, which, at the current marker density (approximately three markers/cM), is a clear sign of typing errors. Second, SNPs genotyped using a highly automated method like the Illumina GoldenGate assay result in a much lower genotyping error rate than that obtained with markers like microsatellites and AFLPs. In addition, SNP markers showing evidence of null-alleles or markers with an excess number of double recombinants were removed from the analyses. Due to this careful error checking, the current map shows a 24% reduction in total length compared with the previous consensus map (Groenen et al. 2000; Schmid et al. 2005).
The current map is comprised of 34 linkage groups, with a total size of 3228 cM. When we consider the fact that at least five of the microchromosomes are not covered by the current map, and given the observation that the genetic size of the microchromosomes, on average, is ∼50–60 cM, it is to be expected that the genetic size of the complete chicken genome is at least 3700 cM. The size of a genetic map reflects the average number of recombination events per meioses. There is a good correlation across species between the number of chromosomes and the length of the genetic map, probably reflecting the need for at least one crossover for each bivalent during meiosis. An even better predictor for the genetic length is the number of chromosomal arms, in particular when the short arms of the acrocentric chromosomes are excluded (Pardo-Manuel de Villena and Sapienza 2001). A plot of the genetic length against the number of chromosome arms for a variety of mammals shows that the genetic length increases by ∼75 cM per chromosomal arm (Coop and Przeworski 2007). If we apply the same rule to chicken, a genetic size of 3700 cM suggests the presence of 48 chromosome arms, which would suggest the majority of the microchromosomes to be acrocentric. This finding is in agreement with the results obtained with chicken synaptonemal complex chromosome spreads (Pigozzi 2007).
The large variation in size between the different chicken chromosomes, and in particular the presence of many small microchromosomes, constitutes a challenge for any attempt to develop a genetic map with full genome-wide coverage. Previous linkage maps (Cheng et al. 1995; Groenen et al. 1998, 2000), physical maps (Wallis et al. 2004), and the sequence map (Hillier et al. 2004) only covered around 30 of the 39 chromosomes, with the smallest microchromosomes missing. Also, the current consensus map still only contains 34 linkage groups and thus does not cover at least five microchromosomes despite the large number of markers used and the inclusion of markers from sequence contigs that had not been assigned to a specific chromosome. Mapping and sequencing results, so far, indicate that specific physical characteristics of some of the microchromosomes are inhibitory to cloning these sequences in E. coli, resulting in the absence of these sequences in libraries used to construct genetic, physical, and sequence maps. A clear example is the difficulty of obtaining sequences that are orthologous to human chromosome 19q (Hillier et al. 2004; Gordon et al. 2007). Based on EST data, many of the chicken orthologs for genes located on human chromosome 19q are known to exist, yet sequences of these genes were virtually absent from the whole-genome shotgun data and completely absent from all BAC libraries available for chicken. Because AFLP markers do not require any cloning step in E. coli, the presence of two small linkage groups that are comprised (almost) completely of AFLP markers is in agreement with these observations. The recent development of second generation sequencing technologies like Roche 454, Illumina Solexa, or AB SOLiD (Chi 2008) may circumvent this difficulty and generate future maps that also cover these missing microchromosomes.
A striking difference between the length of the male and female maps exists in many species (Lenormand and Dutheil 2005). In some species (e.g., Drosophila melanogaster), recombination is even restricted to only one of the sexes, and in those cases it is the homogametic sex where recombination takes place. Higher recombination rate in the homogametic sex is referred to as the Haldane–Huxley rule (Haldane 1922; Huxley 1928). Although the Haldane–Huxley rule often is generalized to apply to cases where both sexes recombine, many exceptions to this rule exist, with longer genetic maps in the heterogametic sex (Lenormand and Dutheil 2005). In chicken, the female is the heterogametic sex (ZW) and, on average, there is hardly any difference in total size between the male and female maps. However, many chromosomes exhibit sex differences, such as chromosomes 9, 10, 11, 12, 13, and 14, where the female map is more than 20% longer or chromosomes 7, 19, 23, 27, and LG1, where the male maps appear to be longer. To further examine whether these differences reflect true underlying biological and genetic differences between the sexes and the different chromosomes, we also calculated the maps for the individual populations that were used (Table 1). The results indicate that there is no significant difference in size between the male and female maps when the maps are calculated for the individual WU and UPP populations. Furthermore, for several chromosomes, the size differences for specific chromosomes are not in agreement between the WU and UPP maps. Clear examples are chromosome 1 and chromosome 8. For WU, the male map of chromosome 1 is 13% longer than the female map, whereas the opposite is found for UPP with the male map being 7.5% smaller than the female map. The opposite is observed for chromosome 8, where for WU the male map is 11% smaller than the female map, whereas for UPP it is 71% larger than the female map.
Because the EL population is a backcross population, in which an F1 male was crossed with the females of an inbred parental line, only a male map could be produced for this population. The total size of the map produced using only the EL population is 20% smaller than that of the WU population and 4% smaller than the UPP map. Because the EL map only affects the male map in the consensus linkage map, it will result in a smaller size of the male consensus map compared with the female consensus map. The consensus male linkage map is, in fact, 5% smaller than the female map. Hence, we conclude that there is no difference in total map length between sexes. Although we cannot rule out small regional differences in recombination along the different chromosomes, the most likely explanation for these differences is that these are due to differences in the data structure, i.e., the information content of the different markers within the different populations used in the analysis. We have observed several regions where markers are only informative in either the male or female parents, thereby causing local differences in the size of the genetic map at those locations.
Contrary to the lack of an apparent difference in recombination between the two sexes, a significant difference exists between populations. As mentioned above, the EL map is 20% and 4% smaller than the WU and UPP maps, respectively. The clearest difference is between the WU map on the one side and the EL and UPP maps on the other. Interestingly, both the EL and UPP populations are crosses between Red Jungle fowl and White Leghorn breeds, whereas the WU map is based on a cross between two broiler dam lines originating from White Plymouth Rock. Variation in recombination rates have been observed in many populations in animals (Andersson and Sandberg 1984; Broman et al. 1998; Koehler et al. 2002) as well as in plants (Williams et al. 1995). Kong et al. (2004) have estimated that the heritability of total recombination in human is around 30%. Local rearrangements such as inversion polymorphisms can result in lower recombination rates (Giglio et al. 2001; Stefansson et al. 2005). Although currently no information is available about inversions in chicken, the fact that a lower recombination rate is observed in the two crosses between the more divergent breeds (Red Jungle fowl × White Leghorn) suggests that chromosomal rearrangements could potentially contribute to the observed difference. An alternative explanation is that there has been selection for a higher recombination rate during chicken domestication, as recombination may remove unfavorable genetic correlations and thereby increase the selection response. A comparison of the genetic maps for the individual populations did not show any clear evidence for chromosomal rearrangements and indicate that the latter explanation is the most likely (Supplemental Fig. S1). In fact, Burt and Bell (1987) have reported that domestic animals deviate from natural populations by having unusually high chiasmata counts. Our data showing that the F1 Red Jungle fowl/White Leghorn males and females from the UPP and EL populations showed significantly lower recombination rates compared with the WU broiler population is consistent with a higher recombination rate in purebred domestic animals under strong artificial selection. This interesting observation needs to be further investigated by examining average recombination rates in additional purebred domestic as well as purebred Red jungle fowl populations.
The rate of recombination varies considerably between different genomic regions, which is most evident between the macro- and microchromosomes. This increased recombination rate on the smaller chromosomes has been observed in previous studies (Rodionov 1996; Hillier et al. 2004) and is thought to reflect the requirement of at least one chiasmata per bivalent during meiosis (Rodionov 1996). Because of the large variation in chromosome size in chicken, this effect is very clear but can also be observed in human where the recombination rate on the smallest chromosome (HSA22) is twofold higher than that of the largest chromosome (HSA1). However, differences in recombination rate are also observed at a finer resolution along the chromosomes. The variation in recombination rate using a sliding window of 5 Mb is shown in Figure 2 for all linkage groups assigned to a specific chromosome. Similar to what has been observed in mammals, on average, recombination rates tend to be elevated toward telomeres and lower close to the centromeres. However, some exceptions are observed, in particular, GGA2 and GGAZ, showing higher recombination rates at only one of the telomeres. One explanation for this observation would be that the ends of these chromosomes are not yet covered by the sequence map in the current genome build (WASHUC2).
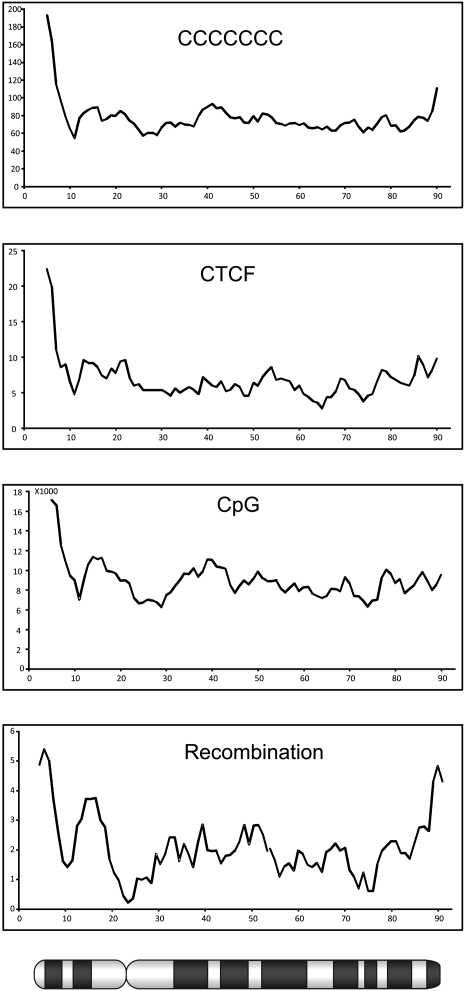
Analysis of the sequence composition at the recombination hot and cold spots clearly indicated a strong correlation between high recombination rates and GC-rich sequences. This observation is in agreement with observations in human and mouse, where a highly significant correlation was observed between recombination and the density of the GC-rich elements CCTCCCT and CCCACCCC (Myers et al. 2005; Shifman et al. 2006). It was suggested that in human there might be a link to the THE1A- and THE1B-type retrovirus-like transposons, but the absence of this transposable element in mouse, as well as in chicken, shows that the observed increased recombination rate cannot be attributed to this transposable element only. Furthermore, in chicken, the highest correlation was found with the motif CCCCCCC. Not surprisingly, the GC-rich cohesin binding site (CCNCCNGGNGG) also showed a high correlation with recombination hot spots. Comparing the ratio of the density of these sequence elements in recombination deserts to recombination jungles, the cohesin binding sites stand out from the other GC-rich sequences with a 3.4-fold higher density in recombination jungles. The different studies in human, mouse, and chicken indicate a correlation between several features, including high recombination rate, high GC content, high gene density, and high density of cohesin binding sites. In addition, a correlation is seen between these features and the size of the chromosome, which, because of the large variation in chromosome size, is particularly apparent in the chicken genome (Fig. 3). Although the correlations between recombination frequency, GC content, and density of cohesin binding sites are most clearly seen in the chromosome plots shown in Figure 3, the correlation exists both between, as well as within the different chromosomes. A special case is represented by chromosome 4 where the highest recombination rate, GC content, and cohesion density is observed within the p-arm of this chromosome. Interestingly, a comparison between the karyotypes of chicken, turkey, and other birds indicates that chicken chromosome 4 is the result of a fusion between a macrochromosome and a microchromosome in the lineage leading to chicken (Griffin et al. 2008). The density plots for chromosome 4 shown in Figure 4 provide further support for the microchromosomal origin of the p-arm of this chromosome.
Figure 4.
Distribution of recombination rates and sequence composition across chicken chromosome 4. The scale on the _x_-axis is in megabases, whereas the _y_-axis shows the number of elements per megabase or recombination rates in centimorgan/megabase. An ideogram of chromosome 4 is depicted below the _x_-axis.
It is difficult to distinguish between causative and secondary effects shaping the pattern of recombination, and it even is likely that some or all of these features work synergistically. One explanation for the observations is that high densities of cohesin binding sites will increase the chance of formation of the synaptonemal complex in those regions and, consequently, would result in a higher rate of recombination. Given the fact that proper segregation of the chromosomes during meioses requires at least one chiasma, even for the microchromosomes, selection for a high density of cohesin binding sites on the microchromosomes would ensure efficient formation of chiasmata. Conversely, recombination is closely linked to gene conversion, which has been show to be biased toward elevating the GC content (Marais 2003). Consequently, the increased GC content might, on average, have a positive effect on the expression of the genes within that region, favoring the accumulation of highly expressed housekeeping genes over the larger more complex genes involved in development and transcriptional regulation (Gordon et al. 2007).
Methods
Mapping populations
The animals used in the current study were derived from three different populations previously used for linkage mapping. Briefly, the original East Lansing mapping population (EL) (Crittenden et al. 1993) consisted of 52 animals derived from a backcross (BC1) between a partially inbred Red Jungle fowl line and a highly inbred White Leghorn line. In the current study, the number of genotyped animals was extended to 88. The Wageningen University (WU) population (Groenen et al. 1998) consisted of 92 F2 animals in two full-sib families from a cross between two broiler dam lines originating from White Plymouth Rock. The population consisted of 10 families with a total of 456 F2 offspring; however, in the current study only two of the 10 families were genotyped. The third population used is maintained at the Uppsala University (UPP) and consists of 55 animals (47 F2, four F1, four F0) from a Red Jungle fowl × White Leghorn intercross (Kerje et al. 2003; Wahlberg et al. 2007).
SNP selection
The SNP markers were selected from dbSNP versions 122 and 123. SNPs were selected independently within three different projects with different overall objectives. (1) SNP panel 1 consisted of 3072 SNPs evenly spaced throughout the chicken genome and were selected from the 2.8 million SNP data set previously identified by International Chicken Polymorphism Map Consortium (2004). Toward this end, the genome sequence (build WASHUC1) was divided into 3072 bins, taking into account the recombination rate per chromosome. For each bin, three SNPs were selected and preference was given to high-confidence SNPs located within genes, especially those judged to be tolerant nonsynonymous SNPs (cnSNPs). All SNPs were evaluated for assay suitability, and the best SNP in each bin was selected. In addition, 34 SNPs in genes of interest were evaluated (Muir et al. 2008). (2) A further 657 SNPs (SNP panel 2) were selected and represented SNPs not yet assigned to a chromosomal location on build WASHUC1. (3) Finally, 9216 SNPs (SNP panel 3) were selected evenly spaced throughout the chicken genome, also taking into account the sequence contigs that were not yet assigned a chromosomal location on build WASHUC1 (Wahlberg et al. 2007).
SNP genotyping
All selected SNPs were genotyped using the Illumina GoldenGate assay. DNA was isolated from blood using standard procedures and used at a concentration of 50 ng/μL. Genotyping was done at three different locations. SNP panel 1 was typed on the EL and WU populations at Illumina. Genotyping of SNP panel 2 was done on the EL and WU populations using a manually operated BeadLab facility at the University of Utrecht. Genotyping of SNP panel 3 on the WU and UPP populations was done at the Center National de Génotypage (CNG) in a fully automated BeadLab.
Linkage analysis and map construction
Linkage maps were constructed with a modified version of the CRI-MAP software (Green et al. 1990). The modified version allowed the simultaneous analysis of much larger numbers of markers than the original program. The modified version of CRI-MAP was kindly provided by Drs. Liu and Grosz of Monsanto. The modified version of CRI-MAP allowed us to simultaneously analyze all markers on each chromosome. All markers were checked for non-Mendelian inheritance errors using the option “prepare.” Initially, markers were assembled in separate chromosome-specific files based on previous linkage information (Groenen et al. 2000) and based on the chicken genome sequence (Hillier et al. 2004; genome build WASHUC1). Subsequently, correct assignment of the markers to a specific chromosome was checked by performing an all-to-all comparison of the loci using the CRI-MAP option “twopoint.” Loci that did not show linkage to multiple markers within the same file were removed from the analysis and stored in a separate file with other unassigned markers. Finally, all unassigned markers were compared against all other markers using the CRI-MAP “twopoint” option, and in case of clear linkage (LODscore > 4) to assigned markers, moved to that specific chromosome file. New linkage groups were only constructed in case individual markers were linked with a LOD score higher than 4.
The linkage map was subsequently constructed in a number of iterative rounds using the “build” option within CRI-MAP, starting with a threshold of LOD = 5 with subsequent stepwise lowering of the LOD threshold until LOD = 0.2. Closely linked markers not separated by recombination events were ordered according to their location on the sequence map (build WASHUC2). The location of all markers on the WASHUC2 build was determined using BLAT (Kent 2002). Markers were included in the growing linkage map according to the number of informative meiosis, with the most informative markers being included first. The order of markers in the final map was verified using the “flips” option. Finally, potential typing errors were recognized as unlikely double recombinants using the “chrompic” option. Regions showing excess recombination or where the male and female maps deviated considerably were checked in detail by removing markers from the map and evaluating the effect on the size of the male and female linkage maps. Differences in recombination frequencies between populations and sexes were based on normalized recombination frequencies as the ratio of the population-specific value over the average for all populations. These values were subsequently fitted to a simple linear model (normalized recombination frequency ∼ population, “aov” in R). Confidence intervals on the differences between the means of recombination per population pair were calculated using the Tukey Honest Significant Differences (“TukeyHSD” on “aov” fit in R).
Recombination rates in relation to sequence motifs
The distribution of sequence motifs were calculated per 1-Mb bins using the exact marker positions as the borders of the bins. The calculation of the distribution of LINEs, LTRs, Simple repeats, and low-complexity sequences within the chicken genome were based on the position of these elements downloaded from the UCSC genome browser. The distribution of the sequence motifs CCTCCCT, CTCTCCC, CCCCCCC, CG, and the CTCF consensus sequence CCNCCNGGNGG were calculated using specific perl scripts. Correlations between recombination frequency and abundance of different sequence motifs within 1-Mb bins were tested using Pearson's product moment correlation coefficient as implemented in the “cor.test” function in R (www.r-project.org).
Acknowledgments
This work was supported in part by funding from the USDA NRICGP (#2004-05434 to H.H.C., M.A.M.G., G.K.S.W., and W.M.M.) and from the Swedish Foundation for Strategic Research and The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (to L.A.).
Footnotes
References
- Andersson L., Sandberg K. Genetic linkage in the horse. II. Distribution of male recombination estimates and the influence of age, breed and sex on recombination frequency. Genetics. 1984;106:109–122. doi: 10.1093/genetics/106.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Murray J.C., Sheffield V.C., White R.L., Weber J.L. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., Bell G. Mammalian chiasma frequencies as a test of two theories of recombination. Nature. 1987;326:803–805. doi: 10.1038/326803a0. [DOI] [PubMed] [Google Scholar]
- Cheng H.H., Levin I., Vallejo R.L., Khatib H., Dodgson J.B., Crittenden L.B., Hillel J. Development of a genetic map of the chicken with markers of high utility. Poult. Sci. 1995;74:1855–1874. doi: 10.3382/ps.0741855. [DOI] [PubMed] [Google Scholar]
- Chi K.R. The year of sequencing. Nat. Methods. 2008;5:11–15. doi: 10.1038/nmeth1154. [DOI] [PubMed] [Google Scholar]
- Coop G., Przeworski M. An evolutionary view of human recombination. Nat. Rev. Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Crittenden L.B., Provencher L., Santangelo L., Levin I., Abplanalp H., Briles R.W., Briles W.E., Dodgson J.B. Characterisation of a Red Jungle Fowl by White Leghorn backcross reference population for molecular mapping of the chicken genome. Poult. Sci. 1993;72:334–348. [Google Scholar]
- Daw E.W., Thompson E.A., Wijsman E.M. Bias in multipoint linkage analysis arising from map misspecification. Genet. Epidemiol. 2000;19:366–380. doi: 10.1002/1098-2272(200012)19:4<366::AID-GEPI8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Fan J.B., Chee M.S., Gunderson K.L. Highly parallel genomic assays. Nat. Rev. Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- Fingerlin T.E., Abecasis G.R., Boehnke M. Using sex-averaged genetic maps in multipoint linkage analysis when identity-by-descent status is incompletely known. Genet. Epidemiol. 2006;30:384–396. doi: 10.1002/gepi.20151. [DOI] [PubMed] [Google Scholar]
- Giglio S., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L., Yang S., Tran-Gyamfi M., Baggott D., Christensen M., Hamilton A., Crooijmans R., Groenen M., Lucas S., Ovcharenko I., et al. Chicken chromosome 28 and the dynamic evolutionary history of gene-rich vertebrate genomic regions. Genome Res. 2007;17:1603–1613. doi: 10.1101/gr.6775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P., Falls K., Crooks S. Washington School of Medicine; St. Louis, MO: 1990. Documentation for CRI-MAP, version 2.4. [Google Scholar]
- Griffin D.K., Robertson L.B., Tempest H.G., Vignal A., Fillon V., Crooijmans R.P.M.A., Groenen M.A.M., Deryusheva S., Gaginskaya E., Carré W., et al. Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genomics. 2008;9:168. doi: 10.1186/1471-2164-9-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M.A.M., Crooijmans R.P.M.A. Structural genomics: Integrating linkage, physical and sequence maps. In: Muir W.M., Aggrey S.E., editors. Poultry genetics, breeding and technology. CAB International; Cambridge, MA: 2003. [Google Scholar]
- Groenen M.A., Crooijmans R.P., Veenendaal A., Cheng H.H., Siwek M., van der Poel J.J. A comprehensive microsatellite linkage map of the chicken genome. Genomics. 1998;49:265–274. doi: 10.1006/geno.1998.5225. [DOI] [PubMed] [Google Scholar]
- Groenen M.A., Cheng H.H., Bumstead N., Benkel B.F., Briles W.E., Burke T., Burt D.W., Crittenden L.B., Dodgson J., Hillel J., et al. A consensus linkage map of the chicken genome. Genome Res. 2000;10:137–147. doi: 10.1101/gr.10.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J.B.S. Sex-ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Huxley J.S. Sexual difference of linkage in Gammarus chereuxi. J. Genet. 1928;20:145–156. [Google Scholar]
- International Chicken Polymorphism Map Consortium. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson L., Park H.-B., Wahlberg P., Jiang S., Siegel P.B., Andersson L. Assignment of 14 microsatellite markers to the chicken linkage map. Poult. Sci. 2004;83:1825–1831. doi: 10.1093/ps/83.11.1825. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S., Carlborg Ö., Jacobsson L., Schütz K., Hartmann C., Jensen P., Andersson L. The two-fold difference in adult size between the red jungle fowl and White Leghorn chickens is largely explained by a limited number of QTLs. Anim. Genet. 2003;34:264–274. doi: 10.1046/j.1365-2052.2003.01000.x. [DOI] [PubMed] [Google Scholar]
- Koehler K.E., Cherry J.P., Lynn A., Hunt P.A., Hassold T.J. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics. 2002;162:297–306. doi: 10.1093/genetics/162.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Barnard J., Gudbjartsson D.F., Thorleifsson G., Jonsdottir G., Sigurdardottir S., Richardsson B., Jonsdottir J., Thorgeirsson T., Frigge M.L., et al. Recombination rate and reproductive success in humans. Nat. Genet. 2004;36:1203–1206. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- Lenormand T., Dutheil J. Recombination difference between sexes: A role for haploid selection. PLoS Biol. 2005;3:e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. Biased gene conversion: Implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- Matise T.C., Chen F., Chen W., De La Vega F.M., Hansen M., He C., Hyland F.C.L., Kennedy G.C., Kong X., Murray S.S., et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir W.M., Wong G.K.-S., Zhang Y., Wang J., Groenen M.A.M., Crooijmans R.P.M.A., Megens H.-J., Zhang H.M., McKay J.C., McLeod S., et al. Review of the initial validation and characterization of a 3K chicken SNP array. World Poul. Sci. J. 2008;64:219–225. [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome. 2001;12:318–322. doi: 10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- Pigozzi M.I. Localization of single-copy sequences on chicken synaptonemal complex spreads using fluorescence in situ hybridization (FISH) Cytogenet. Genome Res. 2007;119:105–112. doi: 10.1159/000109626. [DOI] [PubMed] [Google Scholar]
- Rodionov A.V. Micro vs. macro: Structural-functional organization of avian micro- and macrochromosomes. Genetika. 1996;32:597–608. [PubMed] [Google Scholar]
- Schmid M., Nanda I., Hoehn H., Schartl M., Haaf T., Buerstedde J.M., Arakawa H., Caldwell R.B., Weigend S., Burt D.W., et al. Second report on chicken genes and chromosomes 2005. Cytogenet. Genome Res. 2005;109:415–479. doi: 10.1159/000084205. [DOI] [PubMed] [Google Scholar]
- Shifman S., Bell J.T., Copley R.R., Taylor M.S., Williams R.W., Mott R., Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C.C.A., Deloukas P., Hunt S., Mullikin J., Myers S., Silverman B., Donnelly P., Bentley D., McVean G. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:1375–1385. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H., Helgason A., Thorleifsson G., Steinthorsdottir V., Masson G., Barnard J., Baker A., Jonasdottir A., Ingason A., Gudnadottir V.G., et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- Wahlberg P., Strömstedt L., Tordoir X., Foglio M., Heath S., Lechner D., Hellström A.R., Tixier-Boichard M., Lathrop M., Gut I.G., et al. A high-resolution linkage map for the Z chromosome in chicken reveals hot spots for recombination. Cytogenet. Genome Res. 2007;117:22–29. doi: 10.1159/000103161. [DOI] [PubMed] [Google Scholar]
- Wallis J.W., Aerts J., Groenen M.A., Crooijmans R.P., Layman D., Graves T.A., Sheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., et al. A physical map of the chicken genome. Nature. 2004;432:761–764. doi: 10.1038/nature03030. [DOI] [PubMed] [Google Scholar]
- Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Williams C.G., Goodman M.M., Stuber C.W. Comparative recombination distances among Zea mays L. inbreds, wide crosses and interspecific hybrids. Genetics. 1995;141:1573–1581. doi: 10.1093/genetics/141.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]