Role of p54 RNA Helicase Activity and Its C-terminal Domain in Translational Repression, P-body Localization and Assembly (original) (raw)
Abstract
The RNA helicase p54 (DDX6, Dhh1, Me31B, Cgh-1, RCK) is a prototypic component of P-(rocessing) bodies in cells ranging from yeast to human. Previously, we have shown that it is also a component of the large cytoplasmic polyadenylation element-binding protein translation repressor complex in Xenopus oocytes and that when tethered to the 3′ untranslated region, Xp54 represses reporter mRNA translation. Here, we examine the role of the p54 helicase activity in translational repression and in P-body formation. Mutagenesis of conserved p54 helicase motifs activates translation in the tethered function assay, reduces accumulation of p54 in P-bodies in HeLa cells, and inhibits its capacity to assemble P-bodies in p54-depleted cells. Similar results were obtained in four helicase motifs implicated in ATP binding and in coupling ATPase and RNA binding activities. This is accompanied by changes in the interaction of the mutant p54 with the oocyte repressor complex components. Surprisingly, the C-terminal D2 domain alone is sufficient for translational repression and complete accumulation in P-bodies, although it is deficient for P-body assembly. We propose a novel RNA helicase model, in which the D2 domain acts as a protein binding platform and the ATPase/helicase activity allows protein complex remodeling that dictates the balance between repressors and an activator of translation.
INTRODUCTION
The DEAD-box p54 RNA helicase is a member of a helicase SF2/DDX6 subfamily, highly conserved from clams and trypanosomes to humans, with homologues in Xenopus (Xp54), Drosophila (Me31B), Caenorhabditis elegans (CGH-1), and Saccharomyces cerevisiae (Dhh1) (for review, see Weston and Sommerville, 2006; Rajyaguru and Parker, 2009). DEAD-box helicases are involved in splicing, ribosome biogenesis, RNA transport, degradation, and translation, although their precise contribution to most of these processes is not known. Their catalytic core is composed of two RecA-like domains with nine conserved domains, including the eponymous DEAD motif and the more recently identified Q motif, with roles in catalysis and substrate binding. Helicases are understood to use NTP (usually ATP) binding and hydrolysis to remodel RNA, or RNA–protein complexes, resulting in double-stranded RNA unwinding, and/or in displacement of proteins from RNA (for review, see Cordin et al., 2006; Bleichert and Baserga, 2007; Pyle, 2008).
The p54 family of RNA helicases is particularly remarkable because of its critical involvement in widespread biological processes, including sexual development of the protozoan Plasmodium, regulation of multiple virulence-associated genes in Cryptococcus neoformans, germline apoptosis, and embryonic cytokinesis regulation in C. elegans and viral replication (for review, see Weston and Sommerville, 2006; Beckham and Parker, 2008; Rajyaguru and Parker, 2009).
The unifying themes of its action at the molecular level encompass roles in translational repression (Minshall et al., 2001, 2007; Nakamura et al., 2001; Minshall and Standart, 2004; Coller and Parker, 2005), including that mediated by microRNAs (Chu and Rana, 2006), enhancing decapping (Coller et al., 2001), and being a component of very large ribonucleoprotein (RNP) complexes. In Xenopus oocytes, Xp54 helicase is a component of 2- to 3-MDa RNP that contain cytoplasmic polyadenylation element-binding protein (CPEB), a well characterized regulator of translation in early development and in neurons. CPEB has a dual role—it represses translation in the oocyte and activates translation via cytoplasmic polyadenylation in meiotically matured eggs (Richter, 2007; Radford et al., 2008). The large CPEB RNP repressor complex contains, in addition to p54, Pat1; Rap55; the eukaryotic initiation factor (eIF) 4E-binding protein 4E-T; and eIF4E1b, a close homologue of the canonical cap-binding protein (Minshall et al., 2007). We and others have found that tethering of some of these proteins, including Xp54, Rap55, and 4E-T, to the 3′ untranslated region (UTR) of reporter mRNA represses its translation in Xenopus oocytes (Tanaka et al., 2006; Minshall et al., 2007). Interestingly, all known components of the CPEB complex are also found in P-(rocessing) or GW-bodies.
P-bodies are distinct cytoplasmic granules that are sites of reversible mRNA repression and mRNA decay. In addition to mRNAs and microRNAs, they contain RNA decay enzymes responsible for decapping, deadenylation, and 5′-3′ degradation, such as Dcp1/2, Ccr4, and Xrn1, factors that enhance decapping (some of which have also been shown to act as translational repressors), and this class includes p54, Pat1, Lsm1–7, and Edc3, proteins involved in microRNA-mediated silencing such as Argonautes and GW182; other translational repressors including CPEB, Rap55, and Y-box proteins; and eIF4E and 4E-T. However, they do not contain other initiation factors or ribosomal proteins (for reviews, see Eulalio et al., 2007a; Parker and Sheth, 2007). Recent studies indicate that formation of microscopically visible P-bodies is a consequence, not the cause, of gene silencing, implying that mRNAs may be repressed initially by simple association with the factors in the absence of aggregation (Decker et al., 2007; Eulalio et al., 2007b). In mammalian cells, several P-body components seem to be involved in P-body assembly because their depletion by RNA interference leads to their disappearance, including Lsm4, GW182, and CPEB1. However, P-bodies can be readily reinduced by arsenite treatment in the absence of these proteins, indicating that they are not involved in the mechanism of assembly per se (Kedersha et al., 2005; Serman et al., 2007). One exception is p54, whose depletion triggers P-body disappearance and prevents their induction by arsenite (Serman et al., 2007). Although this clearly indicates a central role of p54 in the mechanism of P-body assembly, it is not known whether this reflects its function as a translational repressor, decapping enhancer, RNA helicase, or a combination. In yeast, the p54 homologue Dhh1, not alone but in combination with Pat1, also acts in P-body assembly after glucose deprivation (Coller and Parker, 2005).
We set out to investigate the role of the p54 RNA helicase in the regulation of gene expression by focusing on three major aspects: its ability to repress translation when tethered to reporter RNA and its binding to partner proteins in Xenopus oocytes, as well as its ability to localize and assemble P-bodies in mammalian cells.
METHODS AND MATERIALS
p54 Mutagenesis and Cloning
MS2-Xp54 wild type and DQAD (E246Q) and HRIGQ (R423Q) mutants were described previously (Minshall et al., 2001). DILAAAA (R138A/K140A) and AAA (S276A/T278A) mutations were introduced using a Stratagene (La Jolla, CA) QuikChange site-directed mutagenesis kit with double-stranded oligonucleotide 1 and 2, respectively (Supplemental Table 1). The MS2-Xp54-D1 (amino acids [aa] 1-301) was cloned by insertion of a stop codon over aa 302 followed by an NheI site by using site-directed mutagenesis with oligonucleotide 3. This was digested with NheI and religated to create the MS2-Xp54-D2 (aa 304-481). The HRIGQ mutation was subsequently incorporated into the D2 domain as described for full-length Xp54.
The expression vector for RFP-tagged p54 contains the full open reading frame of human p54 followed by the first 824 nt of 3′UTR sequence, cloned in DsRed2-C1 plasmid (Clontech, Mountain View, CA). DILAAAA, DQAD, and HRIGQ mutations were created as described above using oligonucleotide 4, 5, and 6, respectively (Supplemental Table 1). For expression after si-p54 transfection, the 3′UTR was removed from these plasmids by PCR. In RFP-p54-D1, p54 sequence stops at an internal EcoRI restriction site, so that the last p54 aa is M288. In RFP-p54-D2, p54 sequence starts at M288.
MS2 Tethering
The MSP vector and MS2-ePAB, Fluc-MS2, and CSFV-Fluc-MS2 reporter cDNAs were supplied by Nicola Gray (Gray et al., 2000; Gorgoni et al., 2005). MS2 tethering using nonadenylated Fluc-MS2 reporter mRNA and a control Rluc mRNA was performed as described previously, with lysates from five pools of five to 10 oocytes for each mRNA assayed by the Dual-Luciferase reporter assay system (Promega, Madison, WI) (Minshall et al., 2001; Minshall and Standart, 2004).
Immunoprecipitation and Western Blot Analysis
Immunoprecipitation experiments using lysates prepared from oocytes injected with MS2-fusion protein mRNAs, with mouse monoclonal MS2 antibodies and protein G-Sepharose were described previously (Minshall et al., 2007).
Xenopus protein samples were separated by 10 or 15% SDS-polyacrylamide gel electrophoresis gels and then used for silver staining or Western blot analysis using enhanced chemiluminescence (Minshall et al., 2007) or the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). Antibodies included mouse monoclonal CPEB (Wilczynska et al., 2005) and rabbit antibodies against Xp54 (Minshall and Standart, 2004), ePAB (Voeltz et al., 2001), RAP55 (Pepling et al., 2006), XPat1 (Minshall et al., 2007), and human eIF4E1 (Minshall et al., 2007). HeLa cell lysates were separated on a NuPage 4–12% gel (Invitrogen, Carlsbad, CA), transferred to a nitrocellulose membrane (Optitran Ba-S83; Whatman France, Versailles, France), and analyzed with rabbit polyclonal anti-p54 (Bethyl Laboratories, Montgomery, TX) and mouse monoclonal anti-actin (clone AC-40; Sigma-Aldrich, St. Louis, MO) antibodies.
Cell Culture and Transfection
Epithelioid carcinoma HeLa cells were routinely maintained in DMEM, supplemented with 10% fetal calf serum. Cells were plated on 20-mm glass coverslips 1 d before transfection. Transient transfections were performed with 1.5 μg of plasmid DNA or 3 μg of small interfering RNA (siRNA) per 35-mm-diameter dish by a standard calcium phosphate procedure.
The si-p54 sequence was GGAACUAUGAAGACUUAAAdTdT. Control siRNA was a globin siRNA described previously as si-Glo.1 (Serman et al., 2007).
Immunofluorescence
Immunofluorescence was performed as described previously (Ernoult-Lange et al., 2009) by using mouse monoclonal anti-Ge1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) (Stoecklin et al., 2006) on a Leica DMR microscope (Leica, Heidelberg, Germany), with a 63× 1.32 oil immersion objective. Photographs were taken using a Micromax charge-coupled device camera (Princeton Scientific Instruments, Monmouth Junction, NJ) driven by MetaMorph software (Molecular Devices, Sunnyvale, CA).
For the scoring of cells with P-bodies, 100 transfected cells were blindly scored for the presence or absence of P-bodies by visual examination under the 63× objective.
RESULTS
Mutation of Tethered p54 Activates Translation
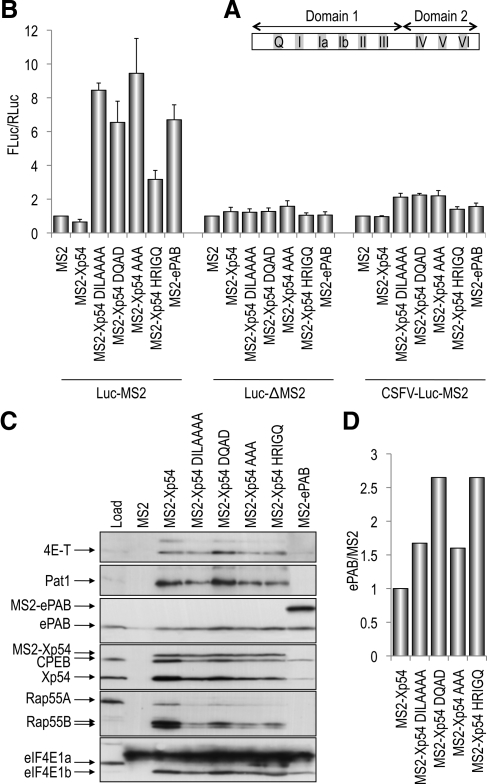
p54 RNA helicase in common with other SF2 DEAD box family members is composed of two linked RecA-like domains, domains 1 and 2. Domain 1 contains the Q motif followed by motifs I–III, and domain 2 contains motifs IV–VI (Figure 1A). Both domains participate in ATP binding (Q, I, II, V, and VI) and RNA binding (Ia, Ib, IV, and V), with motif III forming intramolecular interactions (Cordin et al., 2006).
Figure 1.
Mutagenesis of p54 motifs I, II, III, and VI activate translation in a tethered function assay. (A) Schematic cartoon of p54 RNA helicase. The two RecA-like domains are indicated, as well as the disposition of the conserved motifs Motif Q and I–VI within each domain. Not to scale. (B) Effects of tethered p54 on reporter mRNA translation. mRNAs encoding MS2-fusion proteins as indicated were injected into oocytes, which after 5 h, were reinjected with control Renilla luciferase mRNA and m7GpppG-capped, nonadenylated firefly luciferase-MS2 mRNA (Luc-MS2), or m7GpppG-capped, nonadenylated firefly luciferase mRNA lacking MS2 binding sites in the 3′UTR (Luc-ΔMS2) or ApppG-capped, nonadenylated CSFV firefly luciferase-MS2 mRNA (CSFV-Luc-MS2). Luciferase activities were normalized to MS2. (C) Inactivation of p54 RNA helicase alters the pattern of interacting proteins. Oocytes were injected with MS2-fusion protein mRNAs as indicated, and lysates were immunoprecipitated with MS2 antibodies, and the bead-bound proteins analyzed by Western blotting with indicated antibodies. Load represents the input oocyte lysate from uninjected cells. (D) Western blots were probed with anti-ePAB and anti-MS2 antibodies and relative quantities were analyzed on the Odyssey infrared imaging system. Ratios of ePAB and MS2 were calculated and normalized to MS2-Xp54. Similar data were obtained in an independent experiment.
To assess the importance of the conserved p54 helicase motifs in translational repression, we performed MS2 tether function assays, in which mRNAs encoding bacteriophage MS2-p54 fusion proteins are coexpressed with a firefly luciferase mRNA with three MS2 binding sites in its 3′UTR. Altogether, we mutated single or multiple residues in four conserved helicase domains: I, II, III, and VI. Motif II (DEAD - > DQAD) and VI (HRIGR -> HRIGQ) mutations abrogate ATP binding, RNA binding, and hence helicase activity in several well characterized RNA helicases, including eIF4A and Ded1 (Pause and Sonenberg, 1992; Pause et al., 1993; Cordin et al., 2006). In eIF4A, mutation of motif III (SAT -> AAA) causes a dramatic loss of helicase activity, although only minor effects on ATP binding, ATP hydrolysis, and RNA binding are observed, and it is thus said to uncouple ATPase and helicase activity (Pause and Sonenberg, 1992; Cordin et al., 2006). eIF4A DQAD, HRIGQ, and AAA mutant proteins are all dominant negative in translation in vitro (Pause and Sonenberg, 1992; Pause et al., 1993). In Dhh1, the motif I mutation (DILARAK -> DILAAAA) had reduced RNA binding and a lethal growth phenotype (Cheng et al., 2005).
Xenopus oocytes were injected with mRNA encoding MS2 alone and wild-type and mutant MS2-Xp54 helicases followed by a second injection, several hours later, of m7GpppG capped and nonpolyadenylated firefly luciferase mRNA, and the control Renilla luciferase mRNA. Lysates prepared from pools of injected oocytes were first checked for the level of expression of MS2 fusion proteins, found to be approximately constant (Supplemental Figure S1). Normalizing the ratio between firefly and Renilla enzyme to 1 in the coinjection of MS2 alone, we show that tethering wild-type Xp54 represses translation approximately twofold, whereas DQAD, HRIGQ, AAA, and DILAAAA mutations all activate translation three- to ninefold (Figure 1B; Minshall et al., 2001). The degree of stimulation of translation by mutant Xp54 proteins is comparable with that of ePAB and PABP (Figure 1B; data not shown), well characterized activators of translation when tethered on nonpolyadenylated reporter mRNA in Xenopus oocytes (Gray et al., 2000; Wilkie et al., 2005). Both translational repression and activation by Xp54/PABP proteins require MS2 binding sites in the firefly mRNA 3′UTR (Figure 1B), and they do not reflect changes in reporter mRNA levels (Supplemental Figure S2).
To address whether repression/activation was dependent on the cap, we used a reporter mRNA which contained the classical swine fever virus (CSFV) internal ribosome entry sequence (IRES) upstream of firefly luciferase, and which was capped with ApppG. In Xenopus oocytes, ApppG-CSFV-luciferase mRNA is translated as robustly as m7GpppG-luciferase mRNA. However, neither translational repression nor activation of reporter mRNA by tethered p54 proteins/ePAB is supported by internal initiation mediated by the CSFV IRES (Figure 1B). Initiation on the CSFV IRES is determined by its ability to bind independently to 40S subunits and eIF3, and it does not require eIF4E, eIF4G, eIF4A, eIF4B, eIF1, and eIF1A (Pestova et al., 1998). Our results are in contrast to those obtained when translation assays are performed in vitro, which suggest a cap-independent manner of translation inhibition by p54 helicases (Coller and Parker, 2005).
We conclude that inactivating mutations in four conserved motifs in tethered p54 helicase robustly activate translation of reporter mRNA in oocytes, in a cap-dependent manner; in contrast to the wild-type protein, which represses translation.
Activation of Translation by Tethered Mutant p54 Correlates with an Imbalance of Protein Partners
To assess the effect the helicase motif mutations may have on interacting partners of p54, we coimmunoprecipitated binding partners in Xenopus oocytes. We showed previously that ectopically expressed Xp54 interacts with CPEB (Minshall and Standart, 2004). More recently, using both coimmunoprecipitation and gel filtration assays, we demonstrated that CPEB interacts with endogenous Xp54, and with additional RNA-binding proteins, including Pat1 and Rap55B (Minshall et al., 2007). Here, we extend our analysis and show that MS2-Xp54, and endogenous CPEB, also interact with ePAB (Supplemental Figure S3), the maternal/embryonic form of the somatic poly(A)-binding protein PABP1 (Voeltz et al., 2001), in agreement with Kim and Richter (2007).
Oocytes were injected with MS2-Xp54 (wild-type and mutant form) mRNAs and allowed to synthesize proteins before the preparation of lysates for immunoprecipitation with MS2 antibodies and subsequent Western blot analysis. Ectopically expressed wild-type MS2-Xp54 interacted with self, Pat1, CPEB, Rap55B, ePAB, and eIF4E1b. These interactions were due to Xp54 as MS2 alone did not significantly interact with any of the CPEB complex proteins (Figure 1C). When immunoprecipitated mutant p54 helicases were compared with the wild-type protein, the mutations were noted to alter the pattern of binding partners, consistently seen in several experiments (data not shown). One of the clearest effects was observed in Xp54 DQAD, which showed enhanced binding to Pat1, 4E-T, and eIF4E1b and modestly reduced binding to CPEB, endogenous Xp54, and more reduced binding to Rap55 (Figure 1C and Supplemental Figure S3). Xp54-DILAAAA and AAA proteins, in contrast, had approximately wild-type levels of bound 4E-T with reduced levels of bound Pat1, CPEB, Xp54, and eIF4E1b, and very much reduced levels of Rap55. Xp54-HRIGQ interacted with its partners in a similar way to the previous two mutant helicases, except that binding to endogenous Xp54 was further reduced though the levels of bound eIF4E1b were approximately the same as to wild-type MS2-Xp54 (Figure 1C). Interestingly, common to all mutations is a relative increase in the ePAB/MS2-Xp54 ratio compared with the wild-type protein. Acknowledging that Western blotting is at best semiquantitative, we scanned two representative silver-stained gels containing the immunoprecipitation reactions (Supplemental Figure S1) and performed densitometry on the ePAB and MS2-p54 proteins. All mutations increased the balance between a bound activator, ePAB, and Xp54, approximately twofold, relative to wild-type MS2-Xp54, compatible with their activation of translation in the tethered assay (Figure 1B). We confirmed the densitometry data with a quantitative western Odyssey infrared imaging system, by using ePAB and MS2 antibodies. The Odyssey data also indicated a modest but consistent enhanced binding of ePAB by mutant helicases, relative to wild-type MS2-Xp54 (Figure 1D).
Mislocalization of p54 Helicase Mutants in HeLa Cells
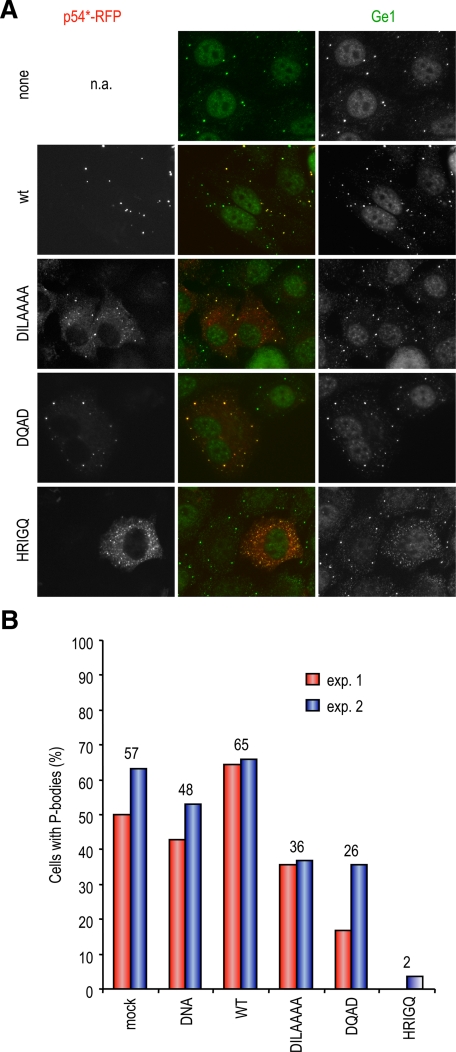
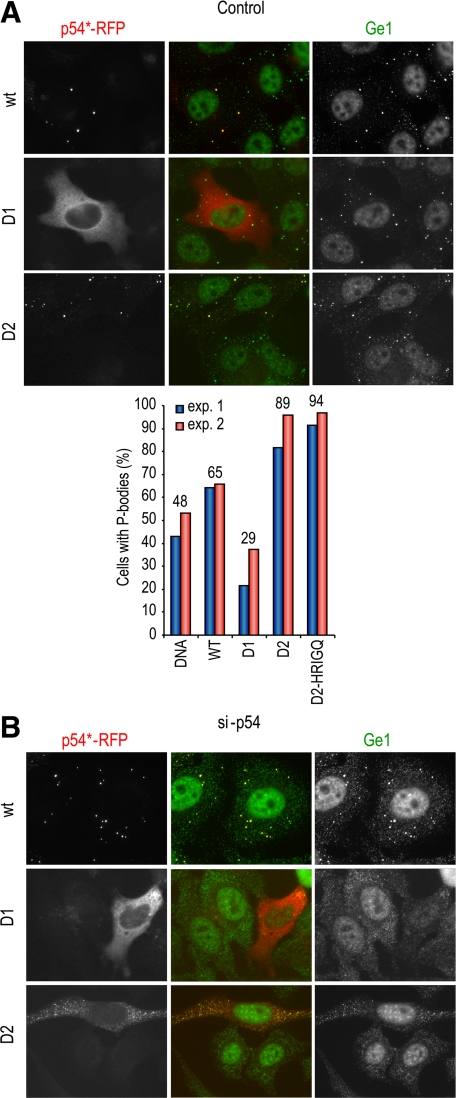
To assess the effect of inactivating p54 helicase activity on its ability to localize to P-bodies, wild-type and mutant human p54/rck helicases fused to RFP were expressed in HeLa cells, and endogenous P-bodies were monitored by immunostaining for Ge-1, a P-body component (Yu et al., 2005). Before transfection, approximately half of the cells contained distinct P-bodies, with the remaining cells showing only a diffuse Ge-1 staining (Figure 2A, top). After transfection, RFP-p54 was strongly concentrated in P-bodies (Figure 2A) or spread throughout the cytoplasm in cells devoid of P-bodies, as observed for endogenous p54 in untransfected cells (data not shown). When mutations in motifs I (DILAAAA) and II (DQAD) were introduced, the mutant protein was still able to localize in P-bodies. However, a significant portion of the protein remained diffuse outside of P-bodies (Figure 2A), indicating a partial defect in P-body localization. When motif VI (HRIGQ) was mutated, the mutant protein had a fine granular localization in most cells, and Ge-1 staining indicated the near complete disappearance of distinct P-bodies in these transfected cells. The same result was obtained when looking at a second P-body marker, Rap55 (data not shown). This p54 mutant was therefore a potent inhibitor of P-body assembly. To evaluate such an effect of the other mutants, we counted for each construct the percentage of transfected cells with normal size P-bodies. The data of two independent experiments are shown in Figure 2B. On average, 57 and 48% of cells contained P-bodies after mock transfection and transfection with a control DNA (sperm DNA), respectively. Wild-type RFP-p54 increased the number of cells with P-bodies to 65%, in agreement with a previous report (Chu and Rana, 2006). DILAAAA and DQAD mutations decreased this number to 36 and 26%, respectively, whereas HRIGQ mutation decreased it to 2%. Although the extent of decrease observed with DILAAA and DQAD mutants was variable from one experiment to the other, as illustrated for the DQAD mutant (Figure 2; data not shown), the effect of the HRIGQ mutation was always near complete. Therefore, in addition to a partial defect in P-body accumulation, the two former mutants have a partially dominant-negative effect on P-body assembly, whereas the latter is a drastic dominant negative mutant.
Figure 2.
Localization of p54 mutants with respect to P-bodies. (A) Mislocalization of mutant p54. HeLa cells were transfected with expression vectors for RFP-tagged wild-type (wt) and mutant human p54, as indicated. After 40 h, cells were fixed, P-bodies were stained with anti-Ge1 antibodies, and cells were observed by fluorescence microscopy. (B) Down-regulation of preexisting P-bodies by mutant p54. Mock-transfected cells and cells transfected with the various p54 constructs were examined for the presence of normal size P-bodies in the experiment described above. The graph represents the percentage of cells containing normal size P-bodies in two representative experiments (exp. 1 and 2). The average percentage for each construct is indicated above.
p54 Helicase Mutants Cannot Assemble P-Bodies
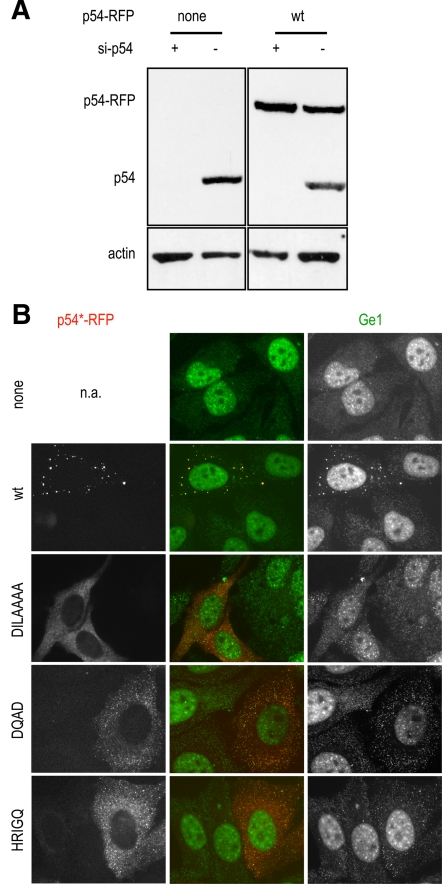
Next, we investigated the capacity of mutant p54 to assemble P-bodies. To this aim, p54 mRNA was first knocked down by RNA interference using a siRNA targeting its 3′UTR. This resulted in efficient p54 depletion (>90%), as judged by Western blot analysis (Figure 3A), and in P-body disappearance, as attested by Ge-1 immunostaining (Figure 3B, top). Twenty-four hours after siRNA transfection, cells were transfected with p54 plasmids lacking the 3′UTR sequence and therefore insensitive to the si-p54 (Figure 3A, right). The wild-type p54 helicase fully complemented the depleted protein and restored P-bodies in most cells, whereas all mutants were defective in P-body assembly (Figure 3B). Only the DILAAAA mutant could occasionally assemble a few Ge1-containing granules (6% of the cells; data not shown).
Figure 3.
P-body assembly by p54 mutants. HeLa cells were successively transfected with si-p54 to deplete endogeneous p54, and, 24 h later, with expression vectors for RFP-tagged wt and mutant p54, as indicated. After 40 h, p54 proteins were analyzed by Western blot (A), and P-bodies were analyzed as described in Figure 2A (B).
Altogether, all mutations alter localization in pre-existing P-bodies and P-body assembly, although their behavior is different in the two assays. Mutations in motifs I and II moderately decrease p54 accumulation in preexisting P-bodies, indicating that the mutant proteins are still able to bind P-body components. In addition, these mutants lead to the partial down-regulation of P-bodies. In contrast, the mutation in motif VI leads to a severe phenotype with a strong dominant negative effect on preexisting P-bodies. This dominant-negative effect over endogenous p54 suggests that the mutant protein sequesters a factor required for P-body assembly, whether endogenous p54 or another partner. Finally, none of the mutants is able to assemble bona fide P-bodies. However, they often assembled numerous tiny granules, which contained Ge-1, but were much smaller than P-bodies. Whether they could support the same function as full size P-bodies is unknown. Nevertheless, it is clear that the ATPase/helicase function is required for normal size P-body assembly.
The C-Terminal Domain of p54 Helicase Alone Is Sufficient for Translational Repression, Binding of Partner Proteins in Xenopus Oocytes, and Accumulation in P-Bodies
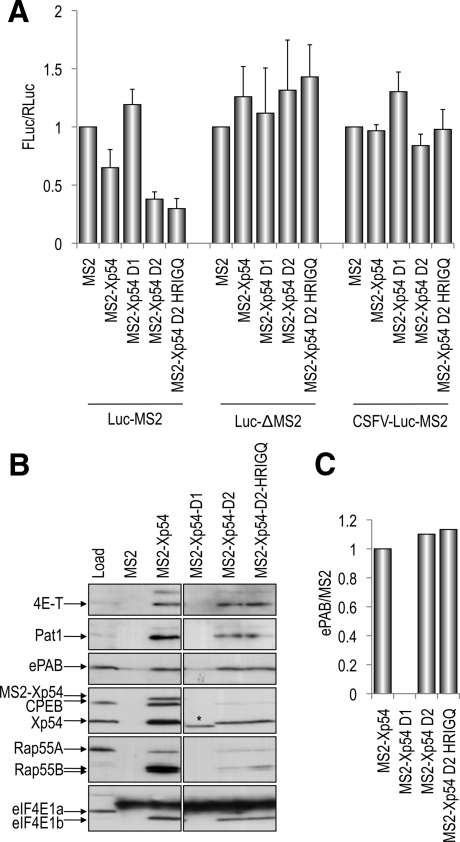
Next, we examined the effect of dividing p54 into its two folded domains in tethering, protein binding, and localization assays. Mutations were introduced into the linker regions of the Xenopus and human cDNAs to enable the fusion to MS2 or RFP as appropriate of the N-terminal (D1) and C-terminal (D2) domains of p54 (Figure 1A). First, in the tethering assay we observed that p54-D1 had no effect on reporter mRNA, like MS2 alone, whereas, surprisingly, p54-D2 repressed as well as the full-length protein (Figure 4A). As with the full-length protein, repression by p54-D2 required 3′UTR MS2 binding sites in a capped reporter mRNA (Figure 4A) and did not reflect changes in reporter RNA levels (Supplemental Figure S2). In the MS2 coimmunoprecipitation assay, p54-D1 did not bind any of the tested repressor complex proteins, whereas, in contrast, p54-D2 interacts with all the partners of full-length MS2-Xp54, albeit to differing extents. This striking difference was not due to varying levels of expression of the two halves of the protein (Supplemental Figure S1). p54-D2 interacted with 4E-T and ePAB to about the same extent as the full-length p54 protein, whereas binding relatively less Pat1, Xp54, and eIF4E1b, and with very reduced binding to Rap55 and CPEB (Figure 4B). Quantitating the binding of ePAB to full-length and truncated MS2-Xp54 proteins by using the Odyssey system showed that p54 D2 binds ePAB in similar proportions to the full-length helicase (Figure 4C), in contrast to the mutant helicases that bind more ePAB (Figure 1D).
Figure 4.
The role of p54 domain 1 and 2 in translational repression. (A) The p54 D2 domain represses translation when tethered as well as the full-length protein. Tethering assays were performed as described in Figure 1, with MS2, MS2-Xp54 full-length, and MS2-Xp54–D1 and MS2-Xp54-D2 encoding mRNAs. (B) The p54 D2 domain interacts with components of the translation repressor complex. Binding assays were performed as described in Figure 1C, by using oocytes injected with MS2, MS2-Xp54 full-length and MS2-Xp54 –D1and MS2-Xp54-D2 mRNAs, and subsequent immunoprecipitation with MS2 antibodies. The asterisk (*) indicates the MS2-Xp54 D2 protein. (C) Western blots were analyzed using the Odyssey system as described in Figure 1D.
When transiently transfected into HeLa cells, the RFP-p54-D1 protein distributed homogenously in the cytoplasm and was absent from P-bodies (Figure 5A). Its expression decreased the number of cells with P-bodies to 26% (Figure 5A, right), indicating a dominant-negative effect similar to the one of DILAAAA and DQAD mutants (Figure 2B). By contrast, RFP-p54-D2 was highly concentrated in P-bodies such as full-length p54. In addition, it strongly enhanced the number of cells with P-bodies to 89%, versus 65% for cells transfected with wild-type p54. After the depletion of endogenous p54, the D1 domain predictably did not assemble P-bodies (Figure 5B). Surprisingly, despite its enhancing effect on preexisting P-bodies, the D2 domain was also unable to assemble P-bodies.
Figure 5.
The C-terminal domain of p54 helicase alone is sufficient for accumulation in P-bodies but not for their assembly. (A) Localization of p54 domains 1 and 2. HeLa cells were transfected with indicated p54 constructs, in the presence of endogenous p54 protein, as described in Figure 2. The percentage of cells containing normal size P-bodies is plotted on the right panel. (B) Activity of p54 domains 1 and 2 in P-body assembly. Indicated constructs were transfected in cells depleted of endogenous p54 by si-p54 transfection, as described in Figure 3.
Next, we tested the effect of the HRIGQ mutation in the p54-D2 protein. We found that this mutation, the only mutation that resides in the C-terminal D2 domain, had no effect on D2 in tethering, binding, or localization assays (Figures 4 and 5A, right). In all three assays, the behavior of D2 and D2-HRIGQ proteins was indistinguishable. Thus, the mutation that in the context of the full-length protein, activated translation, dispersed endogenous P-bodies and had an altered balance of partner proteins, was inactive in the absence of the N-terminal moiety of the protein.
Altogether, we have shown that the D2 domain of p54 is sufficient to perform the function of the full-length protein in terms of translational repression and P-body localization. However, it is inactive in P-body assembly.
DISCUSSION
We propose a model for p54 helicase based on three principal sets of observations. First, mutagenesis of conserved p54 helicase motifs activates translation in the tethered function assay, reduces accumulation of p54 in P-bodies, and inhibits its capacity to assemble P-bodies in HeLa cells. Similar results were obtained in the case of four helicase motifs, I, II, III, and VI, implicated in ATP binding and in coupling ATPase and RNA binding activities. Strikingly, we also show that the C-terminal Rec A-like D2 domain of p54 alone, although presumably devoid of ATPase activity, is sufficient for translational repression and complete accumulation in P-bodies. However, it is deficient for P-body assembly. Last, we document changes in interactions with protein partners, in response to mutations or truncations, which presumably underlie the functional differences observed in translation and localization assays.
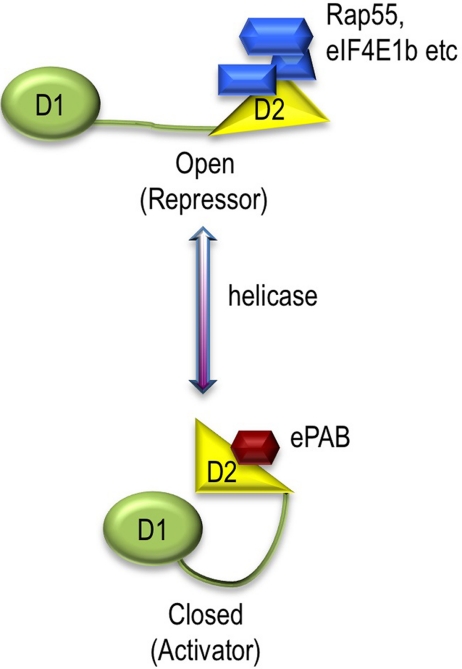
In a primary model (Figure 6), the full-length helicase would have two conformations—open and closed—modulated by ATP hydrolysis/helicase activity. Within the full-length protein, the repressor complex would bind to the D2 domain, provided that the protein is in the open conformation. Mutations inhibiting ATPase activity would prevent opening of the helicase, and hence efficient binding of this complex. In contrast, the D2 domain expressed alone assembles a complex that is sufficient for translational repression. We are aware that this property of the D2 domain was observed in the presence of endogenous p54, but because Xp54 is a very abundant protein, it is not possible to deplete it in the nondividing oocytes. However, full-length mutants bind similar amount of endogenous Xp54 to D2 and yet have the opposite effect on translation. We propose therefore that in addition to the D2 platform, the ATPase/helicase activity of p54 is required for allowing opening/closing dynamics necessary for recycling the repressor complex. This model is strengthened by our observation that the HRIGQ mutation in the C-terminal motif VI, although strongly dominant negative in the full-length protein, has no effect on translational repression when harbored in the D2 domain.
Figure 6.
Model for p54 RNA helicase function. See text.
Our analysis of p54 partners indicates that helicase mutations reduce, to varying extents, depending on the partner and on the mutation, binding of repressor complex proteins in oocytes, and enhance binding of an activator, ePAB. In a modification of the initial model, p54 helicase activity would be required to allow efficient binding of repressor proteins, and, concomitantly, reduced interaction with ePAB. Translational activation by helicase mutants would result from an altered balance between bound repressor and activator partner proteins. The behavior of the D2 domain of p54 helicase, which can functionally replace the full-length protein in tethering assays, can also be accounted for by this model. Interactions with several repressor proteins is reduced, but there is no concomitant increase in binding of ePAB, compared with the full-length protein, overall resulting in a balance of factors still tipped toward repression. Importantly, though, no helicase activity is apparently involved in maintaining the repressor complex. At this stage, our analysis of the p54 complex may be still incomplete. By monitoring altogether seven oocyte p54 partners, we have revealed complex and subtle changes, depending on mutations and truncations, which require further investigation. In addition, we acknowledge that there may be further p54 regulatory factor(s), although from examining the silver-stained gels of coimmunoprecipitation reactions (Supplemental Figures S1 and S3), it is evident that this putative additional/alternative factor is not abundant. Moreover, we cannot exclude the possibility that the mutations in p54 helicase influence the manner in which it interacts with mRNA, in addition to effecting changes in protein interactions.
Elements of our model are echoed by recent observations of yeast, Drosophila, and C. elegans p54 helicases. Decker et al. (2007) showed that the C-terminal domain of Dhh1 interacts with overexpressed decapping enzyme and enhancer, Dcp2 and Edc3, whereas the D2 domain of Me31B binds either ectopically expressed Dcp1, Dcp2 and Edc3, or Tral (Rap55), Cup (4E-T), and Dcp1 (Tritschler et al., 2008). Based on these reports and our study, it is tempting to speculate that p54 helicases can be found in two types of subcomplexes, one subcomplex engaged in decapping and the other in translational repression. Because decapping cannot be detected in Xenopus until zygotic transcription is activated (Gillian-Daniel et al., 1998; Zhang et al., 1999), Xp54 may only be found in the translational repressor complex in oocytes. In line with this proposal, C. elegans CGH-1 is present in so-called storage bodies or germ granules during oogenesis and in P-bodies in somatic tissues; and these distinct RNA granules interact in embryos. Interestingly, the germline CGH-1 RNP contain PAB-1(ePAB) and CAR-1(Rap55), whereas the somatic RNP contains DCAP-1 and 2 (Dcp1/2) (Boag et al., 2008; Gallo et al., 2008; \?\Jud et al., 2008; Noble et al., 2008; reviewed in Rajyaguru and Parker, 2009). In HeLa cells, p54 may distribute in both decapping and repressive complexes, although they cannot be distinguished by immunofluorescence.
These investigations also highlight the critical role of the C-terminal domain of p54 helicases to interact with protein partners. This conserved function is very likely because of the folded D2 domain, rather than to sequences flanking the catalytic core of the helicase, because these differ in the yeast, fly, and vertebrate orthologues. Although Dhh1 possesses a Q/P-rich C-terminal extension, shown to contribute to P-body localization in yeast (Reijns et al., 2008), neither Me31B nor vertebrate p54 helicases contain such sequences at their C termini.
This model also provides a frame for better understanding of p54 function in P-bodies. All mutants have some defect of accumulation in the P-bodies, except the truncated D2 domain that concentrates in P-bodies such as the endogenous protein. The localization of p54 mutant proteins outside P-bodies therefore correlates with their stimulating activity in the tethered assay, and suggests that this fraction of the protein is in the “closed” conformation. In addition, all full-length mutants have a partial to complete dominant-negative effect on the number of P-bodies, establishing for the first time the role of the helicase activity of p54 in P-body maintenance, as well as strengthening the conclusion that p54 plays a primary role in P-body assembly in mammals. Yet, with the exception of p54-HRIGQ, a significant fraction of the ATPase mutants are detected in the P-bodies, and particularly so the D2 domain, which even enhances the number of cells with P-bodies despite its obvious lack of helicase activity. This demonstrates that the helicase activity is dispensable for localization in preexisting P-bodies and that the interaction with other P-body components is sufficient for such localization. However, when endogenous Rck/p54 was silenced in HeLa cells, none of the mutants could assemble P-bodies de novo. In the case of the full-length mutants, it could be simply explained by their inability to repress translation, as judged from tethered assays. As to the D2 domain, although it may bind RNA inefficiently, several of its partners have RNA binding domains, such as Rap55 and Pat1 (Pilkington and Parker, 2008), which should complement this defect. This therefore suggests that P-body assembly requires all functions of p54, including both binding of a repressor complex and helicase activity.
In summary, we provide evidence that the helicase activity of p54 enhances P-body localization and is strictly required for P-body assembly. Although helicase activity is dispensable for translational repression when RNA binding is forced by tethering, we show that it modulates the ability of the C-terminal domain to interact with partner proteins. Our current understanding of the interactions of p54 helicase and its partner proteins strongly suggests that the helicase activity contributes to a protein complex remodeling that dictates the balance between repressors and an activator of translation.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We are grateful to Niki Gray (MRC HRSU, Edinburgh, United Kingdom), Joan Steitz (Howard Hughes Medical Institute, Yale University, New Haven, CT), and Jim Wilhelm (University of California, San Diego, La Jolla, CA) for gifts of antibodies and constructs. We thank Vanessa Philippot for help in plasmid preparation and Patrick Linder for discussions. This work was supported by the Biotechnology and Biological Sciences Research Council, Centre National de la Recherche Scientifique, and the Agence Nationale pour la Recherche.
Footnotes
REFERENCES
- Beckham C. J., Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F., Baserga S. J. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Boag P. R., Atalay A., Robida S., Reinke V., Blackwell T. K. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 2008;182:543–557. doi: 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Coller J., Parker R., Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. Y., Rana T. M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J. M., Tucker M., Sheth U., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N. K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M., Wilczynska A., Harper M., Aigueperse C., Dautry F., Kress M., Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1-nucleolar-bodies. Mol. Biol. Cell. 2009;20:176–187. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause of RNA-mediated gene silencing. Mol. Cell. Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C. M., Munro E., Rasoloson D., Merritt C., Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Gillian-Daniel D. L., Gray N. K., Astrom J., Barkoff A., Wickens M. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol. Cell. Biol. 1998;18:6152–6153. doi: 10.1128/mcb.18.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B., Andrews S., Schaller A., Schumperli D., Gray N. K., Muller B. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. doi: 10.1261/rna.7281305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N., Coller J., Dickson K., Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jud M. C. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 2008;318:38–51. doi: 10.1016/j.ydbio.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Richter J. D. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Reiter M. -H., Weil D., Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- Minshall N., Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Thom G., Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Amikura R., Hanyu K., Kobayashi S. Me31B silences translation of oocyte-localising RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Noble S. L., Allen B. L., Goh L. K., Nordick K., Evans T. C. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 2008;182:559–572. doi: 10.1083/jcb.200802128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pause A., Méthot N., Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling M. E., Wilhelm J. E., O'Hara A. L., Gephardt G. W., Spradling A. C. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc. Natl. Acad. Sci. USA. 2006;104:187–192. doi: 10.1073/pnas.0609923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington G. R., Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A. M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Radford H. E., Meijer H. A., de Moor C. H. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P., Parker R. CGH-1 and the control of maternal mRNAs. Trends Cell Biol. 2009;19:24–28. doi: 10.1016/j.tcb.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Reijns M.A.M., Alexander R. D., Spiller M. P., Beggs J. D. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Serman A., Le Roy F., Aigueperse C., Kress M., Dautry F., Weil D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007;35:4715–4727. doi: 10.1093/nar/gkm491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. J., Ogawa K., Takagi M., Imamoto N., Matsumoto K., Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 2006;281:40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- Tritschler F., Eulalio A., Helms S., Schmidt S., Coles M., Weichenrieder O., Izaurralde E., Truffault V. A similar mode of interaction enables Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell. Biol. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G. K., Ongkasuwan J., Standart N., Steitz J. A. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A., Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between Dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Wilkie G. S., Gautier P., Lawson D., Gray N. K. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol. Cell. Biol. 2005;25:2060–2071. doi: 10.1128/MCB.25.5.2060-2071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Yang W. H., Gulick T., Bloch K. D., Bloch D. B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Williams C. J., Wormington M., Stevens A., Peltz S. W. Monitoring mRNA decapping activity. Methods. 1999;17:46–51. doi: 10.1006/meth.1998.0706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]