Therapeutic Potential of Splice-Switching Oligonucleotides (original) (raw)
Abstract
Alternative splicing enables a single pre-messenger RNA transcript to yield multiple protein isoforms, making it a major contributor to the diversity of the proteome. While this process is essential for normal development, aberrations in alternative splicing are the cause of a multitude of human diseases. Methods for manipulating alternative splicing would thus be of therapeutic value. Chemically modified antisense oligonucleotides that alter alternative splicing by directing splice site selection have been developed to achieve this end. These splice-switching oligonucleotides (SSOs) have been applied to correct aberrant splicing, induce expression of a therapeutic splice variant, or induce expression of a novel therapeutic splice variant in a number of disease-relevant genes. Recently, in vivo efficacy of SSOs has been reported using animal disease models, as well as in results from the first clinical trial.
Introduction
Alternative splicing is an essential biological process with important implications for human disease. Early work from this laboratory demonstrated that antisense oligonucleotides could be used to correct aberrantly spliced human β-globin pre-messenger RNA (pre-mRNA) in cell-free extracts and in cultured erythroid cells from β-thalassemia patients (Dominski and Kole, 1993; Sierakowska et al., 1996; KOLE, 1997; Lacerra et al., 2000). Since then, we and others have employed splice-switching oligonucleotides (SSOs) in numerous applications targeting many disease-relevant genes (Kole et al., 2004). Recently, several groups have reported advances using SSOs in animal models of human disease. Importantly, clinical trials using SSOs to treat Duchenne muscular dystrophy have generated positive results in patients. This review will cover recent developments in the application of SSOs to therapeutically relevant targets with an emphasis on disease models (Table 1).
Table 1.
Overview of Potential Therapeutic Targets for SSOs
| Disease | Gene | SSO modification | Study | Referencesa |
|---|---|---|---|---|
| Duchenne muscular dystrophy (DMD) | DMD | 2′OMePS | Local and systemic dystrophin induction in mdx mice | (Mann et al., 2001; Lu et al., 2003, 2005) |
| A Clinical trial in DMD patients by intramuscular injections | (van Deutekom et al., 2007) | |||
| PMO | Functional level of dystrophin induction in body-wide skeletal muscles in mdx mice after systemic delivery of PMO | (Alter et al., 2006) | ||
| A dose-escalating clinical trial in DMD patients by a single intramuscular injection. | Imperial College of London, UK (in progress) | |||
| PPMO | Effective dystrophin restoration in body-wide muscles including cardiac muscle of mdx mice | (Jearawiriyapaisarn et al., 2008) | ||
| β-Thalassemia | β-globin | PPMO | Restoration of HbA production in erythroid cells from peripheral blood of thalassemic patients by free uptake of PMO | (Suwanmanee et al., 2002a, 2002b) |
| Spinal muscular atrophy (SMA) | SMN2 | MOE-PS | Induction of exon 7 inclusion in human SMN2 transgenic mice | (Hua et al., 2008) |
| Inflammatory diseases | TNFR2 | LNA | Potent and persistent anti-TNF-α effects by induction of novel splice variant Δ7TNFR2 in mice | (Graziewicz et al., 2008) |
| MyD88 | MOE-PS | Anti-inflammatory effects by induction of MyD88S in mice | (Vickers et al., 2006) | |
| Cancer | HER2 | MOE-PS | Triggering of apoptosis by induction of novel splice variant Δ15HER2 triggered in breast cancer cells | (Wan et al., 2008) |
| Dystrophia myotonica type 1 | ClC-1 | PMO | Induction of exon 7a skipping in ClC-1 in HSALR and Mbn1ΔE3/ΔE3 mice | (Wheeler et al., 2007) |
| Menkes disease | ATP7A | PMO | Rescue of Menkes phenotype by correction of aberrant ATP7A splicing in a zebrafish disease model | (Madsen et al., 2008) |
| Ataxia telangiectasia | ATM | PMO | Production of functional ATM protein by correction of aberrant ATM splicing in cell culture | (Du et al., 2007) |
| Atherosclerosis | APOB | 2′OMePS | Induction of novel splice variant APOB87skip27 in cells in culture | (Khoo et al., 2007) |
| Propionic and methylmalonic acidemias | PCCA, PCCB, and MUT | PMO | Full recovery of PCC and MUT enzymatic activity in fibroblasts from patients | (Ugarte et al., 2007) |
Alternative pre-mRNA splicing
Alternative splicing is a biological process that regulates gene expression in higher eukaryotes by generating multiple protein isoforms with diverse functions from single pre-mRNA transcripts (Matlin et al., 2005). It therefore contributes significantly to the diversity of the proteome. Indeed, up to 70% of human genes are predicted to undergo alternative splicing (Mironov et al., 1999; Johnson et al., 2003). Importantly, up to 50% of human genetic diseases arise from mutations that affect splicing; this emphasizes the need for drugs that manipulate splicing (Cartegni et al., 2002; Faustino and Cooper, 2003; Pagani and Baralle, 2004; Pagenstecher et al., 2006).
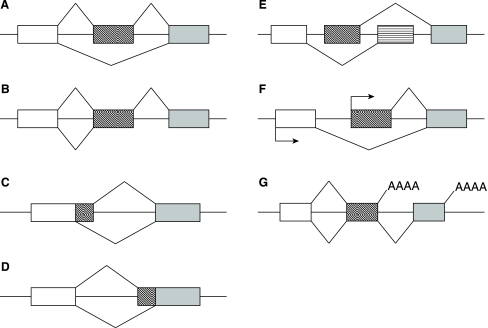
Pre-mRNA splicing is mediated by the spliceosome, a dynamic complex of proteins and RNAs, which is assembled de novo at every splicing event. Sequence specificity and simultaneous plasticity of alternative splicing is achieved by tissue dependent variability of spliceosome composition (Pollard et al., 2000, 2002) and loose conservation of essential sequence elements in pre-mRNA. These elements include splice sites, branch points, polypyrimidine tracts, and auxiliary exonic and intronic sequence elements known as splicing enhancers (ESEs and ISEs, respectively) and silencers (ESSs and ISSs, respectively). Splicing is thought to be controlled by competition among splice sites and splicing elements for splicing factors and the spliceosome. Based on splice site usage, a single pre-mRNA transcript can be alternatively spliced to produce multiple mature mRNA splice variants, which are then translated into variable protein isoforms. This is accomplished by various means, including exon exclusion, intron retention, exon shuffling, alternative 5′ and 3′ splice sites, and alternative promoter and alternative polyadenylation sites (Fig. 1) (Matlin et al., 2005). Alternative splicing can also downregulate gene expression through introduction of premature stop codons leading to nonsense-mediated decay of the transcript (Lewis et al., 2003). The dynamic nature and the gene and tissue dependent variability of the splicing machinery and its interaction with pre-mRNA suggest a richness of potential targets for drugs that interfere with redirect alternative splicing pathways for therapeutic purposes.
FIG. 1.
Patterns of alternative splicing. (A) Exon skipping. (B) Intron retention. (C) Alternative 5′ splice site. (D) Alternative 3′ splice site. (E) Mutually exclusive exons. (F) Alternative start sites. (G) Alternative polyadenylation sites.
Direction of splicing by SSOs
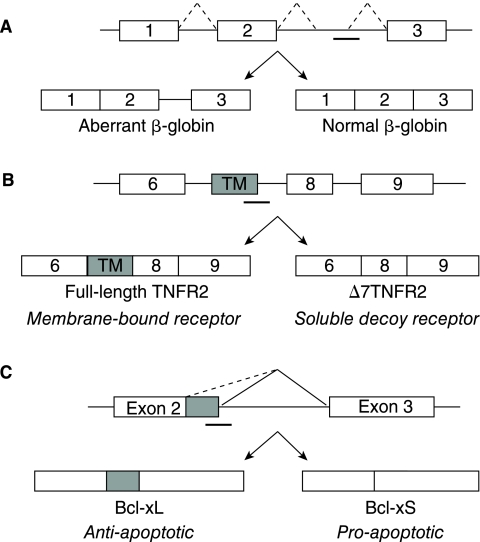
Splice-switching oligonucleotides direct pre-mRNA splicing by binding sequence elements and blocking access to the transcript by the spliceosome and other splicing factors. They can be applied to (1) restore correct splicing of an aberrantly spliced transcript, (2) produce a novel splice variant that is not normally expressed, or (3) manipulate alternative splicing from one splice variant to another (Fig. 2). Through the latter mechanism, SSOs therefore downregulate a deleterious transcript while simultaneously upregulating expression of a preferred transcript. Notably, their activity is enhanced with increased target gene expression because this enables increased production of the preferred splice variant (Mercatante et al., 2002). This is in contrast to traditional anti-sense approaches and small-interfering RNA, which exhibit decreased potency with increased target gene expression.
FIG. 2.
Applications of splice-switching oligonucleotides. (A) Correction of aberrant splicing of human β-globin leads to production of functional protein β-globin. (B) Production of a novel splice variant, Δ7TNFR2, which is a decoy receptor antagonist of TNF-α. (C) Manipulation of Bcl-x alternative splicing switches production from anti-apoptotic Bcl-xL to pro-apoptotic Bcl-xS.
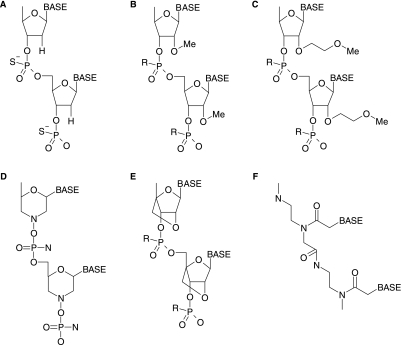
Direction of splicing by SSOs requires that the targeted pre-mRNA duplexed with these compounds is not degraded by RNase H. This requirement distinguishes the SSO mechanism from those of traditional antisense and RNAi, where degradation of mRNA and downregulation of gene expression is desirable. It is achieved primarily by altering the oligonucleotide sugar-phosphate backbones. Examples include phosphorodiamidate morpholinos (PMOs), peptide nucleic acids (PNA), locked nucleic acids (LNA), and the 2′O-methyl (2′OMe) and 2′-O-methoxyethyl (MOE) ribose modifications (Fig. 3) (KURRECK, 2003). In addition to RNase H resistance, these SSO modifications impart increased target affinity and unique and exploitable functional distribution profiles in vivo (Sazani et al., 2002). For example, LNA SSOs demonstrate potent and persistent activity confined almost exclusively to the liver, small intestine, and colon (Roberts et al., 2006) while PMO SSOs exhibit peptide-dependent functional biodistribution in a large number of tissues (Jearawiriyapaisarn et al., 2008). Splice-switching oligonucleotides may also incorporate phosphorothioate (PS) linkages, which do not confer RNase H resistance, but contribute to increased serum stability and bioavailability.
FIG. 3.
Synthetic oligonucleotide chemistries. (A) DNA phosphorothioate. (B) 2′-O-methyl (2′OMe) oligoribonucleotide. (C) 2′-O-methoxyethyl (MOE) oligoribonucleotide. (D) Phosphorodiamidate morpholino (PMO). (E) Locked nucleic acid (LNA). (F) Peptide nucleic acid (PNA). R denotes H or S (for phosphorothioate).
SSO direction of alternative splicing is not only a therapeutic approach but also provides a means to assay and improve cell culture and in vivo activity of the SSOs. Particularly useful reporter systems in which either luciferase or enhanced green fluorescent protein (EGFP) coding sequences are interrupted by an aberrantly spliced human β-globin intron have been developed in this laboratory (Kang et al., 1998; Sazani et al., 2001; Sazani et al., 2002). Functional luciferase or EGFP proteins are expressed only when SSO successfully enters the cell, accesses nuclear pre-mRNA and binds sequence elements in the intron, prompting correct splicing and exclusion of the intron from the mRNA transcript. The EGFP-654 transgenic mouse provides a functional, positive-readout platform for assessing SSO efficacy in vivo (Sazani et al., 2002; Roberts et al., 2006). These reporter systems have also been used to evaluate various enhancers of SSO delivery including cell-penetrating peptides (Jearawiriyapaisarn et al., 2008). Other groups employed the luciferase-based cell systems to evaluate receptor ligands (Alam et al., 2008), nanoparticle carriers (Liu and Franzen, 2008), and SSO uptake (Resina et al., 2007a, 2007b; Wu et al., 2007; Lebleu et al., 2008).
Clinical Trials in Duchenne Muscular Dystrophy
Duchenne muscular dystrophy (DMD) is a fatal genetic disease characterized by severe and progressive muscle wasting. It is caused by deletions and other mutations in the DMD gene, which cause premature termination of translation and result in lack of dystrophin protein. Deletions that maintain the reading frame in DMD gene produce internally deleted but partially functional dystrophin, causing the milder Becker muscular dystrophy (BMD) (Monaco et al., 1988). It has been reported that rare, naturally occurring dystrophin-positive (revertant) fibers are found in the muscle sections from mdx mouse, a mouse model of DMD (Hoffman et al., 1990), and in DMD patients (Nicholson et al., 1989). The dystrophin in these revertant fibers is internally deleted dystrophin generated by the removal of the mutated exon and restoration of translational reading frame of DMD transcripts through alternative splicing pathway of dystrophin pre-mRNA (Klein et al., 1992; Lu et al., 2000). A goal of DMD research has been to mimic this natural phenomenon to convert the DMD phenotype to the BMD phenotype by removing one or more exons to create shortened but in-frame dystrophin transcripts generating partially functional dystrophin. This approach was validated using SSOs that blocked pre-mRNA splicing elements, thereby excluding the mutation-containing exon and restoring the reading frame in DMD transcripts in cell-free extracts, (Takeshima et al., 1995), human lymphoblastoid cells (Pramono et al., 1996), and primary mdx myoblast cultures (Dunckley et al., 1998; Wilton et al., 1999).
The use of SSOs to mediate exon skipping in DMD has recently progressed from cell culture to animal models and clinical trials. The SSO chemistry first used in DMD studies was 2′OMe phosphorothioate (2′OMePS), which was tested in primary myoblasts derived from DMD patients carrying different deletions and a nonsense mutation (van Deutekom et al., 2001; Aartsma-Rus et al., 2003). In mdx mice, which carry a nonsense mutation in exon 23 of the DMD gene, dystrophin expression was restored by intramuscular (i.m.) injections of 2′OMePS SSO targeted to the 5′ splice site of intron 23 complexed with liposome (Mann et al., 2001), or the nonionic block copolymer F127 (Lu et al., 2003). In the latter case, SSO treatment led to functional improvement of the treated muscle. Since body-wide muscles are affected by DMD, it is important to induce dystrophin expression in all muscles (skeletal, smooth, and cardiac muscles). Intravenous (i.v.) injections of 2′OMePS complexed with F127 induced dystrophin expression in body-wide skeletal muscles, but not in cardiac muscle, in mdx mice with no detectable toxicity (Lu et al., 2005).
In addition to 2′OMePS, PMO has also been employed in mdx mice. The PMO annealed to a negatively charged DNA sense strand (“leash”) and complexed with liposome effectively induced exon skipping in mdx mice by local (Gebski et al., 2003; Fletcher et al., 2006) and systemic injections (Fletcher et al., 2006). A step forward was achieved in mdx mice i.v. injected with 2 mg uncomplexed PMO SSO at weekly intervals, which led to functional improvement of tibialis anterior muscles after three injections (Alter et al., 2006). After seven weekly injections, levels of dystrophin expression in gastrocnemius and quadriceps of treated mdx mice reached >50% of normal levels, and the level of serum creatinine kinase (CK) was dramatically reduced, suggesting that the integrity of muscle sarcolemma was improved by the SSO-induced dystrophin production. Administration of uncomplexed PMO, which is simple, would be ideal for clinical application. The potential of PMO in dystrophin induction has also been shown in canine X-linked muscular dystrophy (CXMD), a clinically severe canine model of DMD. Systemic infusions of PMO cocktails inducing skipping of exons 6 and 8 restored dystrophin expression in skeletal muscles and ameliorated exercise ability in CXMD canines (Yokota et al., 2008). Other chemistries such as LNA, ethylene bridged nucleic acid (ENA), PNA have been tested in cell culture and in vivo but they do not seem to be particularly useful in treatment of DMD (Aartsma-Rus et al., 2004b; Surono et al., 2004; Yin et al., 2008).
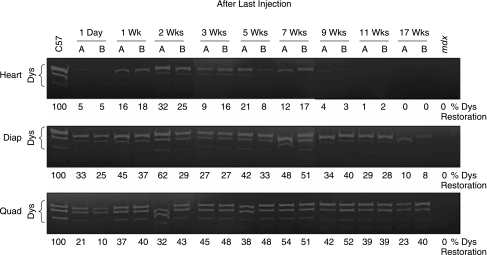
A major advance in oligomer-induced exon skipping in DMD has been application of cell-penetrating-peptide-conjugated PMOs (PPMOs). These compounds have shown enhanced efficacy in exon 23 removal in mdx mice compared to unconjugated PMO. A PPMO carrying (RXR)4XB pep-tide, (where R is arginine, X is 6-aminohexanoic acid, and B is β-alanine), induced normal levels of dystrophin expression in diaphragm and low levels in colon, gut, and skeletal muscles of mdx mice treated as neonates with four weekly intraperitoneal (i.p.) injections at 5 mg/kg, a significantly lower dose than that used for unconjugated PMO (Fletcher et al., 2007; Moulton et al., 2007). However, none of the above studies in animal models showed restoration of dystrophin expression in cardiac muscle. Intracardiac injections of PMO in mdx mice induced only very low level of dystrophin expression (Vitiello et al., 2008). Because cardiomyopathy is one of the major causes of death in DMD patients, induction of dystrophin expression in cardiac muscle is critical for DMD treatment. This laboratory recently demonstrated for the first time that systemic treatment of mdx mice with PPMO-B carrying (RXRRBR)2XB peptide induced in cardiac muscles high levels of exon 23–skipped mRNA and produced 20%–30% of normal dystrophin protein. This treatment resulted in reduced inflammatory cell infiltration in the heart (Jearawiriyapaisarn et al., 2008). Exon 23–skipped mRNA and restored production of dystrophin protein remained detectable for at least 2–3 months after treatment, especially in diaphragm and quadriceps, where 100% exon skipping was maintained. This suggested that the PPMO-B/PMO is very stable in muscle tissues, because the half-life of dystrophin mRNA is only ~16 hours (Tennyson et al., 1996). The sustained dystrophin expression, not only in the heart but also in body-wide muscles (Fig. 4), led to a decrease of serum CK to near wild-type levels, confirming that the rescued dystrophin protein is functional. Sustained dystrophin expression induced by PPMO-B SSO would be desirable for clinical application due to the potential for infrequent readministration. Therefore, splice-switching with PPMO-B holds great potential for the treatment of DMD.
FIG. 4.
Sustained dystrophin expression in the heart, diaphragm, and quadriceps of mdx mice following 4 daily i.v. injections with PPMO-B targeted to 5′ splice site of mouse dystrophin exon 23 at 12 mg/kg/day. Dystrophin from total protein at indicated time points was detected by In-gel immunodetection with mouse monoclonal antibody against C-terminus of dystrophin. (Reprinted from Jearawiriyapaisarn et al., Molecular Therapy, 2008.)
Pre-clinical study has been extended to test sequence specificity and dosing regimens of SSOs in transgenic hDMD mice, which carry full-length human DMD gene (t Hoen et al., 2008). Intramuscular injection of 2′OMePS SSOs excluded targeted human exons without affecting the endogenous mouse dystrophin transcripts, suggesting that high sequence specificity was obtained (Bremmer-Bout et al., 2004).
The pre-clinical results described above have led to three clinical trials for oligonucleotide-induced exon-skipping in DMD patients. First, a PS oligodeoxynucleotide was evaluated in a 10-year-old DMD boy, who carries an out-of-frame DMD transcripts caused by the deletion of exon 20 (Takeshima et al., 2006). This compound, targeted to a splicing enhancer sequence in exon 19, was reported to exclude exon 19, generating shortened, in-frame transcripts. Low level of transcripts lacking exons 19–20 and positive dystrophin muscle fibers were identified in a muscle biopsy 1 week after four weekly i.v. infusions at 0.5 mg/kg. This observation was surprising since oligodeoxynucleotides induce RNase H mediated RNA degradation and are not capable of splicing modulation. Perhaps this was one of the reasons why this treatment did not improve muscle function. In a more robust, well-designed clinical trial, a 2′OMePS SSO, PRO051, was targeted to an internal sequence of exon 51 to induce exon 51 skipping (van Deutekom et al., 2007). This study demonstrated that a single local i.m. injection of 0.8 mg PRO051 specifically excluded exon 51 in four patients with DMD, restoring the translational reading frame. This led to production of truncated dystrophin at 3%–12% normal levels in total protein lysates extracted from muscle biopsies taken 4 weeks after injection. Functional improvement of injected muscles was not observed. Finally, a dose-escalating clinical trial of a PMO SSO, AVI-4658, designed to skip exon 51, is underway. This study is intended to establish the efficacy and safety of a single i.m. injection of AVI-4658 in nine boys with DMD. Systemic administration of AVI-4658 is also being developed in pre-clinical studies for future phase II/III clinical trials (http://clinicaltrials.gov).
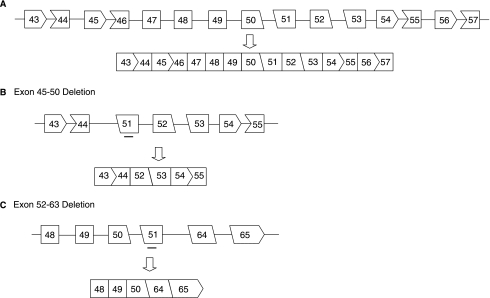
Skipping of exon 51 could rescue DMD patients who suffer from exon 45–50, 47–50, 48–50, 49–50, 50, 52, 52–63 deletions (Fig. 5) and a recently reported inversion of exon 49–50 (Madden et al., 2008), which affect an estimated 16% of DMD patients. The limitation of the exon skipping strategy in DMD is that skipping one exon could restore the translational reading frame only for particular deletions. Optimization of SSOs for different deletions, or personalized molecular medicine, may be necessary (HOFFMAN, 2007); however, the current regulatory hurdles required for each individual drug make this approach prohibitively expensive for commercial development. To overcome this limitation, multiple-exon skipping to increase the number of deletion cases that might benefit from this strategy was explored (Aartsma-Rus et al., 2004a, 2006). It is predicted that multiple-exon skipping from exons 45 through 55 could be applied for up to 63% of DMD patients (Beroud et al., 2007).
FIG. 5.
Schematic representation of reading frame restoration by SSO-targeted exon 51 in different deletion cases. (A) Molecular mechanism of splicing generating in-frame transcript between exon 43–57 of DMD gene. (B and C) Restoration of reading frame of the dystrophin mRNA by SSO-induced exon 51 skipping in patients carrying, for example, exon 45–50 (B) and exon 52–63 deletions (C). Deletions between exon 45 and 50 or 52 and 63 result in a shift of the reading frame abrogating functional dystrophin production. Exclusion of exon 51 maintains the triplet codon, generating an in-frame transcript. Rectangular boxes represent exons ending with full codon triplets. Other shapes represent exons in which the intron separates terminal codon triplet in two-one or one-two nucleotides. Lines represent introns, thick bars represent SSOs.
Still unanswered questions, regarding SSOs bioavailability, therapeutic index, and long-term effects, are being addressed in ongoing pre-clinical and clinical studies. As a result it is likely that among therapeutic applications of SSOs, exon skipping for DMD will be the first to be useful in the clinic.
Advances in Genetic and Other Diseases
β-Globin
β-Thalassemia is a genetic blood disorder characterized by a deficiency of β-globin chains, leading to the reduction or absence of adult hemoglobin A (HbA). Most β-globin gene mutations alter pre-mRNA splicing and are found in intron 1 (IVS1-5, IVS1-6, IVS1-110) and intron 2 (IVS2-654, IVS2-705, IVS2-745). The three intron 2 mutations, IVS2-654 (C>T), IVS2-705 (T>G), and IVS2-745 (C>G), create an aberrant 5′ splice sites and activate a common cryptic 3′ splice site at position 579 in the intron 2. Use of these aberrant splice sites generates an mRNA transcript that includes an intronic sequence encoding a stop codon, resulting in translation of truncated, nonfunctional β-globin protein. These mutations have been most extensively studied and targeted with SSOs in cell-free extracts and model cell lines stably expressing βIVS2 mutants (Sierakowska et al., 1996; Schmajuk et al., 1999). In erythroid progenitor cells isolated from peripheral blood of patients carrying IVS2-654 or −745 mutations, a single treatment with PMOs targeted to aberrant 5′ splice sites by syringe loading resulted in correction of β-globin pre-mRNA splicing and production of HbA (Lacerra et al., 2000). Moreover, the PMOs were freely taken up by erythroid progenitor cells isolated from bone marrow of β-thalassemic mice carrying the human βIVS2-654 mutation (Lewis et al., 1998) and from peripheral blood of patients carrying IVS2–654 mutation, leading to effective splicing correction (Suwanmanee et al., 2002b). Significant HbA upregulation was achieved in the treated human thalassemic erythroid precursors, target cells for thalassemia treatment. Free uptake of PMOs was also shown to be effective in splicing correction and HbA restoration in progenitor cells isolated from patients carrying HbE mutation, which activates a cryptic 5′ splice site in exon 1 (Suwanmanee et al., 2002a). These results suggested that PMO would be applicable in vivo, in mouse models of β-thalassemia.
SMN2
Spinal muscular atrophy (SMA) is characterized by the progressive degeneration of the motor neurons leading to a generalized muscle weakness. Spinal muscular atrophy is caused by homozygous mutations or deletions in the survival of motor neuron 1 (SMN1) gene, encoding the ubiquitously expressed SMN protein which is necessary for assembly of snRNPs involved in splicing, an essential process for cell survival (Wan et al., 2005). In humans there is a second, SMN gene copy, SMN2, which differs from SMN1 in the C-to-T transition at position 6 of SMN2 exon 7. It has been proposed that this transition disrupts an ESE sequence (Cartegni and Krainer, 2002), creates an ESS sequence (Kashima and Manley, 2003), or strengthens an inhibitory RNA stem loop at the 5′ end of exon 7 (Singh et al., 2004), leading to exon 7 exclusion in most of SMN2 transcripts that yields an unstable truncated SMN protein. SMN2 is a modifying gene in SMA patients, as the number of copies of SMN2 is inversely correlated to the severity of SMA (Mailman et al., 2002). Therefore, a strategy to include exon 7 and upregulate expression of full-length, fully functional SMN protein from the intact copies of SMN2 present in SMA patients can offer a therapeutic approach for SMA. Indeed, masking the 3′ splice site of SMN2 intron 7 using a 2′OMePS SSO impaired splice site recognition and forced the splicing machinery to re-couple the 5′ and 3′ splice sites in intron 6, leading to increased inclusion of exon 7 in cell culture (Lim and Hertel, 2001).
Targeting exon 7 by SSOs either linked to serine/arginine-rich splicing factors or including a “tail” that serves as a splicing factor–binding site, called exon specific–splicing enhancement by small chimeric effectors (ESSENCE) (Cartegni and Krainer, 2003) and targeted oligonucleotide enhancer of splicing (TOES) (Skordis et al., 2003), respectively, have also been shown to promote exon 7 inclusion. A synthetic ESSENCE molecule containing a PNA SSO targeted to exon 7 coupled to 10 arginine-serine (RS) repeats significantly induced exon 7 inclusion in a cell free–splicing system. Although the PNA SSO lacking the RS repeats was active, efficacy was much lower than the ESSENCE molecule. The TOES was designed to have two components; the first was complementary to exon 7 and second was a noncomplementary sequence that was designed to mimic an ESE to recruit _trans_-splicing factors. This bifunctional SSO increased SMN2 exon 7 inclusion in vitro and in SMA patient fibroblasts, which led to partial restoration of gem number, an indicator of SMN protein increase, suggesting that SSO bound to exon 7 in the transcript did not interfere with translation.
In a subsequent study, SMN2 exon 7 was saturated with overlapping MOE SSOs (Hua et al., 2007). The cell culture experiments revealed that the most effective SSOs, which promoted exon 7 inclusion and increased full-length SMN protein in SMA patient fibroblasts, were targeted to two putative ESSs close to the 3′ and 5′ splice sites, respectively. These results imply that blocking of the ESS is sufficient to induce exon inclusion and that ESSENCE or TOES modifications, although most likley helpful, are not essential.
SSO targeted to a putative ISS in intron 6 identified also prompted an increase in exon 7 inclusion in the context of a SMN2 minigene (Miyajima et al., 2002). The ISS, called ISS-N1, was subsequently identified immediately downstream of the 5′ splice site of intron 7 (Singh et al., 2006). An extensive SSO screen of sequences downstream of the 5′ splice site in intron 7 identified the same ISS (Hua et al., 2008). Recently, MOE-PS SSO targeted to ISS-N1 was administered to human SMN2 (hSMN2) transgenic mice by biweekly i.v. injections at 25 mg/kg. Significant exon 7 inclusion was observed in the liver and kidney, but not in the spinal cord of treated mice (Hua et al., 2008). In order to achieve the therapeutic effect of this strategy in SMA, the SSO would need to cross the blood–brain barrier or be delivered directly to the central nervous system (CNS); this remains a major obstacle in gene therapy for CNS targets.
TNFR2
The cytokine tumor necrosis factor-α (TNF-α) plays an important role in inflammatory diseases such as rheumatoid arthritis and hepatitis. TNF-α signaling is mediated by two membrane-bound receptors, TNFR1 and TNFR2, which trimerize upon ligand binding, leading to downstream activation of the transcription factor nuclear factor (NF)-κB. Approved anti-TNF-α macromolecular biologic drugs etanercept (a dimerized TNFR2 receptor:Fc fusion protein), and infliximab and adalimumab (anti-TNF-α monoclonal antibodies), bind TNF-α in circulation and block its inflammatory effects.
Researchers in this laboratory hypothesized that exclusion of the transmembrane domain-encoding exon 7 would create a novel soluble protein, Δ7TNFR2, capable of antagonizing TNF-α signaling (Fig. 2B). Screen of 16-mer SSOs consisting of alternating LNA-DNA nucleotides with PS linkages in L929 mouse cells identified SSO3274, which efficiently blocked the 5′ splice site of exon 7 in TNFR2 pre-mRNA, inducing exon skipping in a sequence-specific and dose-dependent manner. SSO3274-induced exon skipping resulted in the expression of soluble Δ7TNFR2 protein, which was detected in the culture medium and which exhibited anti-TNF activity (Graziewicz et al., 2008).
Locked nucleic acid SSOs are exceptionally potent in the liver, where TNFR2 is highly expressed (Roberts et al., 2006). This led to an approach to utilize the liver as a “protein factory,” producing soluble Δ7TNFR2 for release into circulation to antagonize TNF-α. As expected, SSO3274 induced sequence-specific, dose-dependent, persistent splice switching in the mouse liver after 5 daily i.p. injectons at 25mg/kg/day. Δ7TNFR2 protein was detected in the serum of SSO3274-treated animals at 8,000–10,000 pg/mL, exhibited potent anti-TNF activity and was detectable in the circulation up to 35 days after the last injection. Interestingly, SSO-induced Δ7TNFR2 from mouse serum was 10-fold more potent than that from mice treated with etanercept. Evidently etanercept was subject to in vivo degradation that reduced its activity, while SSO3274 continually induced additional Δ7TNFR2 protein, replenishing degraded or inactivated protein in circulation. Importantly, no SSO-induced toxicity was reported. Because SSOs induce the expression of endogenous protein, the likelihood of immune toxicity is very low. The ability of SSOs to produce therapeutic splice variants, such as Δ7TNFR2, for extended time periods and with a low likelihood of immune toxicity is a distinct advantage over biological drugs such as etanercept.
In a mouse model of inflammatory liver disease, SSO3274 injected i.p. for 10 days at 25 mg/kg/day protected the liver from TNF-α insult and prevented liver damage. The observed anti-inflammatory effects were likely enhanced by the ability of SSO3274 to simultaneously downregulate TNFR2 while inducing Δ7TNFR2 in the liver. Importantly, etanercept administered at a dose comparable to the amount of Δ7TNFR2 induced by SSO3274 elicited no such protection. The 10-day dosing regimen also delayed the course of disease in a mouse model of collagen-induced rheumatoid arthritis. These results are notable given that 30% of rheumatoid arthritis patients do not respond to currently available drugs treatments (Pincus et al., 1999; Olsen and Stein, 2004).
MyD88
MyD88 is an adapter protein that mediates pro-inflammatory cytokine signaling through IL-1 and Toll receptors. Upon receptor activation, MyD88 interacts with the receptor intracellular domain and recruits IL-1R-associated kinase-1 (IRAK-1) and IRAK-4, leading to the phosphorylation of IRAK-1 and the subsequent activation of the transcription factor NF-κB. A naturally occurring splice variant of MyD88, MyD88S, results from exon 2 exclusion. MyD88S retains its receptor-binding function but is defective in its ability to recruit IRAK-4 and induce IRAK-1 activation. As a result, MyD88S functions as a dominant-negative regulator of IL-1R and Toll receptor signaling (Janssens et al., 2002). Vickers et al. screened 20-mer MOE-PS SSOs targeted to the 5′ and 3′ splice sites of exon 2 and selected ISIS 337846, an SSO that switched splicing from MyD88 to MyD88S in mouse and human cells. ISIS 337846 induced redirection of MyD88 mRNA splicing in the liver, adipose tissue, and intestine of mice i.p. injected three times per week for 3 weeks with 50 mg/kg SSO, although no MyD88S protein was detected. This regimen abrogated pro-inflammatory IL-1β signaling in the liver (Vickers et al., 2006).
ClC-1
Dystrophia myotonica type 1 (DM1), a common muscular dystrophy among adults, is caused by amplified CTG repeats in the 3′ untranslated region of the dystrophia myotonia protein kinase (DMPK) gene (Brook et al., 1992). DMPK transcripts containing amplified CUG repeats (CUGamp) cause abnormal regulation of alternative splicing, which selectively affects a specific group of genes that includes chloride channel 1 (ClC-1) (Mankodi et al., 2002). Interestingly, misregulated splicing of ClC-1, causing inclusion of exon 7a, was found to be necessary for the development of myotonia in two mouse models of DM1 (HSALR and Mbnl1ΔE3/ΔE3 mice) (Wheeler et al., 2007). Inclusion of exon 7a in ClC-1 caused the introduction of a premature stop codon and translation of a nonfunctional truncated protein. Wheeler et al. screened 25-mer PMO SSOs targeting the 5′ and 3′ splice sites of exon 7a to restore normal ClC-1 splicing. The SSOs were simultaneously injected (5 μg each) into the tibialis anterior muscle of HSALR mice, accompanied by voltage pulses to electroporate muscle fibers and improve uptake. A single injection of the SSOs induced sequence-specific exon 7a skipping that persisted for at least 3 weeks. Splice-switching oligonucleotide–induced splice switching of ClC-1 increased the level of wild-type ClC-1 protein, rescued ClC-1 channel function, and reversed myotonia in skeletal muscle. Similar results were observed in Mbnl1ΔE3/ΔE3 mice (Wheeler et al., 2007). This SSO-based strategy is noteworthy in that it affects the proposed underlying mechanism of the disease, whereas currently available drugs for DM1 provide only partial relief of the symptoms (Trip et al., 2006).
ATP7A
Menkes disease is an X-linked recessive disorder caused by generalized copper deficiency characterized by growth retardation and neurodegeneration, leading to death in early infancy (Menkes et al., 1962). It is caused by loss-of-function mutations in the ATP7A gene encoding a P-type ATPase necessary for copper absorption and homeostasis (Lutsenko et al., 2007). Splicing defects in the zebrafish ortholog of ATP7A produce calamity, a zebrafish model of the Menkes phenotype (Mendelsohn et al., 2006). The defect in calamityvu69 is caused by the creation of an aberrant 3′ splice site in exon 9 of ATP7A. By screening 25-mer PMO SSOs targeted to the aberrant splice site, Madsen et al. identified two SSOs that restore normal splicing and wild-type protein production in calamityvu69 embryos (Madsen et al., 2008). Importantly, a single injection of PMO SSO achieved full rescue of the mutant phenotype that persisted for at least 6 days after fertilization, when 75% of untreated calamity embryos were dead or dysmorphic (Madsen et al., 2008).
Newly Identified SSO Targets
HER2
HER2 belongs to the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases and is amplified or overexpressed in many human cancers, including 25%–30% of breast cancers. Increased HER2 expression is associated with increased tumor aggressiveness and decreased survival. Although it has no known ligand, HER2 forms constitutively active homodimers in HER2-overexpressing cells, signaling through the mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways. In addition, on ligand-induced activation, other EGFR family members preferentially recruit HER2 and form activated heterodimers. Herstatin, a naturally occurring splice variant of HER2, is produced by intron 8 inclusion and functions as a negative regulator of HER2 signaling (Doherty et al., 1999).
To develop a Herstatin-like antagonist of HER2 this laboratory screened a series of 20-mer MOE-PS SSOs designed to modulate splicing of HER2 pre-mRNA. SSO111 induced exon 15 skipping in a sequence-specific, dose-dependent manner, thereby downregulating full-length HER2 while producing a novel splice variant lacking the transmembrane domain, Δ15HER2. In SK-BR-3 human breast cancer cells, which highly express HER2, SSO111-induced splice switching potently inhibited cell growth and induced apoptosis. This effect was less pronounced in MCF7 cells, in which HER2 expression is 100-fold lower, despite the fact that splice switching was still observed, confirming that SSO111-induced growth inhibition was HER2-dependent. This was consistent with the previously reported correlation of SSO potency with increased target gene expression (Mercatante et al., 2002). It also indicates that tumors highly expressing HER2 would be more sensitive to SSO treatment than surrounding healthy tissue. While Δ15HER2 protein encoded by the SSO-induced Δ15HER2 mRNA could not be detected, it was demonstrated that Δ15HER2-His protein potently downregulated HER2 protein expression and inhibited HER3 activation in a dose-dependent manner when added exogenously to SK-BR-3 cells (Wan et al., 2008).
ATM
Ataxia-telangiectasia (A-T) is an autosomal recessive disorder characterized by neurodegeneration, immune defects and predisposition to malignancy. Ataxia-telangiectasia is caused by mutations in the A-T mutated (ATM) gene, half of which disrupt splicing (Teraoka et al., 1999). Recently, 25-mer PMO SSOs were used to block the activation of cryptic splice sites in cell lines representing three types of A-T splicing defect: a 5′ exonic cryptic splice site in TAT[C] cells, a 3′ exonic cryptic splice site in IRAT9 cells, and a pseudoexon inclusion in AT203LA cells (Du et al., 2007). In each case dose-dependent, sequence-specific splice correction was achieved, leading to upregulation of functional ATM protein. These findings are especially promising in light of evidence that only a small amount of functional protein (5%–20% wild-type levels) is necessary to significantly ameliorate the disease phenotype (Gilad et al., 1998).
Apolipoprotein B
Apolipoprotein B (APOB) is the primary apolipoprotein in the low-density lipoprotein (LDL), very low–density lipoprotein (VLDL), intermediate-density lipoprotein (IDL), lipoprotein (a) [Lp(a)], and chylomicron lipoprotein particles. Apolipoprotein B is expressed as two protein isoforms, APOB100 and APOB48. APOB100 results from the inclusion of all 29 exons from APOB pre-mRNA and necessary for synthesis of LDL and Lp(a) particles in the liver, which contribute to atherogenesis. Thus, targeting APOB100 for down-regulation is a strategy for lowering LDL and cholesterol levels in the treatment and prevention of cardiovascular disease (Soutschek et al., 2004; Zimmermann et al., 2006). APOB48 is a C-terminally truncated protein that results from tissue-specific RNA editing in exon 26 that introduces a premature stop codon. Expressed only in the intestine, APOB48 is necessary for chylomicron assembly and intestinal fat transport.
In order to downregulate APOB100 without affecting APOB48 expression, Khoo et al. designed a 2′OMe SSO aimed at skipping exon 27 to create a novel splice variant, APOB87skip27, that could mimic APOB48 function. Interestingly, SSOs targeted to either the 5′ or 3′ splice site of exon 27, or the branch point of intron 26 were ineffective at inducing exon 27 skipping. However, SSO targeting two of these sequences simultaneously did achieve exon 27 skipping in a dose-dependent, sequence-specific manner, as did SSOs targeted to putative exonic splicing enhancer motifs within exon 27, albeit to a lesser degree (Khoo et al., 2007). The authors further showed that SSO-induced skipping of exon 27 resulted in translation of the APOB87skip27 isoform and that this protein was secreted by cells in culture (Khoo et al., 2007).
PCCA, PCCB, MUT
Propionic and methylmalonic acidemias, the two most frequent organic acidemias, are characterized by the lack of propionyl coenzyme A carboxylase (PCC) and methylmalonylCoA mutase (MCM), respectively, which are the enzymes involved in the catabolism of amino acids, odd-chain fatty acids, and cholesterol. The PCC-α subunit (PCCA) and PCC-β subunit (PCCB) genes encode the two subunits of PCC enzyme, and the MUT gene encodes MCM enzyme. An IVS14-1416 (A>G) mutation in PCCA intron 14 creates novel ESE sequences favoring cryptic 3′ and 5′ splice sites of a pseudoexon. The IVS6-462 (A>G) mutation in PCCB in-tron 6 and the IVS11-891 (C>A) mutation in MUT intron 11 increase the cryptic 5′ splice site score of the pseudoexon. These three mutations in the intron promote the inclusion of the pseudoexon in the mRNAs, resulting in a frameshift mutation. This generates transcripts containing a premature termination codon, which leads to the production of truncated, nonfunctional protein.
Blocking either the cryptic 3′ or 5′ splice sites by PMO complexed with a peptide carrier (Endo-Porter; Gene Tools) restored almost 100% correct splicing in fibroblasts from patients carrying mutations in PCCA, PCCB, or MUT (Ugarte et al., 2007). The correctly spliced MUT mRNA persisted for 15 days after treatment, confirming that the PMO was very stable in the cells. The enzymatic activities of PCC and MCM were fully recovered after treatment. This result is notable given that only 30%–40% of normal enzymatic activity is necessary for therapeutic effect. Since there are several oligonucleotide modifications that are efficiently delivered to liver, which is the target tissue for these diseases, there are multiple potential avenues for therapeutic application in vivo.
Conclusion
The number of clinically relevant targets of SSOs has expanded since this technology was pioneered by our laboratory (Dominski and Kole, 1993). Among these targets, DMD, which is currently being tested in clinical trials, is the most promising. Recent results from DMD patients in work by van Deutekom et al. suggest that SSOs constitute a realistic approach for therapeutic application. The advantage of this approach is that SSOs target the pre-mRNA transcribed from native genes in their normal regulatory environment, thereby avoiding the problem of inappropriate transgene expression in conventional gene therapy. While delivery remains a major obstacle, cell-penetrating peptides have been shown to significantly enhance SSO delivery without toxicity (Jearawiriyapaisarn et al., 2008). Because up to 50% of human disease-causing mutations affect splicing, the SSO approach is emerging as a promising alternative to gene therapy.
Acknowledgments
J.B. was partially supported by the American Heart Association Pre-doctoral Fellowship. N.J. was partially supported by the Royal Golden Jubilee (RGJ) scholarship from the Thailand Research Fund.
References
- AARTSMA-RUS A. JANSON A.A. KAMAN W.E. BREMMER-BOUT M. DEN DUNNEN J.T. BAAS F. VAN OMMEN G.J. VAN DEUTEKOM J.C. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- AARTSMA-RUS A. JANSON A.A. KAMAN W.E. BREMMER-BOUT M. VAN OMMEN G.J. DEN DUNNEN J.T. VAN DEUTEKOM J.C. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am. J. Hum. Genet. 2004a;74:83–92. doi: 10.1086/381039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AARTSMA-RUS A. KAMAN W.E. BREMMER-BOUT M. JANSON A.A. DEN DUNNEN J.T. VAN OMMEN G.J. VAN DEUTEKOM J.C. Comparative analysis of antisense oligonucleotide analogs for targeted DMD exon 46 skipping in muscle cells. Gene Ther. 2004b;11:1391–1398. doi: 10.1038/sj.gt.3302313. [DOI] [PubMed] [Google Scholar]
- AARTSMA-RUS A. KAMAN W.E. WEIJ R. DEN DUNNEN J.T. VAN OMMEN G.J. VAN DEUTEKOM J.C. Exploring the frontiers of therapeutic exon skipping for Duchenne muscular dystrophy by double targeting within one or multiple exons. Mol. Ther. 2006;14:401–407. doi: 10.1016/j.ymthe.2006.02.022. [DOI] [PubMed] [Google Scholar]
- ALAM M.R. DIXIT V. KANG H. LI Z.B. CHEN X. TREJO J. FISHER M. JULIANO R.L. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36:2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALTER J. LOU F. RABINOWITZ A. YIN H. ROSENFELD J. WILTON S.D. PARTRIDGE T.A. LU Q.L. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- BEROUD C. TUFFERY-GIRAUD S. MATSUO M. HAMROUN D. HUMBERTCLAUDE V. MONNIER N. MOIZARD M.P. VOELCKEL M.A. CALEMARD L.M. BOISSEAU P. BLAYAU M. PHILIPPE C. COSSEE M. PAGES M. RIVIER F. DANOS O. GARCIA L. CLAUSTRES M. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum. Mutat. 2007;28:196–202. doi: 10.1002/humu.20428. [DOI] [PubMed] [Google Scholar]
- BREMMER-BOUT M. AARTSMA-RUS A. DE MEIJER E.J. KAMAN W.E. JANSON A.A. VOSSEN R.H. VAN OMMEN G.J. DEN DUNNEN J.T. VAN DEUTEKOM J.C. Targeted exon skipping in transgenic hDMD mice: a model for direct preclinical screening of human-specific antisense oligonucleotides. Mol. Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- BROOK J.D. MCCURRACH M.E. HARLEY H.G. BUCKLER A.J. CHURCH D. ABURATANI H. HUNTER K. STANTON V.P. THIRION J.P. HUDSON T. ET AL. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- CARTEGNI L. CHEW S.L. KRAINER A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- CARTEGNI L. KRAINER A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- CARTEGNI L. KRAINER A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- DOHERTY J.K. BOND C. JARDIM A. ADELMAN J.P. CLINTON G.M. The HER-2/neu receptor tyrosine kinase gene encodes a secreted autoinhibitor. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10869–10874. doi: 10.1073/pnas.96.19.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMINSKI Z. KOLE R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DU L. POLLARD J.M. GATTI R.A. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6007–6012. doi: 10.1073/pnas.0608616104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUNCKLEY M.G. MANOHARAN M. VILLIET P. EPERON I.C. DICKSON G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribo-nucleotides. Hum. Mol. Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- FAUSTINO N.A. COOPER T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- FLETCHER S. HONEYMAN K. FALL A.M. HARDING P.L. JOHNSEN R.D. STEINHAUS J.P. MOULTON H.M. IVERSEN P.L. WILTON S.D. Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Mol. Ther. 2007;15:1587–1592. doi: 10.1038/sj.mt.6300245. [DOI] [PubMed] [Google Scholar]
- FLETCHER S. HONEYMAN K. FALL A.M. HARDING P.L. JOHNSEN R.D. WILTON S.D. Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J. Gene Med. 2006;8:207–216. doi: 10.1002/jgm.838. [DOI] [PubMed] [Google Scholar]
- GEBSKI B.L. MANN C.J. FLETCHER S. WILTON S.D. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum. Mol. Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- GILAD S. CHESSA L. KHOSRAVI R. RUSSELL P. GALANTY Y. PIANE M. GATTI R.A. JORGENSEN T.J. SHILOH Y. BAR-SHIRA A. Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am. J. Hum. Genet. 1998;62:551–561. doi: 10.1086/301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRAZIEWICZ M.A. TARRANT T.K. BUCKLEY B. ROBERTS J. FULTON L. HANSEN H. ORUM H. KOLE R. SAZANI P. An endogenous TNF-alpha antagonist induced by splice-switching oligonucleotides reduces inflammation in hepatitis and arthritis mouse models. Mol. Ther. 2008;16:1316–1322. doi: 10.1038/mt.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFFMAN E.P. Skipping toward personalized molecular medicine. N. Engl. J. Med. 2007;357:2719–2722. doi: 10.1056/NEJMe0707795. [DOI] [PubMed] [Google Scholar]
- HOFFMAN E.P. MORGAN J.E. WATKINS S.C. PARTRIDGE T.A. Somatic reversion/suppression of the mouse mdx phenotype in vivo. J. Neurol. Sci. 1990;99:9–25. doi: 10.1016/0022-510x(90)90195-s. [DOI] [PubMed] [Google Scholar]
- HUA Y. VICKERS T.A. BAKER B.F. BENNETT C.F. KRAINER A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUA Y. VICKERS T.A. OKUNOLA H.L. BENNETT C.F. KRAINER A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANSSENS S. BURNS K. TSCHOPP J. BEYAERT R. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr. Biol. 2002;12:467–471. doi: 10.1016/s0960-9822(02)00712-1. [DOI] [PubMed] [Google Scholar]
- JEARAWIRIYAPAISARN N. MOULTON H.M. BUCKLEY B. ROBERTS J. SAZANI P. FUCHAROEN S. IVERSEN P.L. KOLE R. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol. Ther. 2008;16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON J.M. CASTLE J. GARRETT-ENGELE P. KAN Z. LOERCH P.M. ARMOUR C.D. SANTOS R. SCHADT E.E. STOUGHTON R. SHOEMAKER D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- KANG S.H. CHO M.J. KOLE R. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry. 1998;37:6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- KASHIMA T. MANLEY J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- KHOO B. ROCA X. CHEW S.L. KRAINER A.R. Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB. BMC Mol. Biol. 2007;8:3. doi: 10.1186/1471-2199-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN C.J. COOVERT D.D. BULMAN D.E. RAY P.N. MENDELL J.R. BURGHES A.H. Somatic reversion/suppression in Duchenne muscular dystrophy (DMD): evidence supporting a frame-restoring mechanism in rare dystrophin-positive fibers. Am. J. Hum. Genet. 1992;50:950–959. [PMC free article] [PubMed] [Google Scholar]
- KOLE R. Modification of pre-mRNA splicing by antisense oligonucleotides. Acta Biochim. Pol. 1997;44:231–237. [PubMed] [Google Scholar]
- KOLE R. VACEK M. WILLIAMS T. Modification of alternative splicing by antisense therapeutics. Oligonucleotides. 2004;14:65–74. doi: 10.1089/154545704322988067. [DOI] [PubMed] [Google Scholar]
- KURRECK J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- LACERRA G. SIERAKOWSKA H. CARESTIA C. FUCHAROEN S. SUMMERTON J. WELLER D. KOLE R. Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9591–9596. doi: 10.1073/pnas.97.17.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEBLEU B. MOULTON H.M. ABES R. IVANOVA G.D. ABES S. STEIN D.A. IVERSEN P.L. ARZUMANOV A.A. GAIT M.J. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv. Drug Deliv. Rev. 2008;60:517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS B.P. GREEN R.E. BRENNER S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. U.S.A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS J. YANG B. KIM R. SIERAKOWSKA H. KOLE R. SMITHIES O. MAEDA N. A common human beta globin splicing mutation modeled in mice. Blood. 1998;91:2152–2156. [PubMed] [Google Scholar]
- LIM S.R. HERTEL K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- LIU Y. FRANZEN S. Factors determining the efficacy of nuclear delivery of antisense oligonucleotides by gold nano-particles. Bioconjug. Chem. 2008;19:1009–1016. doi: 10.1021/bc700421u. [DOI] [PubMed] [Google Scholar]
- LU Q.L. MANN C.J. LOU F. BOU-GHARIOS G. MORRIS G.E. XUE S.A. FLETCHER S. PARTRIDGE T.A. WILTON S.D. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat. Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- LU Q.L. MORRIS G.E. WILTON S.D. LY T. ARTEM′YEVA O.V. STRONG P. PARTRIDGE T.A. Massive idiosyncratic exon skipping corrects the nonsense mutation in dystrophic mouse muscle and produces functional revertant fibers by clonal expansion. J. Cell. Biol. 2000;148:985–996. doi: 10.1083/jcb.148.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU Q.L. RABINOWITZ A. CHEN Y.C. YOKOTA T. YIN H. ALTER J. JADOON A. BOU-GHARIOS G. PARTRIDGE T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUTSENKO S. BARNES N.L. BARTEE M.Y. DMITRIEV O.Y. Function and regulation of human copper-transporting ATPases. Physiol. Rev. 2007;87:1011–1046. doi: 10.1152/physrev.00004.2006. [DOI] [PubMed] [Google Scholar]
- MADDEN H.R. FLETCHER S. DAVIS M.R. WILTON S.D. Characterization of a complex Duchenne muscular dystrophy-causing dystrophin gene inversion and restoration of the reading frame by induced exon skipping. Hum. Mutat. 2008. (Epub ahead of print). [DOI] [PubMed]
- MADSEN E.C. MORCOS P.A. MENDELSOHN B.A. GITLIN J.D. In vivo correction of a Menkes disease model using antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3909–3914. doi: 10.1073/pnas.0710865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAILMAN M.D. HEINZ J.W. PAPP A.C. SNYDER P.J. SEDRA M.S. WIRTH B. BURGHES A.H. PRIOR T.W. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- MANKODI A. TAKAHASHI M.P. JIANG H. BECK C.L. BOWERS W.J. MOXLEY R.T. CANNON S.C. THORNTON C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- MANN C.J. HONEYMAN K. CHENG A.J. LY T. LLOYD F. FLETCHER S. MORGAN J.E. PARTRIDGE T.A. WILTON S.D. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl. Acad. Sci. U.S.A. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATLIN A.J. CLARK F. SMITH C.W. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- MENDELSOHN B.A. YIN C. JOHNSON S.L. WILM T.P. SOLNICA-KREZEL L. GITLIN J.D. Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell. Metab. 2006;4:155–162. doi: 10.1016/j.cmet.2006.05.001. [DOI] [PubMed] [Google Scholar]
- MENKES J.H. ALTER M. STEIGLEDER G.K. WEAKLEY D.R. SUNG J.H. A sex-linked recessive disorder with retardation of growth, peculiar hair, and focal cerebral and cerebellar degeneration. Pediatrics. 1962;29:764–779. [PubMed] [Google Scholar]
- MERCATANTE D.R. MOHLER J.L. KOLE R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J. Biol. Chem. 2002;277:49374–49382. doi: 10.1074/jbc.M209236200. [DOI] [PubMed] [Google Scholar]
- MIRONOV A.A. FICKETT J.W. GELFAND M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIYAJIMA H. MIYASO H. OKUMURA M. KURISU J. IMAIZUMI K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- MONACO A.P. BERTELSON C.J. LIECHTI-GALLATI S. MOSER H. KUNKEL L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- MOULTON H.M. FLETCHER S. NEUMAN B.W. MCCLOREY G. STEIN D.A. ABES S. WILTON S.D. BUCHMEIER M.J. LEBLEU B. IVERSEN P.L. Cell-penetrating peptide-morpholino conjugates alter pre-mRNA splicing of DMD (Duchenne muscular dystrophy) and inhibit murine coronavirus replication in vivo. Biochem Soc Trans. 2007;35:826–828. doi: 10.1042/BST0350826. [DOI] [PubMed] [Google Scholar]
- NICHOLSON L.V. DAVISON K. JOHNSON M.A. SLATER C.R. YOUNG C. BHATTACHARYA S. GARDNER-MEDWIN D. HARRIS J.B. Dystrophin in skeletal muscle. II. Immunoreactivity in patients with Xp21 muscular dystrophy. J. Neurol. Sci. 1989;94:137–146. doi: 10.1016/0022-510x(89)90224-4. [DOI] [PubMed] [Google Scholar]
- OLSEN N.J. STEIN C.M. New drugs for rheumatoid arthritis. N. Engl. J. Med. 2004;350:2167–2179. doi: 10.1056/NEJMra032906. [DOI] [PubMed] [Google Scholar]
- PAGANI F. BARALLE F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- PAGENSTECHER C. WEHNER M. FRIEDL W. RAHNER N. ARETZ S. FRIEDRICHS N. SENGTELLER M. HENN W. BUETTNER R. PROPPING P. MANGOLD E. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum. Genet. 2006;119:9–22. doi: 10.1007/s00439-005-0107-8. [DOI] [PubMed] [Google Scholar]
- PINCUS T. O'DELL J.R. KREMER J.M. Combination therapy with multiple disease-modifying antirheumatic drugs in rheumatoid arthritis: a preventive strategy. Ann. Intern. Med. 1999;131:768–774. doi: 10.7326/0003-4819-131-10-199911160-00009. [DOI] [PubMed] [Google Scholar]
- POLLARD A.J. KRAINER A.R. ROBSON S.C. EUROPE-FINNER G.N. Alternative splicing of the adenylyl cyclase stimulatory G-protein G alpha(s) is regulated by SF2/ASF and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and involves the use of an unusual TG 3′-splice Site. J. Biol. Chem. 2002;277:15241–15251. doi: 10.1074/jbc.M109046200. [DOI] [PubMed] [Google Scholar]
- POLLARD A.J. SPAREY C. ROBSON S.C. KRAINER A.R. EUROPE-FINNER G.N. Spatio-temporal expression of the trans-acting splicing factors SF2/ASF and heterogeneous ribonuclear proteins A1/A1B in the myometrium of the pregnant human uterus: a molecular mechanism for regulating regional protein isoform expression in vivo. J. Clin. Endocrinol. Metab. 2000;85:1928–1936. doi: 10.1210/jcem.85.5.6537. [DOI] [PubMed] [Google Scholar]
- PRAMONO Z.A. TAKESHIMA Y. ALIMSARDJONO H. ISHII A. TAKEDA S. MATSUO M. Induction of exon skipping of the dystrophin transcript in lymphoblastoid cells by transfecting an antisense oligodeoxynucleotide complementary to an exon recognition sequence. Biochem. Biophys. Res. Commun. 1996;226:445–449. doi: 10.1006/bbrc.1996.1375. [DOI] [PubMed] [Google Scholar]
- RESINA S. ABES S. TURNER J.J. PREVOT P. TRAVO A. CLAIR P. GAIT M.J. THIERRY A.R. LEBLEU B. Lipoplex and peptide-based strategies for the delivery of steric-block oligonucleotides. Int. J. Pharm. 2007a;344:96–102. doi: 10.1016/j.ijpharm.2007.04.039. [DOI] [PubMed] [Google Scholar]
- RESINA S. KOLE R. TRAVO A. LEBLEU B. THIERRY A.R. Switching on transgene expression by correcting aberrant splicing using multi-targeting steric-blocking oligonucleotides. J. Gene Med. 2007b;9:498–510. doi: 10.1002/jgm.1044. [DOI] [PubMed] [Google Scholar]
- ROBERTS J. PALMA E. SAZANI P. ORUM H. CHO M. KOLE R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol. Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- SAZANI P. GEMIGNANI F. KANG S.H. MAIER M.A. MANOHARAN M. PERSMARK M. BORTNER D. KOLE R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- SAZANI P. KANG S.H. MAIER M.A. WEI C. DILLMAN J. SUMMERTON J. MANOHARAN M. KOLE R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001;29:3965–3974. doi: 10.1093/nar/29.19.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMAJUK G. SIERAKOWSKA H. KOLE R. Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J. Biol. Chem. 1999;274:21783–21789. doi: 10.1074/jbc.274.31.21783. [DOI] [PubMed] [Google Scholar]
- SIERAKOWSKA H. SAMBADE M.J. AGRAWAL S. KOLE R. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH N.N. ANDROPHY E.J. SINGH R.N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- SINGH N.K. SINGH N.N. ANDROPHY E.J. SINGH R.N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SKORDIS L.A. DUNCKLEY M.G. YUE B. EPERON I.C. MUNTONI F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOUTSCHEK J. AKINC A. BRAMLAGE B. CHARISSE K. CONSTIEN R. DONOGHUE M. ELBASHIR S. GEICK A. HADWIGER P. HARBORTH J. JOHN M. KESAVAN V. LAVINE G. PANDEY R.K. RACIE T. RAJEEV K.G. ROHL I. TOUDJARSKA I. WANG G. WUSCHKO S. BUMCROT D. KOTELIANSKY V. LIMMER S. MANOHARAN M. VORNLOCHER H.P. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- SURONO A. VAN KHANH T. TAKESHIMA Y. WADA H. YAGI M. TAKAGI M. KOIZUMI M. MATSUO M. Chimeric RNA/ethylene-bridged nucleic acids promote dystrophin expression in myocytes of duchenne muscular dystrophy by inducing skipping of the nonsense mutation-encoding exon. Hum. Gene Ther. 2004;15:749–757. doi: 10.1089/1043034041648444. [DOI] [PubMed] [Google Scholar]
- SUWANMANEE T. SIERAKOWSKA H. FUCHAROEN S. KOLE R. Repair of a splicing defect in erythroid cells from patients with beta-thalassemia/HbE disorder. Mol. Ther. 2002a;6:718–726. doi: 10.1006/mthe.2002.0805. [DOI] [PubMed] [Google Scholar]
- SUWANMANEE T. SIERAKOWSKA H. LACERRA G. SVASTI S. KIRBY S. WALSH C.E. FUCHAROEN S. KOLE R. Restoration of human beta-globin gene expression in murine and human IVS2–654 thalassemic erythroid cells by free uptake of antisense oligonucleotides. Mol. Pharmacol. 2002b;62:545–553. doi: 10.1124/mol.62.3.545. [DOI] [PubMed] [Google Scholar]
- T HOEN P.A. DE MEIJER E.J. BOER J.M. VOSSEN R.H. TURK R. MAATMAN R.G. DAVIES K.E. VAN OMMEN G.J. VAN DEUTEKOM J.C. DEN DUNNEN J.T. Generation and characterization of transgenic mice with the full-length human DMD gene. J. Biol. Chem. 2008;283:5899–5907. doi: 10.1074/jbc.M709410200. [DOI] [PubMed] [Google Scholar]
- TAKESHIMA Y. NISHIO H. SAKAMOTO H. NAKAMURA H. MATSUO M. Modulation of in vitro splicing of the upstream intron by modifying an intra-exon sequence which is deleted from the dystrophin gene in dystrophin Kobe. J. Clin. Invest. 1995;95:515–520. doi: 10.1172/JCI117693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKESHIMA Y. YAGI M. WADA H. ISHIBASHI K. NISHIYAMA A. KAKUMOTO M. SAKAEDA T. SAURA R. OKUMURA K. MATSUO M. Intravenous infusion of an antisense oligonucleotide results in exon skipping in muscle dystrophin mRNA of Duchenne muscular dystrophy. Pediatr. Res. 2006;59:690–694. doi: 10.1203/01.pdr.0000215047.51278.7c. [DOI] [PubMed] [Google Scholar]
- TENNYSON C.N. SHI Q. WORTON R.G. Stability of the human dystrophin transcript in muscle. Nucleic Acids Res. 1996;24:3059–3064. doi: 10.1093/nar/24.15.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERAOKA S.N. TELATAR M. BECKER-CATANIA S. LIANG T. ONENGUT S. TOLUN A. CHESSA L. SANAL O. BERNATOWSKA E. GATTI R.A. CONCANNON P. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRIP J. DROST G. VAN ENGELEN B.G. FABER C.G. Drug treatment for myotonia. Cochrane Database Syst. Rev. 2006:CD004762. doi: 10.1002/14651858.CD004762.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UGARTE M. AGUADO C. DESVIAT L.R. SANCHEZ-ALCUDIA R. RINCON A. PEREZ B. Propionic and methylmalonic acidemia: antisense therapeutics for intronic variations causing aberrantly spliced messenger RNA. Am. J. Hum. Genet. 2007;81 doi: 10.1086/522376. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DEUTEKOM J.C. BREMMER-BOUT M. JANSON A.A. GINJAAR I.B. BAAS F. DEN DUNNEN J.T. VAN OMMEN G.J. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum. Mol. Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- VAN DEUTEKOM J.C. JANSON A.A. GINJAAR I.B. FRANKHUIZEN W.S. AARTSMA-RUS A. BREMMER-BOUT M. DEN DUNNEN J.T. KOOP K. VAN DER KOOI A.J. GOEMANS N.M. DE KIMPE S.J. EKHART P.F. VENNEKER E.H. PLATENBURG G.J. VERSCHUUREN J.J. VAN OMMEN G.J. Local dystrophin restoration with anti-sense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- VICKERS T.A. ZHANG H. GRAHAM M.J. LEMONIDIS K.M. ZHAO C. DEAN N.M. Modification of MyD88 mRNA splicing and inhibition of IL-1beta signaling in cell culture and in mice with a 2′-O-methoxyethyl-modified oligonucleotide. J. Immunol. 2006;176:3652–3661. doi: 10.4049/jimmunol.176.6.3652. [DOI] [PubMed] [Google Scholar]
- VITIELLO L. BASSI N. CAMPAGNOLO P. ZACCARIOTTO E. OCCHI G. MALERBA A. PIGOZZO S. REGGIANI C. AUSONI S. ZAGLIA T. GAMBA P. BARONI M.D. DITADI A.P. In vivo delivery of naked antisense oligos in aged mdx mice: analysis of dystrophin restoration in skeletal and cardiac muscle. Neuromuscul Disord. 2008. [DOI] [PubMed]
- WAN L. BATTLE D.J. YONG J. GUBITZ A.K. KOLB S.J. WANG J. DREYFUSS G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAN J. SAZANI P. KOLE R. Modification of HER2 pre-mRNA alternative splicing and its effects on breast cancer cells. Int. J. Cancer. 2008. [DOI] [PMC free article] [PubMed]
- WHEELER T.M. LUECK J.D. SWANSON M.S. DIRKSEN R.T. THORNTON C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILTON S.D. LLOYD F. CARVILLE K. FLETCHER S. HONEYMAN K. AGRAWAL S. KOLE R. Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscul. Disord. 1999;9:330–338. doi: 10.1016/s0960-8966(99)00010-3. [DOI] [PubMed] [Google Scholar]
- WU R.P. YOUNGBLOOD D.S. HASSINGER J.N. LOVEJOY C.E. NELSON M.H. IVERSEN P.L. MOULTON H.M. Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 2007;35:5182–5191. doi: 10.1093/nar/gkm478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YIN H. LU Q. WOOD M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in mdx mice. Mol. Ther. 2008;16:38–45. doi: 10.1038/sj.mt.6300329. [DOI] [PubMed] [Google Scholar]
- YOKOTA T. LU Q. PARTRIDGE T. KOBAYASHI M. NAKAMURA A. TAKEDA S. HOFFMAN E.P. Body-wide restoration of dystrophin expression and amelioration of pathology in dystrophic dogs using a morpholino cocktail. Mol. Ther. 2008;16(Supp. 1):S143. [Google Scholar]
- ZIMMERMANN T.S. LEE A.C. AKINC A. BRAMLAGE B. BUMCROT D. FEDORUK M.N. HARBORTH J. HEYES J.A. JEFFS L.B. JOHN M. JUDGE A.D. LAM K. MCCLINTOCK K. NECHEV L.V. PALMER L.R. RACIE T. ROHL I. SEIFFERT S. SHANMUGAM S. SOOD V. SOUTSCHEK J. TOUDJARSKA I. WHEAT A.J. YAWORSKI E. ZEDALIS W. KOTELIANSKY V. MANOHARAN M. VORNLOCHER H.P. MACLACHLAN I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]