Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle (original) (raw)
Abstract
There is clear evidence that the phenotypic modulation of smooth muscle cells (SMCs) contributes to the pathophysiology of vascular disease. Phenotypic modulation refers to the unique ability of SMCs to alter their phenotype in response to extracellular stimuli and is hallmarked by the loss of SMC marker gene expression. The transcription factor Krüppel-like factor 4 (KLF4) is a known powerful negative regulator of SMC marker gene expression that works, in part, by decreasing the expression of the serum response factor (SRF) myocardin. KLF4 is not expressed in healthy adult SMCs but is increased in SMCs in response to vascular injury in vivo or PDGF-BB treatment in vitro. The aim of the present study was to determine the molecular mechanisms that regulate the expression of KLF4 in phenotypically modulated SMCs. The results demonstrated that the transcription factor stimulating protein-1 (Sp1) regulated the expression of KLF4 in SMCs. The KLF4 promoter contains three consensus Sp1 binding sites. Using a series of truncated KLF4 promoters, we showed that only fragments containing these Sp1 sites could be activated by PDGF-BB. In addition, overexpression of Sp1 alone was sufficient to increase the activity of the KLF4 promoter. Moreover, inhibiting Sp1 expression with small-interfering RNA attenuated the effects of PDGF-BB on KLF4 expression. Mutation of the three Sp1 sites within the KLF4 promoter abolished both baseline and PDGF-BB-induced activity. Finally, the results demonstrated enhanced Sp1 binding to the KLF4 promoter in SMCs treated with PDGF-BB in vitro and following vascular injury in vivo. Taken together, the results suggest a novel role for Sp1 in increasing the expression of KLF4 in phenotypically modulated SMCs.
Keywords: Krüppel-like factor 4, stimulating protein-1, platelet-derived growth factor-BB, vascular injury
unlike other cells of the myogenic lineage, such as cardiac and skeletal muscle cells, which are terminally differentiated, vascular smooth muscle cells (SMCs) retain the ability to dramatically modulate their phenotype in response to extracellular cues and stimuli. The phenotypic modulation of SMCs plays an important role in normal vascular development as well as the progression of vascular diseases such as atherosclerosis and restenosis (26). Normally, healthy adult SMCs express a distinct repertoire of SMC-specific marker genes including SM α-actin, SM myosin heavy chain (SMMHC), and SM22α. However, in response to vascular injury or disease, SMCs alter their phenotype toward a more dedifferentiated state, characterized by a decreased expression of SMC marker genes and increased rates of proliferation and migration. An accumulation of these phenotypically modulated SMCs within the diseased vessel contributes to plaque formation and the consequent narrowing of the vascular lumen. Our laboratory has had a long-standing interest in unraveling the molecular mechanisms that contribute to the phenotypic modulation of SMCs to prevent or reduce the contribution of these cells to the progression of vascular diseases.
Platelet-derived growth factor-BB (PDGF-BB) is one of the key modulators of the SMC phenotype (26). In cultured SMCs, it represses the expression of the SMC marker genes and increases rates of proliferation and migration (7, 15, 30). In vivo, the expression of PDGF-BB and the PDGF-β receptor are increased at the site of vascular lesions and inhibition of their signaling has been reported to reduce neointimal thickening (3, 10, 20, 35, 36, 39). The PDGF-BB-mediated repression of SMCs genes is mediated through multiple mechanisms including the downregulation of myocardin expression and the disruption of myocardin/megakaryoblastic leukemia factor binding, both of which ultimately result in a disruption of serum response factor (SRF) binding (22, 40). In addition, we recently found that vascular injury in vivo, as well as the treatment of cultured SMCs with PDGF-BB or oxidized phospholipids, was associated with the increased expression of the transcription factor Krüppel-like factor 4 (KLF4), which is normally not expressed in healthy adult SMCs (22, 28). Our laboratory previously identified KLF4 as a potent repressor of SMC marker gene expression that works through several mechanisms including 1) direct binding of KLF4 to SMC marker gene promoters through a transforming growth factor-β control element (TCE); 2) reducing the binding of the transcription factor SRF to CArG-box elements located within SMC marker gene promoters; 3) decreasing expression of myocardin, a potent SRF coactivator; and 4) recruitment of histone deacetylases that induce changes in chromatin structure associated with transcriptional silencing (21, 22, 25, 40, 41). There is also direct evidence demonstrating that KLF4 plays an important role in inducing phenotypic switching of SMCs. For example, we previously reported that small-interfering (si)RNA knockdown of KLF4 in cultured SMCs blocked the oxidized phospholipid-induced repression of endogenous SM α-actin and SMMHC expression. In addition, siRNA-induced suppression of KLF4 attenuated the PDGF-BB-induced repression of SM α-actin promoter-reporter constructs in transient transfection assays, although it has not been determined whether KLF4 also plays a role in mediating the PDGF-BB-mediated repression of endogenous SMC marker genes (22, 28). Finally, we recently showed that conditional KLF4 knockout mice displayed a delayed repression of SMC marker genes following vascular injury (41). Despite the fact that we have identified KLF4 as playing an integral role in SMC marker gene repression, the molecular mechanisms responsible for activating KLF4 expression in phenotypically modulated SMCs are currently unknown. There are three binding sites for the transcription factor stimulating protein-1 (Sp1) located in the 5′ proximal KLF4 promoter that have been reported to play a role in regulating KLF4 expression in other cell types (5). Interestingly, our laboratory has previously shown that Sp1 expression is increased in SMCs in response to vascular injury in vivo or PDGF-BB treatment in vitro (24, 37). Moreover, the siRNA-induced suppression of Sp1 abolished the PDGF-BB-mediated repression of SMC marker gene expression in cultured SMCs (37). Thus we hypothesized that Sp1 may play an important role in regulating the increased KLF4 expression in phenotypically modulated SMCs in vitro and in vivo.
The overall goals of the present study were 1) to determine whether KLF4 plays an important role in mediating the PDGF-BB-induced repression of multiple endogenous SMC marker genes and 2) to determine whether Sp1 is involved in the upregulating expression of KLF4 in phenotypically modulated SMCs.
MATERIALS AND METHODS
Cell culture and PDGF-BB treatment.
Cultured rat aortic SMCs were cultured and stimulated with PDGF-BB (Millipore) or vehicle (10 mM acetic acid + 2 μg/ml fatty acid-free BSA) as previously described with minor modifications (7). Cells were plated at a density of 1.0 × 104 cells/cm2 unless otherwise indicated.
siRNA transfections.
SMCs were transfected with siRNA oligos (MWG Biotech) using Oligofectamine (Invitrogen) according to the manufacturer's protocols with minor modifications. Cells were transfected with 100–300 nM siRNA oligo (as indicated) for 4 h before the addition of insulin-free serum-free media and incubated for an additional 24–72 h (as indicated) before treatment or harvest. Sequences of the siRNA oligos used are as follows: small-interfering green fluorescent protein, 5′-GAACGGCAUCAAGGUGAAC-3′; siKLF4, 5′-GUACAAUGGUUUAUUCCCA-3′; and siSp1, 5′-UUGAGUCACCCAAUGAGAA-3′. siSp1 plasmid transfections were done using the pMighty system as previously described (37).
Transient transfection and luciferase assay.
Cells were transiently transfected with FuGENE6 (Roche) as previously described (28). KLF4-luciferase constructs (containing 2,200, 1,190, 520, or 50 bp of proximal KLF4 promoter region) were a gift from Chi-Chuan Tseng (Boston University School of Medicine). The KLF4 p520 Sp1 mutant luciferase plasmid was generated using basic restriction enzyme cloning methods, and mutations were confirmed by sequencing. Briefly, the KLF4 p520 promoter was excised from pGL3 backbone (_Kpn_I/_Hind_III) and cloned into a pBlueScriptII-KS(−) that had had the intrinsic _Not_I site destroyed by T4 DNA polymerase. The resulting plasmid was digested with _Not_I/_Nco_I to remove a 142-bp region of the KLF4 p520 promoter (from −152 to −10 of the mouse KLF4 promoter) containing the three Sp1 sites. A 142-bp double-stranded oligo containing the _Not_I/_Nco_I region of the mouse KLF4 promoter but harboring GGCGGG to GTTTTG substitutions to mutate the three Sp1 binding sites was next generated (IDT) and cloned into the _Not_I/_Nco_I-digested KLF4 p520 promoter (lacking the region between −152 and −10) in pBlueScript-KS(−) to generate the KLF4 p520 Sp1 mutant promoter fragment. The entire KLF4 p520 Sp1 mutant promoter was then excised (_Kpn_I/_Hind_III) from pBlueScript-KS(−) and cloned back into pGL3 (_Kpn_I/_Hind_III) to make the KLF4 p520 Sp1 mut-luc plasmid. The cytomegalovirus-Sp1 plasmid was a gift from Robert Tijian (Howard Hughes Medical Institute, University of California). Luciferase activity was measured (in relative luciferase units) and normalized to total protein content as previously described (17).
Western blot analysis.
Cells were lysed using modified radioimmunoprecipitation assay buffer and subjected to immunoblotting using standard techniques as previously described using the following antibodies: KLF4 (custom rabbit antibody generated by Chemicon using the peptide ASGPAGREKTLRPAGC, which corresponds to amino acids 15–29 in mouse KLF4 and is conserved in rat), SM α-actin (M2, Sigma), SMMHC (BTI), SM22α (Abcam), β-tubulin (Cell Signaling Technologies), and Sp1 (Santa Cruz Biotechnology) (17).
RNA isolation, cDNA synthesis, and quantitative real-time RT-PCR.
Total RNA was isolated from cultured SMCs using TRIzol reagent (Invitrogen), and cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's protocols. To quantify the expression of KLF4 mRNA, a real-time RT-PCR analysis (iCycler, Bio-Rad) of cDNA was performed using dual-fluorescence-labeled probes (IDT). All results were normalized to 18S rRNA. Specific primers and probes for each gene are as follows (forward primer, reverse primer, and probe): 18S rRNA (rat), 5′-CGGCTACCACATCCAAGGAA-3′, 5′-AGCTGGAATTACCGCGGC-3′, and 5′-TGCTGGCACCAGACTTGCCCTC-3′; and KLF4 mRNA (rat), 5′-CTTTCCTGCCAGACCAGATG-3′, 5′-GGTTTCTCGCCTGTGTGAGT-3′, and 5′-ATTATCAAGAGCTCATGCCACCGGG-3′.
Quantitative chromatin immunoprecipitation assay.
Quantitative chromatin immunoprecipitation (ChIP) assays were performed as previously described (16). Chromatin protein complexes were immunoprecipitated with 3 μg anti-Sp1 antibody (Santa Cruz Biotechnology), whereas negative control/input DNA was immunoprecipitated with no antibody. Real-time PCR was performed to amplify the region of the KLF4 promoter containing the three Sp1 sites using the following primer set: forward, 5′-TTCCCATGGCAGGACTTTCAA-3′; and reverse, 5′-ATGGCAGCTAAATAAACAAACT-3′.
Rat aorta balloon injury model and in vivo ChIP assay.
Male Sprague-Dawley rats (350–400 g; Charles River) were anesthetized and subjected to acute balloon injury to the thoracic aorta and left common carotid artery in accordance with Institutional Animal Care and Use Committee-approved protocols as previously described (6, 25). In vivo ChIP assays were performed on injured and uninjured (sham operated) aortas as previously described (11, 25). Chromatin-protein complexes were immunoprecipitated with 3 μg anti-Sp1 antibody (Santa Cruz Biotechnology), whereas negative control/input DNA was immunoprecipitated with no antibody. Semiquantitative RT-PCR was performed using TaqPCRx DNA Polymerase (Invitrogen) per the manufacturer's protocol for G/C-rich templates to amplify the region of the KLF4 promoter containing the three Sp1 sites using the KLF4 promoter primers described for quantitative ChIP assay.
Statistical analysis.
Unless otherwise indicated, all experiments were performed in triplicate and represent 2–4-independent experiments giving similar results. Data were analyzed for statistical significance using Student's _t_-test or one-way ANOVA with Bonferroni's post hoc testing. Error bars represent means ± SE. P values < 0.05 were considered statistically significant.
RESULTS
siRNA knockdown of KLF4 attenuated PDGF-BB-induced repression of SM α-actin, SMMHC, and SM22α.
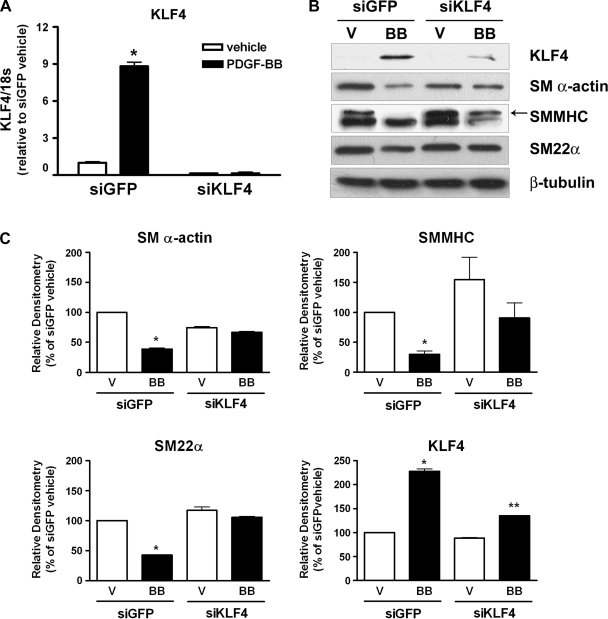
Previous studies from our laboratory showed that siRNA-induced suppression of KLF4 blocked the oxidized phospholipid-induced repression of endogenous SM α-actin and SMMHC expression and attenuated the PDGF-BB-induced repression of SM α-actin promoter activity (22, 28). We extended these studies to examine whether a knockdown of KLF4 expression would prevent the PDGF-BB-induced repression of endogenous SM α-actin, SMMHC, and SM22α expression. Cultured SMCs were transfected with siRNA oligos specific for the knockdown of KLF4 expression. Following knockdown, the cells were treated with PDGF-BB for either 3 h (to show maximal KLF4 induction) or 72 h (to show maximal SMC marker gene repression), and changes in endogenous KLF4 and SMC marker gene expression were analyzed by Western blot analysis. Time points were selected based on our previous results showing that KLF4 induction is both rapid and transient in response to PDGF-BB in vitro or vascular injury in vivo (22, 41). siKLF4 oligos dramatically attenuated PDGF-BB-mediated increases in KLF4 expression as measured by both quantitative real-time RT-PCR (Fig. 1_A_) and Western blot analysis (Fig. 1, B and C). Furthermore, the knockdown of KLF4 attenuated the PDGF-BB-mediated repression of SM α-actin, SMMHC, and SM22α (Fig. 1, B and C). These data provide compelling evidence that KLF4 plays an important role in mediating the PDGF-BB-induced repression of SMC marker genes.
Fig. 1.
PDGF-BB-mediated increase in Krüppel-like factor 4 (KLF4) expression were required for repression of smooth muscle (SM) cell (SMC) marker genes. SMCs were transfected with 300 nM control [small-interfering green fluorescent protein (siGFP)] or KLF4-specific (siKLF4) siRNA oligos and treated with vehicle (V) or PDGF-BB (BB) (30 ng/ml) for 3 h (KLF4) or 72 h (SMC genes). A: changes in KLF4 mRNA were measured by quantitative real-time RT-PCR. B: changes in KLF4, SM α-actin, SM myosin heavy chain (SMMHC), and SM22α proteins were determined by Western blot analysis. β-Tubulin was examined as a control for loading. C: changes in protein levels were quantified using densitometry. All values represent means (normalized to vehicle) ± SE. *P < 0.05 compared with siGFP; **P < 0.05, compared with siKLF4 (n = 3 experiments).
PDGF-BB-induced increases in KLF4 promoter activity required regulatory elements between −520 and −50 bp of the 5′ region of the KLF4 promoter.
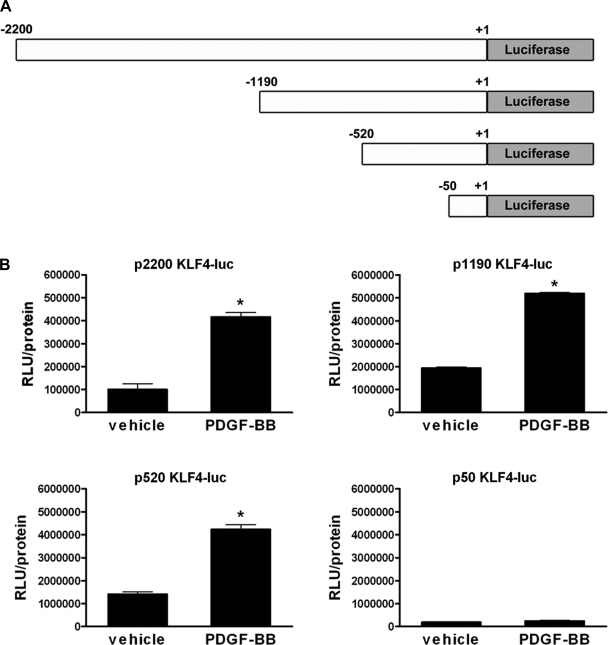
To determine the region of the KLF4 promoter required for increased KLF4 expression in response to PDGF-BB treatment of cultured SMCs, we used a series of promoter-reporter plasmids containing various lengths of the KLF4 promoter (depicted in Fig. 2_A_). Cultured SMCs were transfected with the different KLF4 promoter-reporter plasmids and treated with either vehicle or PDGF-BB. Promoter activity was analyzed by luciferase assay. The results showed that PDGF-BB increased the activity of the 2,200-, 1,190-, and 520-bp promoters but was unable to increase the activity of the 50-bp promoter. These results suggest that the regulatory elements necessary for the PDGF-BB-induced increase in KLF4 expression are found within −520 and −50 bp of the 5′ region of the KLF4 promoter.
Fig. 2.
PDGF-BB-mediated increases in KLF4 promoter activity required sequences between −520 and −50 bp. A: schematic showing the series of KLF4 promoter-reporter plasmids used. B: SMCs were transfected with either the 2,200-, 1,190-, 520-, or 50-bp KLF4 promoter-luciferase plasmid and treated with vehicle or PDGF-BB (30 ng/ml) for 3 h before determining relative changes in luciferase expression. All values represent means ± SE in relative luciferase units (RLUs), normalized to total protein. *P < 0.05 (n = 3 experiments).
The KLF4 promoter contains three Sp1 binding sites, and overexpression of Sp1 alone was sufficient to activate the KLF4 promoter.
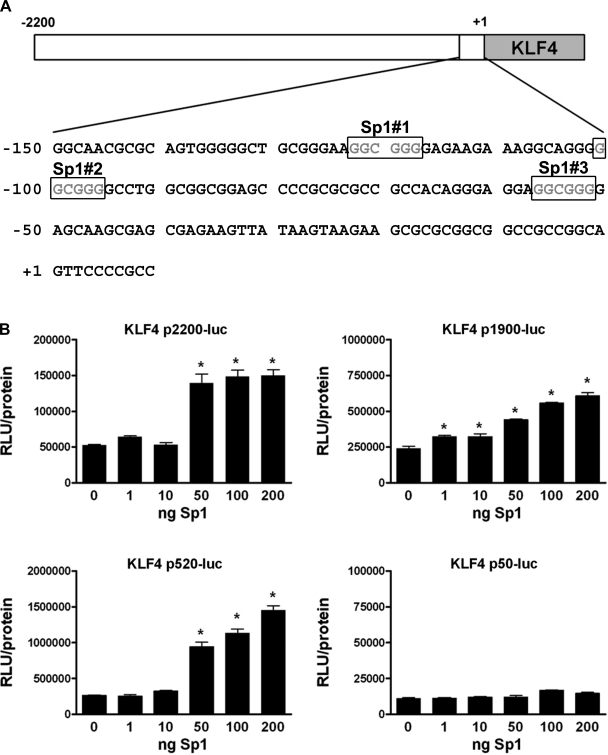
There are three consensus Sp1 binding sites located between −150 and −50 bp of the KLF4 promoter (as depicted in Fig. 3_A_). These sites have been shown to be required for the increased expression of KLF4 in HT-29 colon cancer cells in response to butyrate treatment (5). Interestingly, our laboratory previously reported that Sp1 expression was increased in the neointima following balloon injury of the rat carotid artery (24). In addition, we previously found that the treatment of cultured SMCs with PDGF-BB caused an increased expression, phosphorylation, and nuclear localization of Sp1 (37). Based on this previous data, we hypothesized that Sp1 may regulate KLF4 expression in SMCs. To determine whether the expression of Sp1 alone could increase KLF4 expression in SMCs, we cotransfected SMCs with the various KLF4 promoter-reporter plasmids and increasing amounts of a Sp1 expression plasmid. The results showed that the overexpression of Sp1 alone increased the activity of the 2,200-, 1,190-, and 520-bp KLF4 promoters in a dose-dependent manner (Fig. 3_B_). The 50-bp KLF4 promoter, which does not contain the Sp1 sites, was unaffected by Sp1 overexpression. These results provide evidence that Sp1 may contribute to the regulation of KLF4 expression in cultured SMCs.
Fig. 3.
Overexpression of exogenous stimulating protein-1 (Sp1) increased the activity of the 2,200-, 1,190-, and 520-bp KLF4 promoters but not the 50-bp promoter lacking the 3 Sp1 binding sites. A: sequence of the KLF4 promoter region from −150 to +10 showing the 3 consensus Sp1 binding sites. B: SMCs were cotransfected with the 2,200-, 1,190-, 520-, or 50-bp KLF4 promoter reporter plasmid and increasing amounts of cytomegalovirus-Sp1 plasmid (0–200 ng) for 48 h before determining relative changes in luciferase expression. Values represent means ± SE in RLUs, normalized to total protein. *P < 0.05 (n = 3 experiments).
siRNA-induced suppression of Sp1 prevented PDGF-BB-induced activation of the KLF4 promoter.
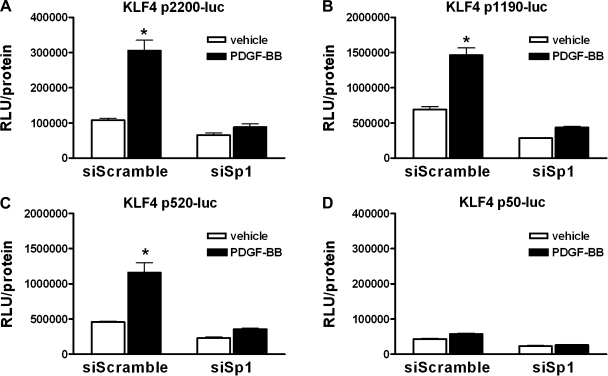
To determine whether Sp1 is required for the PDGF-BB-induced activation of the KLF4 promoter, SMCs were cotransfected with control (siScramble) or Sp1-specific (siSp1) siRNA plasmids (to knockdown Sp1) and one of the KLF4 promoter-reporter plasmids. Following knockdown, cells were treated with vehicle or PDGF-BB and promoter activity was analyzed by luciferase assay. The knockdown of Sp1 prevented PDGF-BB-induced activation of the 2,200-, 1,190-, and 520-bp KLF4 promoters, which contain the Sp1 binding sites (Fig. 4). These data demonstrate that Sp1 is required for the PDGF-BB-mediated activation of the KLF4 promoter in cultured SMCs.
Fig. 4.
siRNA knockdown of Sp1 abolished PDGF-BB-induced activation of the KLF4 promoter. SMCs were cotransfected with the 2,200-bp (A), 1,190-bp (B), 520-bp (C), or 50-bp (D) KLF4 promoter reporter plasmid and either control siRNA plasmid (siScramble) or Sp1-specific siRNA plasmid (siSp1) for 48 h before treatment with vehicle or PDGF-BB (30 ng/ml) for 3 h before determining relative changes in luciferase expression. All values represent means ± SE in RLUs, normalized to total protein. *P < 0.05 (n = 3 experiments).
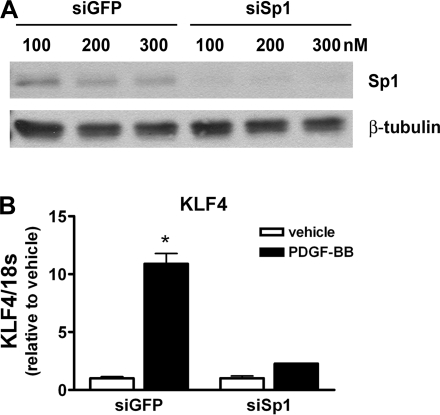
Knockdown of Sp1 prevented PDGF-BB-induced increases in endogenous KLF4 expression.
Thus far, we have used KLF4 promoter-reporter constructs as an indicator of the relative changes in the expression of KLF4 in response to PDGF-BB treatment. To ensure that our results were not merely an artifact of using artificial, truncated, episomal promoter-reporter constructs, we analyzed whether siRNA knockdown of Sp1 could prevent PDGF-BB-induced increases in endogenous KLF4 expression as measured by quantitative real-time RT-PCR. SMCs were transfected with siRNA oligos to knockdown Sp1 expression followed by treatment with vehicle or PDGF-BB. Efficient Sp1 knockdown was confirmed by Western blot analysis (Fig. 5_A_). Quantitative real-time PCR results showed that the knockdown of Sp1 abolished the PDGF-BB-induced increase in KLF4 expression (Fig. 5_B_). These results provide evidence that Sp1 is required for the increased expression of endogenous KLF4 in SMCs treated with PDGF-BB.
Fig. 5.
siRNA knockdown of Sp1 using siRNA oligos abolished PDGF-BB-mediated increases in endogenous KLF4 expression. A: SMCs were transfected with 100, 200, or 300 nM control siRNA (siGFP) or Sp1-specific siRNA (siSp1) oligos for 72 h. Knockdown of Sp1 was confirmed by Western blot analysis. B: SMCs were transfected with 300 nM control siRNA (siGFP) or Sp1-specific siRNA (siSp1) for 72 h and then treated with vehicle or PDGF-BB for 3 h before isolating total RNA. Changes in the expression of KLF4 were determined by quantitative real-time RT-PCR. Values represent means ± SE (normalized to vehicle). *P < 0.05 (n = 3 experiments).
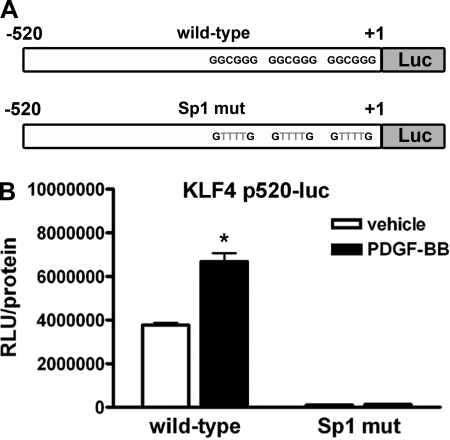
The three Sp1 sites in the KLF4 promoter were required for both baseline and PDGF-BB-induced KLF4 promoter activity.
To directly test whether the three Sp1 sites within the KLF4 promoter were responsible and required for PDGF-BB-induced increase in KLF4 promoter activity, the sequence of each of these sites was mutated from GGCGGG to GTTTTG (as depicted in Fig. 6_A_), which has previously been shown to abolish Sp1 binding (5). To test the effect of these mutations, SMCs were transfected with either the wild-type or Sp1 mutant 520-bp KLF4 promoter-reporter plasmid and treated with vehicle or PDGF-BB. Interestingly, the mutation of the three Sp1 sites drastically decreased the baseline activity of the 520-bp KLF4 promoter compared with wild-type (Fig. 6_B_). In addition, the mutation of the three Sp1 sites prevented PDGF-BB-induced increases in KLF4 promoter activity. Taken together, these data suggest that the three Sp1 sites in the KLF4 promoter are required for both baseline and PDGF-BB-induced KLF4 promoter activity in SMCs.
Fig. 6.
Mutation of the 3 Sp1 binding sites within the 520-bp KLF4 promoter abolished basal and PDGF-BB-induced activity of the promoter. A: 3 consensus Sp1 binding sites in the 520-bp KLF4 promoter were mutated from G_GCGG_G to G_TTTT_G to generate a 520-bp Sp1 mutant (mut) KLF4 promoter-reporter plasmid. B: SMCs were transfected with either wild-type or Sp1 mutant KLF4 promoter-reporter and treated with vehicle or PDGF-BB (30 ng/ml) for 3 h before determining changes in luciferase activity. Values represent means ± SE. *P < 0.05 (n = 3 experiments).
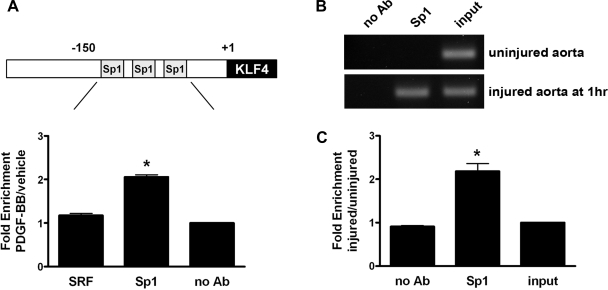
Binding of Sp1 to the endogenous KLF4 promoter is increased in cultured SMCs treated with PDGF-BB in vitro and following vascular injury in vivo.
To determine whether PDGF-BB enhances the binding of Sp1 to the three Sp1 sites in the endogenous KLF4 promoter within intact chromatin, we performed quantitative ChIP analysis on chromatin isolated from SMC treated with vehicle or PDGF-BB. As shown in Fig. 7_A_, Sp1 binding was enhanced twofold in SMCs treated with PDGF-BB compared with no antibody and SRF antibody controls. These results demonstrate that PDGF-BB stimulates the binding of Sp1 to the endogenous KLF4 promoter in cultured SMCs. To further extend these studies, we examined whether Sp1 binding to the KLF4 promoter was enhanced in the rat aorta following vascular injury. For these experiments, we performed in vivo ChIP assays on chromatin isolated from aortic tissue of either sham-operated (uninjured) rats or rats that had been subjected to aortic balloon injury. As shown in Fig. 7, B and C, the Sp1 binding to the endogenous KLF4 promoter was enhanced twofold in response to balloon injury compared with no antibody or input controls. Taken together, these results provide compelling evidence that the binding of Sp1 to the KLF4 promoter plays an important role in mediating an increased expression of KLF4 in phenotypically modulated SMCs in vitro and in vivo.
Fig. 7.
Binding of Sp1 to the endogenous KLF4 promoter within intact chromatin was induced in SMCs in vitro by treatment with PDGF-BB and in vivo following balloon catheter injury of rat aorta. A: SMCs were treated with vehicle or PDGF-BB for 1 h before isolating and harvesting chromatin for quantitative chromatin assay immunoprecipitation (ChIP) analysis. Changes in binding of Sp1 to the region of the KLF4 promoter containing the 3 putative Sp1 sites were determined by quantitative ChIP assay as described in materials and methods. Values represent means ± SE. *P < 0.001 compared with either serum response factor (SRF) or input group (n = 3 experiments). B: rats were subjected to balloon catheter injury, and aortas were harvested at 1-h postinjury for in vivo ChIP analysis. Changes in binding of Sp1 to the region of the KLF4 promoter containing the 3 putative Sp1 sites were determined by semiquantitative in vivo ChIP assay as described in materials and methods. C: quantitative graph showing the change in Sp1 binding to the KLF4 promoter based on densitometry measurements from 2 independent in vivo ChIP assay experiments. Values represent means ± SE. *P < 0.05 compared with either no antibody (Ab) or input group (n = 2 experiments).
DISCUSSION
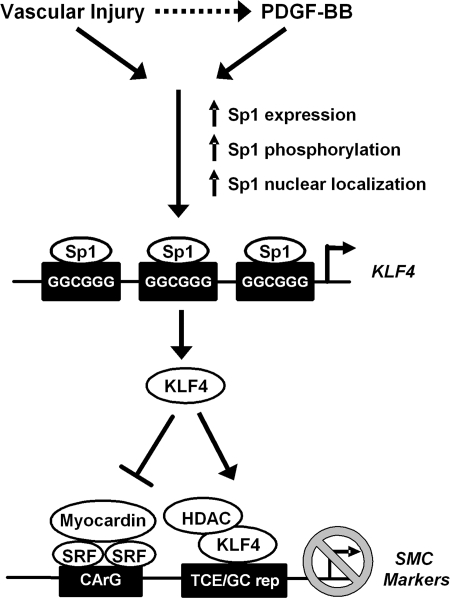
The overall goals of the present study were to determine whether KLF4 plays an important role in mediating a PDGF-BB-induced repression of multiple endogenous SMC marker genes and to determine the molecular mechanism(s) required for an increased expression of KLF4 in phenotypically modulated SMCs. The results showed that KLF4 played a key role in mediating the repressive effects of PDGF-BB on SM α-actin, SMMHC, and SM22α in cultured SMCs. Moreover, we found that KLF4 expression was regulated by a direct binding of Sp1 to the KLF4 promoter not only in cultured SMCs treated with PDGF-BB but also in vivo within the rat aorta following vascular injury. Of major interest, the results of recent studies from our laboratory demonstrated that KLF4 expression is rapidly induced following vascular injury in vivo and that the conditional knockout of KLF4 in mice resulted in a transient delay in the downregulation of multiple SMC marker genes following vascular injury (22, 41). Moreover, it has previously been shown that Sp1 expression is increased in vivo following vascular injury (24). Taken together, these results and those of the present studies provide novel evidence that injury-induced phenotypic switching of SMCs in vivo is mediated, at least in part, through Sp1-dependent increases in KLF4 expression (Fig. 8).
Fig. 8.
Schematic showing the proposed role of Sp1-induced activation of KLF4 expression in repression of SMC phenotypic modulation. HDAC, histone deacetylase; TCE, transforming growth factor-β control element.
Of note, it appears that the suppression of KLF4 expression effectively blocked the PDGF-BB-induced repression of SM α-actin and SM22α but only partially attenuated the PDGF-BB-induced repression of SMMHC (Fig. 1, B and C). This may be due to the fact that we were only able to partially abrogate the PDGF-BB-induced expression of KLF4 at the protein level (Fig. 1, B and C). Interestingly, a suppression of the KLF4 expression also increased baseline SMMHC expression in addition to partially attenuating the PDGF-BB-induced repression of this gene. Taken together, these results suggest that SMMHC may be more sensitive to subtle changes in KLF4 expression and/or that KLF4 may be more important for regulating the basal expression of SMMHC. However, it is also likely that KLF4 works in conjunction or in parallel with other well-documented PDGF-BB-induced repression mechanisms (8, 18, 32, 38). Consistent with this possibility, vascular injury in conditional KLF4 knockout mice resulted in a reduced downregulation of SM α-actin and SM22α expression at 3 days postinjury, but by 7 days postinjury, alternative, presumably KLF4 independent, pathways had compensated.
Indeed, the exact mechanisms whereby KLF4 represses the expression of SMC marker genes are still unclear. We previously presented evidence based on in vitro electrophoretic mobility shift assays showing that KLF4 bound to TCE elements contained within multiple SMC marker genes (1, 14). In addition, we also recently presented ChIP assay results showing increased binding of KLF4 to the TCE-containing region of the endogenous SM α-actin and SM22α promoters in vivo following vascular injury or in cultured SMC overexpressing KLF4 (21, 41). Interestingly, the TCE-containing regions of the SM22α and SMMHC promoters also contain a G/C-repressor element that closely resembles the KLF4 consensus binding site, and we have shown that the SM22α G/C-repressor element is required for the downregulation of expression of this gene during the phenotypic switching of SMCs in vivo in response to wire-induced injury or experimental atherosclerosis in the apolipoprotein E-deficient mouse (31, 37). As such, a key unresolved issue is whether the effects of KLF4 in vivo are mediated through a direct binding of KLF4 to the TCE or the G/C repressor. In addition to directly binding the SMC marker gene promoters to decrease their activity, KLF4 can also inhibit myocardin expression (22). Interestingly, there are several consensus KLF4 binding sites within the recently identified myocardin promoter, although the ability of KLF4 to bind to these sites has not yet been determined, nor have studies been done to determine whether a mutation of the KLF4 sites in the myocardin promoter prevent KLF4-dependent repression of its expression. Finally, KLF4 can decrease SRF binding to SMC marker gene promoters although it is unclear whether this is due to a binding of KLF4 to the SMC marker gene promoters, a decreased myocardin expression, or an as yet undefined mechanism (22). Despite our incomplete knowledge of the precise mechanisms of KLF4-mediated repression, it is clear that it involves multiple mechanisms that significantly contribute to SMC phenotypic modulation.
The results of the present studies showed that PDGF-BB-induced expression of KLF4 was dependent on a binding of Sp1 to the KLF4 promoter. Our initial studies were done using a truncated KLF4 promoter fragment; therefore, it is possible that there are additional unidentified regulatory regions that may also play a role in PDGF-BB-induced KLF4 expression (in addition to Sp1). However, we confirmed that Sp1 does bind to the region of the endogenous KLF4 promoter containing the three Sp1 sites within intact chromatin in response to PDGF-BB treatment of SMCs and that knockdown of Sp1 prevented PDGF-BB-induced increases in endogenous KLF4 expression. These data demonstrate that the effects of Sp1 are not artifacts of using a truncated, episomal KLF4 promoter-reporter plasmid but represent a functionally relevant signaling pathway. Importantly, we have previously shown that Sp1 plays a critical role in the PDGF-BB-induced repression of SMC marker genes in part through the G/C-repressor element. Our current data support and extend our previous findings by identifying an additional mechanism through which Sp1 mediates the repression of SMC marker genes. Interestingly, we have been unable to detect Sp1 bound to the either the SMMHC or SM22α G/C repressor within intact chromatin (23, 37). Based on data presented here, it is possible that the repressive effects of Sp1 are due to an increased expression and binding of KLF4 to the G/C repressor and not through a direct binding of Sp1 to this element. It is also possible that KLF4 mediates Sp1-dependent repression of SMC marker genes through a non-G/C-repressor-dependent mechanism since our previous studies showed that the mutation of the G/C repressor in the SM22α promoter only partially blocked PDGF-BB/Sp1-dependent repression (37). The fact that Sp1 increases KLF4 expression and represses SMC marker gene expression raises the possibility that Sp1 may alter the expression of multiple genes that contribute to phenotypic switching. Indeed, Sp1 has been shown to increase the expression of genes involved in promoting SMC proliferation (such as PDGF-A, -B, and -D), migration (such as matrix metalloproteinase-9), and inflammation (such as ICAM-1) (19, 27, 29, 33, 34). In contrast, others have reported an antiproliferative role for Sp1 in SMCs (2, 12). Interestingly, our laboratory recently reported that conditional KLF4 knockout mice displayed both a transient delay in SMC marker gene repression as well as an increased neointimal formation and SMC proliferation following vascular injury (41). Moreover, we have recently shown that siRNA-mediated suppression of KLF4 can reduce the migration of cultured SMCs stimulated by the proatherogenic oxidized phospholipid POVPC (O. Cherapanova and G. K. Owens, unpublished observations). Taken together, it is possible that Sp1, much like KLF4, may play multiple different roles in regulating the SMC phenotype.
Our laboratory has previously reported that PDGF-BB not only increased Sp1 expression but also induced its nuclear localization and phosphorylation, which is reported to increase its transcriptional activity (29, 34, 37). Interestingly, we have also previously shown that the pretreatment with the protein synthesis inhibitor cyclohexamine had no effect on PDGF-BB-induced KLF4 expression (22). This is consistent with our results demonstrating that the binding of Sp1 to the KLF4 promoter and the increased KLF4 expression occurred very rapidly following PDGF-BB treatment of SMCs (within 30–60 min). Taken together, these data strongly suggest that rapid posttranslational modifications of Sp1 may be important for inducing KLF4 expression. Of interest, it has been reported that Sp1 can be phosphorylated by ERK1/2 (4). Moreover, the activation of ERK1/2 has been shown to play a role in the PDGF-BB-induced repression of SMC marker genes through an Elk-1-dependent mechanism (38). Despite this, we found that the inhibition of the ERK1/2 signaling pathway using PD-98059 had no effect on the PDGF-BB-induced upregulation of KLF4 expression (data not shown). Additionally, the inhibitors to JNK (SP-600126), p38 MAPK (SB-203580), and phosphatidylinositol 3-kinase (LY-294002) were also ineffective at blocking the PDGF-BB-induced upregulation of KLF4 expression (data not shown). Additional studies will be needed to determine the signaling pathway(s) required for activating an Sp1-dependent increase in KLF4 expression in SMCs.
Of critical significance, the results from in vivo ChIP assays showed an increased binding of Sp1 to the KLF4 promoter in rat aorta in response to balloon vascular injury (an in vivo model of phenotypic modulation of SMCs). This strongly suggests that the role of a Sp1-mediated increase in KLF4 expression is not an artifact of cell culture or PDGF-BB treatment but may represent a universal mechanism to increase KLF4 expression resulting in the phenotypic modulation of SMCs associated with vascular disease/injury. Although the results from our in vivo ChIP assay using injured aorta coincide with the results of our in vitro ChIP assays using PDGF-BB-treated cultured SMCs, we cannot definitively determine whether the increased binding of Sp1 to the KLF4 promoter following injury occurs exclusively in the SMC component of the aorta. KLF4 is expressed in endothelial cells, although its reported role is anti-inflammatory and thus its expression would be expected to be downregulated (not upregulated) following vascular injury (13). KLF4 is also increased in macrophages in response to proinflammatory stimuli and therefore would be expected to be upregulated in macrophages following vascular injury (9). Therefore, in theory, it is conceivable that Sp1 binding to the KLF4 promoter may be elevated in chromatin isolated from macrophages following vascular injury. However, masswise, the majority of chromatin isolated from an intact aorta is derived from SMCs, not ECs or macrophages. Thus it is likely that the increased binding of Sp1 to the KLF4 promoter following vascular injury seen in our in vivo ChIP assay occurred primarily in SMCs. It will be interesting to determine whether Sp1-mediated increases in KLF4 expression also occurs in other animal models of vascular disease, including naturally occurring and experimentally induced atherosclerosis.
In summary, we provide evidence that KLF4 plays a critical role in the PDGF-BB-mediated repression of SMC marker genes. PDGF-BB-induced increases in KLF4 expression were mediated through a Sp1-dependent mechanism that involves the direct binding of Sp1 to the KLF4 promoter and requires three consensus Sp1 sites. The knockdown of Sp1 or mutation of the Sp1 binding sites within the KLF4 promoter prevented the PDGF-BB-induced activation of KLF4. Sp1 may also play a critical role in increasing KLF4 expression in vivo in response to vascular injury as Sp1 binding to the KLF4 promoter was increased following aortic balloon injury. Further studies are needed to determine whether the Sp1-dependent induction of KLF4 expression is a universal mechanism involved in the phenotypic modulation of SMCs in vascular diseases, such as experimental and naturally occurring atherosclerosis.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute (NHLBI) Grants PO1-HL-19242, R01-HL-38854, and R37-HL-57353 (to G. K. Owens) and American Heart Association Mid-Atlantic Affiliate postdoctoral Fellowship 0725378U (to R. A. Deaton). R. A. Deaton was also supported by NHLBI Training Grant 5T32-HL-0084-30.
Acknowledgments
We acknowledge the expert technical assistance of Rupande Tripathi and Diane Raines.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Krüppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem 275: 37798–37806, 2000. [DOI] [PubMed] [Google Scholar]
- 2.Andres V, Urena J, Poch E, Chen D, Goukassian D. Role of Sp1 in the induction of p27 gene expression in vascular smooth muscle cells in vitro and after balloon angioplasty. Arterioscler Thromb Vasc Biol 21: 342–347, 2001. [DOI] [PubMed] [Google Scholar]
- 3.Banai S, Wolf Y, Golomb G, Pearle A, Waltenberger J, Fishbein I, Schneider A, Gazit A, Perez L, Huber R, Lazarovichi G, Rabinovich L, Levitzki A, Gertz SD. PDGF-receptor tyrosine kinase blocker AG1295 selectively attenuates smooth muscle cell growth in vitro and reduces neointimal formation after balloon angioplasty in swine. Circulation 97: 1960–1969, 1998. [DOI] [PubMed] [Google Scholar]
- 4.Bonello MR, Khachigian LM. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J Biol Chem 279: 2377–2382, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZY, Rex S, Tseng CC. Krüppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr 134: 792–798, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Clowes AW, Reidy MA, Clowes MM. Kinetics of cellular proliferation after arterial injury. I. Smooth muscle growth in the absence of endothelium. Lab Invest 49: 327–333, 1983. [PubMed] [Google Scholar]
- 7.Dandre F, Owens GK. Platelet-derived growth factor-BB and Ets-1 transcription factor negatively regulate transcription of multiple smooth muscle cell differentiation marker genes. Am J Physiol Heart Circ Physiol 286: H2042–H2051, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, Sato H, Kawai-Kowase K, Tanaka T, Maeno T, Okamoto E, Arai M, Kedes L, Kurabayashi M. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler Thromb Vasc Biol 25: 2328–2334, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, SenBanerjee S, Jain MK. Krüppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem 280: 38247–38258, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Ferns GA, Raines EW, Sprugel KH, Motani AS, Reidy MA, Ross R. Inhibition of neointimal smooth muscle accumulation after angioplasty by an antibody to PDGF. Science 253: 1129–1132, 1991. [DOI] [PubMed] [Google Scholar]
- 11.Gan Q, Yoshida T, Li J, Owens GK. Smooth muscle cells and myofibroblasts use distinct transcriptional mechanisms for smooth muscle α-actin expression. Circ Res 101: 883–892, 2007. [DOI] [PubMed] [Google Scholar]
- 12.Gizard F, Amant C, Barbier O, Bellosta S, Robillard R, Percevault F, Sevestre H, Krimpenfort P, Corsini A, Rochette J, Glineur C, Fruchart JC, Torpier G, Staels B. PPAR alpha inhibits vascular smooth muscle cell proliferation underlying intimal hyperplasia by inducing the tumor suppressor p16INK4a. J Clin Invest 115: 3228–3238, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerszten RE, Edelman ER, Jain MK. Krüppel-like factor 4 regulates endothelial inflammation. J Biol Chem 282: 13769–13779, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Hautmann MB, Madsen CS, Owens GK. A transforming growth factor beta (TGFbeta) control element drives TGFbeta-induced stimulation of smooth muscle alpha-actin gene expression in concert with two CArG elements. J Biol Chem 272: 10948–10956, 1997. [DOI] [PubMed] [Google Scholar]
- 15.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev 79: 1283–1316, 1999. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix JA, Wamhoff BR, McDonald OG, Sinha S, Yoshida T, Owens GK. 5′ CArG degeneracy in smooth muscle α-actin is required for injury-induced gene suppression in vivo. J Clin Invest 115: 418–427, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai-Kowase K, Kumar MS, Hoofnagle MH, Yoshida T, Owens GK. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol Cell Biol 25: 8009–8023, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol 292: C59–C69, 2007. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Liang J, Castrillon DH, DePinho RA, Olson EN, Liu ZP. FoxO4 regulates tumor necrosis factor alpha-directed smooth muscle cell migration by activating matrix metalloproteinase 9 gene transcription. Mol Cell Biol 27: 2676–2686, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Libby P, Warner SJ, Salomon RN, Birinyi LK. Production of platelet-derived growth factor-like mitogen by smooth-muscle cells from human atheroma. N Engl J Med 318: 1493–1498, 1988. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Sinha S, Owens G. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem 278: 48004–48011, 2003. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Krüppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem 280: 9719–9727, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Madsen CS, Hershey JC, Hautmann MB, White SL, Owens GK. Expression of the smooth muscle myosin heavy chain gene is regulated by a negative-acting GC-rich element located between two positive-acting serum response factor-binding elements. J Biol Chem 272: 6332–6340, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Madsen CS, Regan CP, Owens GK. Interaction of CArG elements and a GC-rich repressor element in transcriptional regulation of the smooth muscle myosin heavy chain gene in vascular smooth muscle cells. J Biol Chem 272: 29842–29851, 1997. [DOI] [PubMed] [Google Scholar]
- 25.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest 116: 36–48, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84: 767–801, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Pazdrak K, Shi XZ, Sarna SK. TNFα suppresses human colonic circular smooth muscle cell contractility by SP1- and NF-κB-mediated induction of ICAM-1. Gastroenterology 127: 1096–1109, 2004. [DOI] [PubMed] [Google Scholar]
- 28.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, Leitinger N, Owens GK. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circ Res 101: 792–801, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Rafty LA, Khachigian LM. Sp1 phosphorylation regulates inducible expression of platelet-derived growth factor B-chain gene via atypical protein kinase C-ζ. Nucleic Acids Res 29: 1027–1033, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raines EW PDGF and cardiovascular disease. Cytokine Growth Factor Rev 15: 237–254, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J Clin Invest 106: 1139–1147, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakata Y, Xiang F, Chen Z, Kiriyama Y, Kamei CN, Simon DI, Chin MT. Transcription factor CHF1/Hey2 regulates neointimal formation in vivo and vascular smooth muscle proliferation and migration in vitro. Arterioscler Thromb Vasc Biol 24: 2069–2074, 2004. [DOI] [PubMed] [Google Scholar]
- 33.Santiago FS, Khachigian LM. Ets-1 stimulates platelet-derived growth factor a-chain gene transcription and vascular smooth muscle cell growth via cooperative interactions with Sp1. Circ Res 95: 479–487, 2004. [DOI] [PubMed] [Google Scholar]
- 34.Tan NY, Midgley VC, Kavurma MM, Santiago FS, Luo X, Peden R, Fahmy RG, Berndt MC, Molloy MP, Khachigian LM. Angiotensin II-Inducible platelet-derived growth factor-D transcription requires specific Ser/Thr residues in the second zinc finger region of Sp1. Circ Res 102: e38–e51, 2008. [DOI] [PubMed] [Google Scholar]
- 35.Tanizawa S, Ueda M, van der Loos CM, van der Wal AC, Becker AE. Expression of platelet derived growth factor B chain and beta receptor in human coronary arteries after percutaneous transluminal coronary angioplasty: an immunohistochemical study. Heart 75: 549–556, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchida K, Sasahara M, Morigami N, Hazama F, Kinoshita M. Expression of platelet-derived growth factor B-chain in neointimal smooth muscle cells of balloon injured rabbit femoral arteries. Atherosclerosis 124: 9–23, 1996. [DOI] [PubMed] [Google Scholar]
- 37.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. A G/C element mediates repression of the SM22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ Res 95: 981–988, 2004. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428: 185–189, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Wilcox JN, Smith KM, Williams LT, Schwartz SM, Gordon D. Platelet-derived growth factor mRNA detection in human atherosclerotic plaques by in situ hybridization. J Clin Invest 82: 1134–1143, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. Am J Physiol Cell Physiol 292: C886–C895, 2007. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Krüppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res 102: 1548–1557, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]