Polymerizing the fibre between bacteria and host cells: the biogenesis of functional amyloid fibres (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 1.
Summary
Amyloid fibres are proteinaceous aggregates associated with several human diseases, including Alzheimer’s, Huntington’s and Creutzfeldt Jakob’s. Disease-associated amyloid formation is the result of proteins that misfold and aggregate into β sheet-rich fibre polymers. Cellular toxicity is readily associated with amyloidogenesis, although the molecular mechanism of toxicity remains unknown. Recently, a new class of ‘functional’ amyloid fibres was discovered that demonstrates that amyloids can be utilized as a productive part of cellular biology. These functional amyloids will provide unique insights into how amyloid formation can be controlled and made less cytotoxic. Bacteria produce some of the best-characterized functional amyloids, including a surface amyloid fibre called curli. Assembled by enteric bacteria, curli fibres mediate attachment to surfaces and host tissues. Some bacterial amyloids, like harpins and microcinE492, have exploited amyloid toxicity in a directed and functional manner. Here, we review and discuss the functional amyloids assembled by bacteria. Special emphasis will be paid to the biology of functional amyloid synthesis and the connections between bacterial physiology and pathology.
Amyloid: a convergence of diverse proteins along a common folding pathway
The ability of bacteria to interact with their environment is often mediated by the presence of cell-surface organelles composed of protein polymers. These extracellular protein fibres are implicated in diverse processes like locomotion, attachment to surfaces, natural competence and host–pathogen interactions (Fernandez and Berenguer, 2000; Jonson et al., 2005). The extracellular environment can be a harsh locale with little energy available for protein folding, and dynamic physical conditions like hydrophobicity, salt concentrations, pH fluctuations and bombardment with denaturing chemicals. Therefore, assembly of surface structures in the extracellular environment would be predicted to feature proteins that can self-assemble without assistance from cellular chaperones, have low-energy folding requirements and are resistant to denaturation and chemical perturbation.
One protein folding assembly pathway that satisfies these conditions is amyloidogenesis. Amyloid fibril assembly is a hallmark of diverse neurological diseases, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease and bovine spongiform encephalopathy (mad cow disease). Disease-associated amyloid formation is the result of misfolding and aggregation of peptides into structurally conserved fibres (Ross and Poirier, 2004). However, amyloid formation is also integrated as a critical element of cellular physiology, and many organisms direct amyloid fibre formation to accomplish important biological tasks (Fowler et al., 2007). Whether the product of disease-associated protein misfolding or a dedicated assembly pathway, amyloid fibres are characterized by a ‘cross-β strand’ structure, where the β sheets are oriented anti-parallel to the fibre axis (Sunde et al., 1997; Nelson et al., 2005). This structure gives rise to common physical properties. Amyloid fibres are self-assembling protein polymers that are resistant to chemical and temperature denaturation, and to digestion by proteinases (Nordstedt et al., 1994). The β sheet-rich fibres also have similar tinctorial properties: binding to amyloid-specific dyes like Congo red and thioflavin T is diagnostic of amyloid (Elghetany and Saleem, 1988). Amyloids are abundant in bacterial biofilms, and a recent report suggested that up to 40% of the biomass in activated sludge consists of amyloid-like filaments (Larsen et al., 2007; 2008).
Much of our knowledge concerning the highly adaptive amyloid fold comes from studies of eukaryotic protein-misfolding diseases. Each of these diseases shares a common etiology: the conversion of soluble, non-toxic protein into highly ordered and aggregative fibres. Remarkably, the amyloid-forming proteins of different diseases share little to no sequence homology while the fibres themselves converge on a biochemically similar β sheet-rich conformation. Indeed, a common folding pathway characterizes most amyloid fibres where oligomeric intermediates form that give way to the mature β sheet-rich fibres (Fig. 1). Conformation-specific antibodies have been isolated that recognize the transient oligomeric intermediate, but not soluble monomers or mature fibres (Kayed et al., 2003). Conformationally reactive antibodies raised against oligomers from the Alzheimer’s disease-associated amyloid peptide (Aβ) also recognize oligomers of non-related amyloid proteins. This suggests that the amyloid folding pathway is conserved, even among diverse amyloidogenic proteins (Kayed et al., 2003; 2007; Larsen et al., 2007; 2008; Wang et al., 2007; Yoshiike et al., 2007).
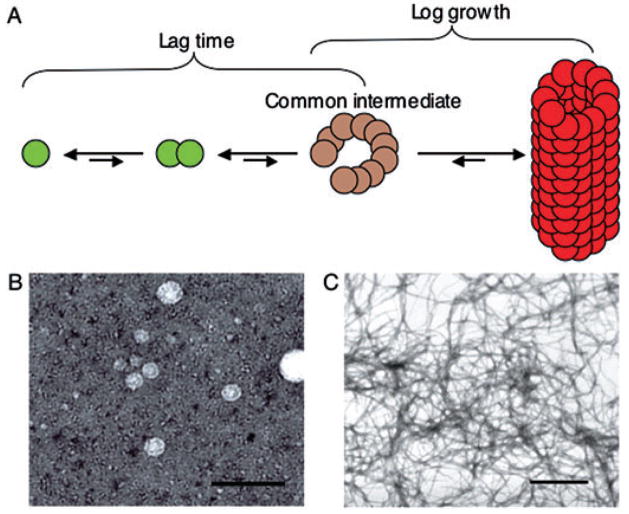
Fig. 1.
The amyloid polymerization pathway proceeds through a common intermediate.
A. Hypothetical amyloid folding pathway. Soluble pre-amyloid peptides (green circles) may accumulate and form a toxic oligomeric common intermediate (tan circles) before the appearance of mature fibres (red circles). Arrow length corresponds to the relative favourability of the folding step.
B. Electron micrograph of biologically active pre-amyloid oligomer formed by the harpin HpaG. Purified HpaG oligomers were negatively stained with uranyl acetate before visualization (Oh et al., 2007). Scale bar is 100 nm.
C. Electron micrograph of negatively stained amyloid fibres generated by polymerization of R1 peptide of CsgA, the major component of curli fibres (Wang et al., 2007). Scale bar is 6 μm.
Evidence is accumulating that suggests that the most toxic species formed by the amyloid folding pathway is the oligomeric intermediate, not the fibres themselves (Caughey and Lansbury, 2003). Further, the conformation of the oligomers as viewed by electron microscopy seems to be quite similar between different amyloid proteins. Indeed, the similar structure of the pre-amyloid oligomers may underlie their common mechanism of toxicity: spherical pre-amyloid oligomers may compromise the integrity of membranes by acting as ion-conducting channels (Caughey and Lansbury, 2003). This model of toxicity has led to the speculation that the mature amyloid fibres may serve a detoxifying function in cells, and so amyloid formation may offer physiological benefits for cells.
Functional amyloids have been identified in mammals, yeast and bacteria (Fowler et al., 2007). In bacteria, these amyloids play diverse physiological roles, from providing the building blocks of the extracellular matrix to mediating toxicity in plants and bacteria – physiological processes that are highly relevant to study in their own right. Functional bacterial amyloids and their disease-associated counterparts converge on a folding pathway that converts soluble proteins to β sheet-rich fibres (Fig. 1). Functional amyloid formation involves a transient oligomeric intermediate species akin to one detected in disease-associated amyloids (Wang et al., 2007), and a pre-amyloid oligomer has been implicated in the functional toxicity of the harpins and microcinE492 (Bieler et al., 2005; Oh et al., 2007). Notably, the properties that are the most exploited, and even beneficial for the organisms that assemble functional amyloids, are precisely those characteristics that are the most detrimental when amyloids form in the context of disease. This review focuses on some of the best-studied bacterial amyloids and the insights on amyloid formation that we can draw from their respective properties. In particular, we examine bacterial amyloids that appear to functionally favour either the mature, non-toxic amyloid fibres (e.g. curli) or the toxic oligomeric intermediate (e.g. MccE492), as well as survey several newly discovered functional bacterial amyloid-like proteins.
Curli amyloid fibres: biology and pathology
Curli are highly aggregative surface fibres assembled by many Enterobacteriaceae, including Escherichia coli (Bokranz et al., 2005). First observed in 1989 in fibronectin-binding E. coli isolates from bovine faecal samples (Olsen et al., 1989), curli fibres have been shown to mediate interactions between individual bacteria, between bacteria and host tissues, and even to inert surfaces that often resist bacterial colonization, like Teflon and stainless steel (Gophna et al., 2001; Zogaj et al., 2003; Ryu et al., 2004; Bokranz et al., 2005; Pawar et al., 2005; Uhlich et al., 2006). Curli stimulate the host inflammatory response and contribute to persistence within the host (Bian et al., 2000; Lawley et al., 2006). Curli are also required for formation of bacterial multicellular communities called biofilms (Olsen et al., 1998; Gophna et al., 2001; Johansson et al., 2001; Uhlich et al., 2002). Curli fibres are 4–6 nm-wide, densely tangled masses of extracellular structures that entangle cells (Fig. 2). Curli, like other amyloid fibres, are resistant to proteinase digestion and are insoluble when boiled in 1% SDS (Olsen et al., 1989; Collinson et al., 1991). Curli fibres display a characteristic β sheet-rich spectrum when analysed by circular dichroism spectroscopy and bind the amyloid-specific dyes Congo red and thioflavin T (Chapman et al., 2002).
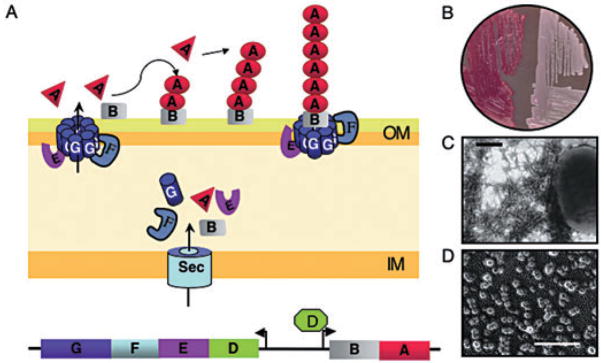
Fig. 2.
A secretion and assembly machine directs amyloid fibre formation in E. coli.
A. Model of curli assembly. Curli biosynthesis requires the products of the divergently transcribed csgBAC and csgDEFG operons. CsgD is a transcriptional activator of the csgBAC operon. CsgB, CsgA, CsgE, CsgF and CsgG have SEC signal sequences that target them across the inner membrane (IM). CsgG, CsgE and CsgF are non-structural proteins that interact at the outer membrane (OM). CsgA and CsgB are secreted across the outer membrane in a CsgG-dependent manner (see text). CsgB interacts with the outer membrane and presents an amyloid-like template to soluble CsgA (red triangles). CsgA adopts the amyloid conformation (red ovals) and becomes anchored to the cell surface, where it can propagate the β sheet-rich amyloid fold onto unpolymerized CsgA monomers. It is unclear if CsgB is anchored directly to the outer membrane or to the assembly complex via a protein–protein interaction (both are shown).
B. Curli fibres bind the amyloid-specific dye Congo red. Curliated bacteria stain red when grown on media containing the diazo dye Congo red (left), but _csg_– bacteria remain white (right).
C. Electron micrograph of curliated bacteria, where the bacteria were grown for 40 h at 26°C prior to analysis (Wang et al., 2007). Scale bar is 1 μm.
D. CsgG is the central component of the curlin secretion complex. Rotary shadow electron micrograph of purified CsgG indicates that it assembles into regular barrel-like structures. Scale bar is 100 nm (Robinson et al., 2006).
The curli biogenesis machine
Curli assembly requires the products of at least two divergently transcribed operons, csgBAC and csgDEFG (Fig. 2). The csgBAC operon encodes the protein subunits of curli fibres, CsgA and CsgB (Arnqvist et al., 1992; Collinson et al., 1996; Johansson et al., 2001; Knobl et al., 2001; La Ragione et al., 2001). A third gene in the operon, csgC, has no described function, nor has a transcript or protein been detected in E. coli. A recent report on AgfC, encoded by the csgC homologue in Salmonella spp., suggested that this protein influences the ultra-structural characteristics of curli (Gibson et al., 2007).
CsgA is secreted from cells in a soluble, unfolded state in the absence of CsgB (Hammar et al., 1996). CsgB, which is associated with the bacterial outer membrane, mediates the conversion of soluble CsgA into insoluble and aggregative cell-surface fibres (Hammar et al., 1996; Bian and Normark, 1997; Chapman et al., 2002; Hammer et al., 2007). Interestingly, CsgA and CsgB need not be expressed from the same cell to display these polymerization activities. Curli fibres assemble on the surface of _csgA_− (CsgB+) cells if grown adjacent to cells secreting CsgA (_csgB_−) (Hammar et al., 1996; Chapman et al., 2002). Interbacterial complementation suggests a model for curli assembly termed ‘nucleation-precipitation’ and it provides an elegant means to control amyloid formation (Fig. 2). By separating the nucleator and polymerization activities into two proteins, the cell can direct amyloid formation by controlling when and where the CsgA and CsgB proteins interact.
The csgDEFG operon encodes non-structural proteins essential for production, stability and secretion of the subunit proteins (Hammar et al., 1995; Loferer et al., 1997; Chapman et al., 2002; Robinson et al., 2006). Efficient secretion and polymerization of CsgA and CsgB requires CsgE, CsgF and CsgG. CsgD is a transcriptional activator of the csgBAC operon, and is also a critical activator of several other operons necessary for biofilm formation, most notably, the cellulose biosynthesis pathway (Brombacher et al., 2003; Gerstel and Romling, 2003; Gerstel et al., 2003). The intergenic region between csgDEFG and csgBAC is one of the largest in E. coli, and the expression of the curli operons is regulated by a complex blend of factors that fine-tune operon expression according to environmental conditions. The interplay between different regulatory factors leads to strain-specific responses to any given condition or set of conditions (Romling, 2005; Barnhart and Chapman, 2006). CsgB, CsgA, CsgE, CsgF and CsgG each have putative Sec translocation sequences that target them for translocation across the cytoplasmic (inner) membrane. CsgG is an outer membrane-localized lipoprotein that is essential for stability and secretion of CsgA and CsgB (Loferer et al., 1997; Robinson et al., 2006). Purified CsgG forms barrel-shaped oligomeric structures (Fig. 2), and its expression is correlated with pore-like properties such as increased sensitivity to erythromycin (Robinson et al., 2006). CsgG must be localized to the outer membrane in order to stabilize CsgA or CsgB, and CsgG variants localized either to the inner membrane or the periplasm are unable to stabilize or secrete the subunits (Robinson et al., 2006). CsgG interacts with a specific domain at the N-terminus of CsgA; this domain can be used to target non-csg proteins to CsgG (Robinson et al., 2006). CsgG has also been demonstrated to co-immunoprecipitate with CsgE and CsgF at the outer membrane. CsgG is sufficient to mediate CsgA secretion out of the cell, but CsgE and CsgF are required for efficient curli fibre assembly under most conditions (Hammar et al., 1995; Chapman et al., 2002). The molecular details of CsgE and CsgF function are unclear, although some clues point to their activities. csgE mutants do not assemble curli fibres, and the stability of the CsgA and CsgB proteins is greatly reduced relative to wild type (Chapman et al., 2002). csgF mutants are deficient in nucleation and exhibit a delay in assembly of curli fibres (Chapman et al., 2002; Hammer et al., 2007). Increased amounts of CsgA are secreted when cells lack CsgF, which suggests that CsgF has a negative effect on CsgA secretion (Chapman et al., 2002; Hammer et al., 2007).
Curli biogenesis is an extremely efficient process, but the mechanism by which assembly is achieved is only beginning to be described. How does the cell prevent internal amyloid assembly (that is, association of CsgA and CsgB prior to their localization to the cell surface)? No accumulation of intracellular intermediates is detected in the absence of CsgG, although the reasons for this remain uncharacterized (Robinson et al., 2006). In addition, the functions of CsgE and CsgF appear to modulate some undefined aspect of CsgG activity, as secretion and stability of CsgA and CsgB are altered in this strain (Chapman et al., 2002; Hammer et al., 2007). Fortunately, E. coli is highly amenable to biochemical and genetic analysis, making possible molecular dissection of nucleation and polymerization.
Molecular dissection of curli nucleation and polymerization
The amino acid sequences of CsgA and CsgB contain three readily identifiable domains: a Sec secretion signal, an N-terminal sequence that, at least for CsgA, targets the protein to CsgG, and an amyloid core domain that is incorporated into amyloid fibres (Collinson et al., 1991; Robinson et al., 2006; Wang et al., 2007). The amyloid core can be further divided into five repeating units. The repeating units are characterized by a fivefold internal symmetry, where each unit is comprised of 19–24 amino acids containing conserved serine, glutamine and asparagine residues (Wang et al., 2007). Interestingly, the CsgA repeating units have different propensities for aggregation, and three of the five units are distinctly amyloidogenic (Olsen et al., 2002; Wang et al., 2007).
CsgA can be readily purified from the culture supernatants of a strain overexpressing both CsgA and CsgG; CsgA is an unstructured, soluble monomer immediately following purification (Chapman et al., 2002). Purified CsgA self-assembles into amyloid fibres several hours after purification (Wang et al., 2007). Amyloid polymerization of CsgA in vitro can be followed in real time by monitoring the fluorescence emitted when the protein is mixed with the amyloid-specific dye thioflavin T (Wang et al., 2007). In vitro kinetic studies on CsgA polymerization suggest that the elongation process can happen in three distinct phases: a lag phase, a fast phase and a stationary phase (Fig. 1). The lag phase of CsgA polymerization can be abolished by adding a small amount of preformed CsgA fibres, or ‘seeds’ (Wang et al., 2007).
The amyloid field has long sought to identify the factor(s) that trigger conversion or nucleation of soluble, pre-amyloid monomers into mature amyloid fibres. The curli system is positioned to lend insight into this process, because CsgB is the only dedicated nucleator protein described. It has recently been shown that the CsgB C-terminus mediates interaction with the outer membrane; truncation of CsgB results in its secretion away from the cells (Hammer et al., 2007). How the C-terminal domain mediates membrane interaction is undefined. The N-terminal domain of CsgB self-assembles into amyloid fibres in vitro and in some conditions can nucleate CsgA into curli fibres in vivo (Hammer et al., 2007). The CsgB N-terminal domain can abolish the lag phase of CsgA polymerization in vitro, suggesting that CsgB-mediated nucleation of CsgA may occur via presentation of an amyloid-like template to soluble CsgA molecules (Hammer et al., 2007).
The ability of CsgA and CsgB to self-assemble into fibres coupled with the ability of the fibres to seed CsgA amyloid polymerization suggests a template-driven mechanism for the assembly of curli fibres (Hammer et al., 2007; Wang et al., 2007). First, CsgA may adopt the amyloid conformation presented to it by CsgB at the cell surface. Second, amyloid CsgA can itself act as a template for the subsequent conversion and incorporation of unpolymerized CsgA monomers to the growing tip of the curli fibre (Fig. 2). It is noteworthy that full-length CsgB does not form fibres in vivo, and thus may be an example of a non-fibre-forming amyloid-like protein. The membrane-associated C-terminus of CsgB may prevent CsgB from polymerizing by anchoring the protein to the membrane, but the exact mechanism underlying this aspect of CsgB behaviour remains to be elucidated. CsgA, on the other hand, seems to be adapted to polymerize into amyloid fibres and yet to only undergo that process, in vivo, when CsgB is available to nucleate it. Elucidation of the molecular details of the CsgB–CsgA interaction and the determinants of CsgA response to nucleation and self-seeding will provide new insights into amyloid assembly.
Amyloid formation of microcin E492: exploitation of toxic oligomers
Microcin E492 (MccE492) of Klebsiella pneumoniae is a toxic, bactericidal protein that assembles into oligomeric pores in the inner membrane of neighbouring bacteria. MccE492, which specifically targets Enterobacter species, is imported across the bacterial outer membrane in a receptor-mediated fashion (Destoumieux-Garzon et al., 2003). MccE492 has been shown to form voltage-independent ion channels in planar lipid bilayers. The molecular weight of MccE492 is 6 kDa, but conductance observed for MccE492 ion channels suggests that an oligomeric species forms this pore (Lagos et al., 1993). The toxicity of the MccE492 peptide changes over the growth cycle of the K. pneumoniae: toxicity is greatest during exponential phase and lowest during stationary phase. No difference in the mass of the peptide, post-translational modification or protein stability can be detected during these different phases (Bieler et al., 2005).
The change in bactericidal activity of MccE492 when cells transition into stationary phase is proposed to be dependent on amyloidogenesis. MccE492 forms amyloid fibres in vivo that correspond to MccE492 toxicity decrease. The assembly of amyloid fibres can be monitored in vitro, and corresponds to a loss of toxicity (Bieler et al., 2005). Therefore, as MccE492 forms toxic oligomeric channels in membranes, the biologically active MccE492 species is presumably a transient pre-amyloid oligomer. A tempting speculation is that the toxic MccE492 species has structural similarity with the toxic, transient oligomeric species formed by disease-associated amyloids. Interestingly, MccE492 has been shown to trigger apoptosis in some human cell lines, reminiscent of the toxic effect observed for disease-associated pre-amyloid oligomers (Hetz et al., 2002). K. pneumoniae produces a small protein localized to the inner membrane, MceB, which confers resistance to MccE492 toxicity (Lagos et al., 1999). The mechanism of MceB-mediated immunity has not been reported.
Emerging bacterial amyloids: harpins, chaplins and MTP
Harpins are virulence factors for plant pathogens such as Xanthomonas, Erwinia and Pseudomonas species. Harpins are substrates of type III secretion, and they elicit ‘hypersensitive response’ in the plant host. Hypersensitive response is the generation of cell death in a localized region of plant tissue – presumably to prevent spread of the pathogen. Hypersensitive response is characterized by changes in ion flux followed by production of reactive oxygen species (Heath, 2000). The mechanism by which harpins induce hypersensitive response is unclear and controversial, although evidence suggests that these proteins may compromise membrane integrity (Lee et al., 2001). A recent report shows that harpins from several species could form amyloid-like fibres in vitro (Oh et al., 2007). Furthermore, harpin amyloid formation can be linked to the hypersensitive response. The amyloid-forming ability of HpaG, a harpin from Xanthomonas, directly correlates with hypersensitive response. A mutant HpaG unable to form amyloid also fails to elicit the hypersensitive response. Interestingly, size exclusion chromatography and visualization by electron microscopy suggests that a tetrameric oligomer is present at the earliest time points that elicit hypersensitive response, and that the oligomer does not form in the non-toxic mutant (Oh et al., 2007). The sequence determinants of HpaG amyloid formation have not yet been fully reported. HpaG possesses a glutamine-rich prion-like domain, but this domain is dispensable for hypersensitive response (Kim et al., 2004).
Streptomyces coelicolor are soil bacteria, akin to filamentous fungi, which produce aerial hyphae to dispense spores. The biogenesis of aerial hyphae is a physical challenge that requires a dramatic change in the surface hydrophobicity of the organism: the subterranean hyphae are hydrophilic while the aerial hyphae and spores are hydrophobic (Elliot et al., 2003). This change in hydrophobicity is a result of the SDS-insoluble ‘surface layer’ on the aerial hyphae and spores. The formation of the surface layer requires a group of proteins called chaplins (Claessen et al., 2003). Chaplins in S. coelicolor are encoded by chpA-H and are necessary for formation of aerial hyphae (Claessen et al., 2003). The chaplins assemble into a network of amyloid fibres in the surface layer; the fibres self-assemble to β sheet-rich fibres in vitro and bind amyloid-specific dyes. The amyloid properties of chaplins might impart a hydrophobic nature to the surface layer and cause a decrease in the surface tension at the air–water interface. The hyphae then could be freed from the soil and continue to grow up into the air (Claessen et al., 2004). The mechanism by which chaplin amyloids assemble and the individual contributions of the eight chaplin proteins to the amyloid network remain largely undefined. Remarkably, addition of crude chaplin-containing cell extract restores formation of aerial hyphae to mutant S. coelicolor filaments lacking some of the chp genes, an observation reminiscent of the interbacterial complementation phenotype of curliated bacteria (Claessen et al., 2003). Further, there may be a temporal regulation influencing the expression of the genes encoding chaplins (Claessen et al., 2003), suggesting that a co-ordinated amyloid biosynthesis pathway is at work in the assembly of chaplin amyloids.
A recent report describes the presence of thin, highly aggregative protein fibres on the surface of Mycobacterium tuberculosis (Alteri et al., 2007). Electron microscopic analysis of the M. tuberculosis pili (MTP) shows fibres that are strikingly similar to curli fibres. MTP are SDS-insoluble and bind Congo red, suggesting that MTP indeed may be an amyloid fibre. The significance of MTP fibres for M. tuberculosis physiology is unknown, but more than half of recently examined tuberculosis patients possess antiserum to MTP, suggesting that MTP production may be part of M. tuberculosis pathogenesis (Alteri et al., 2007).
In conclusion: protein misfolding done right
The discovery of proteinaceous plaques in the brains of deceased dementia patients launched a hundred-year exploration for the factors that mediate the conversion of soluble, non-toxic proteins into aggregative amyloid fibres. Despite this effort, the exact nature of amyloid toxicity and the initiation of amyloid protein aggregation remains unclear. Bacteria exploit amyloid folding pathways to accomplish a variety of tasks, from attachment and colonization to toxicity and pathogenesis. Bacteria apparently utilize the toxic intermediate species (in the cases of MccE492 and the harpins) as well as the mature amyloid fibre (as in curli and the chaplins) in positive ways. The study of bacterial amyloid fibres may hold the key to understanding the methods nature has evolved to harness the power of amyloid formation and direct it to beneficial uses.
Acknowledgments
The authors thank Dr Ingyu Hwang for providing the electron micrograph of HpaG oligomers in Fig. 1. We also gratefully acknowledge members of the Chapman lab, especially Xuan Wang and Neal Hammer, for critical evaluation of this manuscript. The authors were supported by the NIH National Research Service Award T32GMO7544 to E.A.E. and by NIH RO1A1073847 to M.R.C.
References
- Alteri CJ, Xicohtencatl-Cortes J, Hess S, Caballero-Olin G, Giron JA, Friedman RL. Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci USA. 2007;104:5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist A, Olsen A, Pfeifer J, Russell DG, Normark S. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol. 1992;6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- Bieler S, Estrada L, Lagos R, Baeza M, Castilla J, Soto C. Amyloid formation modulates the biological activity of a bacterial protein. J Biol Chem. 2005;280:26880–26885. doi: 10.1074/jbc.M502031200. [DOI] [PubMed] [Google Scholar]
- Bokranz W, Wang X, Tschape H, Romling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Dorel C, Zehnder AJ, Landini P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology. 2003;149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Stokroos I, Deelstra HJ, Penninga NA, Bormann C, Salas JA, et al. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol Microbiol. 2004;53:433–443. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destoumieux-Garzon D, Thomas X, Santamaria M, Goulard C, Barthelemy M, Boscher B, et al. Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol Microbiol. 2003;49:1031–1041. doi: 10.1046/j.1365-2958.2003.03610.x. [DOI] [PubMed] [Google Scholar]
- Elghetany MT, Saleem A. Methods for staining amyloid in tissues: a review. Stain Technol. 1988;63:201–212. doi: 10.3109/10520298809107185. [DOI] [PubMed] [Google Scholar]
- Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LA, Berenguer J. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol Rev. 2000;24:21–44. doi: 10.1111/j.1574-6976.2000.tb00531.x. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid – from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Romling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Park C, Romling U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol. 2003;49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- Gibson DL, White AP, Rajotte CM, Kay WW. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella Enteritidis. Microbiology. 2007;153:1131–1140. doi: 10.1099/mic.0.2006/000935-0. [DOI] [PubMed] [Google Scholar]
- Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci USA. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44:321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- Hetz C, Bono MR, Barros LF, Lagos R. Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proc Natl Acad Sci USA. 2002;99:2696–2701. doi: 10.1073/pnas.052709699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Nilsson T, Olsen A, Wick MJ. The influence of curli, a MHC-I-binding bacterial surface structure, on macrophage–T cell interactions. FEMS Immunol Med Microbiol. 2001;30:21–29. doi: 10.1111/j.1574-695X.2001.tb01545.x. [DOI] [PubMed] [Google Scholar]
- Jonson AB, Normark S, Rhen M. Fimbriae, pili, flagella and bacterial virulence. Contrib Microbiol. 2005;12:67–89. doi: 10.1159/000081690. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Jeon E, Oh J, Moon JS, Hwang I. Mutational analysis of Xanthomonas harpin HpaG identifies a key functional region that elicits the hypersensitive response in nonhost plants. J Bacteriol. 2004;186:6239–6247. doi: 10.1128/JB.186.18.6239-6247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobl T, Baccaro MR, Moreno AM, Gomes TA, Vieira MA, Ferreira CS, Ferreira AJ. Virulence properties of Escherichia coli isolated from ostriches with respiratory disease. Vet Microbiol. 2001;83:71–80. doi: 10.1016/s0378-1135(01)00403-5. [DOI] [PubMed] [Google Scholar]
- La Ragione RM, Coles KE, Jorgensen F, Humphrey TJ, Woodward MJ. Virulence in the chick model and stress tolerance of Salmonella enterica serovar Orion var. 15+ Int J Med Microbiol. 2001;290:707–718. doi: 10.1016/S1438-4221(01)80011-4. [DOI] [PubMed] [Google Scholar]
- Lagos R, Wilkens M, Vergara C, Cecchi X, Monasterio O. Microcin E492 forms ion channels in phospholipid bilayer membrane. FEBS Lett. 1993;321:145–148. doi: 10.1016/0014-5793(93)80096-d. [DOI] [PubMed] [Google Scholar]
- Lagos R, Villanueva JE, Monasterio O. Identification and properties of the genes encoding microcin E492 and its immunity protein. J Bacteriol. 1999;181:212–217. doi: 10.1128/jb.181.1.212-217.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Otzen D, Nielsen PH. Amyloid-like adhesins in floc-forming and filamentous bacteria in activated sludge. Appl Environ Microbiol. 2008;74:1517–1526. doi: 10.1128/AEM.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Klusener B, Tsiamis G, Stevens C, Neyt C, Tampakaki AP, et al. HrpZ (Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc Natl Acad Sci USA. 2001;98:289–294. doi: 10.1073/pnas.011265298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstedt C, Naslund J, Tjernberg LO, Karlstrom AR, Thyberg J, Terenius L. The Alzheimer A beta peptide develops protease resistance in association with its polymerization into fibrils. J Biol Chem. 1994;269:30773–30776. [PubMed] [Google Scholar]
- Oh J, Kim JG, Jeon E, Yoo CH, Moon JS, Rhee S, Hwang I. Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J Biol Chem. 2007;282:13601–13609. doi: 10.1074/jbc.M602576200. [DOI] [PubMed] [Google Scholar]
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Olsen A, Wick MJ, Morgelin M, Bjorck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Herwald H, Wikstrom M, Persson K, Mattsson E, Bjorck L. Identification of two protein-binding and functional regions of curli, a surface organelle and virulence determinant of Escherichia coli. J Biol Chem. 2002;277:34568–34572. doi: 10.1074/jbc.M206353200. [DOI] [PubMed] [Google Scholar]
- Pawar DM, Rossman ML, Chen J. Role of curli fimbriae in mediating the cells of enterohaemorrhagic Escherichia coli to attach to abiotic surfaces. J Appl Microbiol. 2005;99:418–425. doi: 10.1111/j.1365-2672.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 (Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Kim H, Frank JF, Beuchat LR. Attachment and biofilm formation on stainless steel by Escherichia coli O157: H7 as affected by curli production. Lett Appl Microbiol. 2004;39:359–362. doi: 10.1111/j.1472-765X.2004.01591.x. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Uhlich GA, Cooke PH, Solomon EB. Analyses of the red-dry-rough phenotype of an Escherichia coli O157: H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol. 2006;72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich GA, Keen JE, Elder RO. Variations in the csgD promoter of Escherichia coli O157: H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect Immun. 2002;70:395–399. doi: 10.1128/IAI.70.1.395-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiike Y, Kayed R, Milton SC, Takashima A, Glabe CG. Pore-forming proteins share structural and functional homology with amyloid oligomers. Neuromolecular Med. 2007;9:270–275. doi: 10.1007/s12017-007-0003-6. [DOI] [PubMed] [Google Scholar]
- Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]