Ligand-dependent PDGF receptor-alpha activation sensitizes rare lung cancer and sarcoma cells to PDGF receptor kinase inhibitors (original) (raw)
. Author manuscript; available in PMC: 2010 May 1.
Abstract
Platelet-derived growth factor receptors (PDGFR) and their ligands play critical roles in several human malignancies. Sunitinib is a clinically-approved multi-targeted tyrosine kinase inhibitor which inhibits VEGFR, c-KIT, and PDGFR, and has demonstrated clinical activity in various solid tumors. Activation of PDGFR signaling has been described in gastrointestinal stromal tumors (GISTs) (PDGFRA mutations) as well as in chronic myeloid leukemia (BCR-PDGFRA translocation), and sunitinib can yield clinical benefit in both settings. However, the discovery of PDGFR activating mutations or gene rearrangements in other tumor types could reveal additional patient populations who might benefit from treatment with anti-PDGFR therapies, such as sunitinib. Using a high-throughput cancer cell line screening platform, we found that only 2 out of 637 tested human tumor-derived cell lines demonstrate significant sensitivity to single-agent sunitinib exposure. These two cell lines (a NSCLC and a rhabdomyosarcoma) demonstrated expression of highly phosphorylated PDGFRA. In the sunitinib-sensitive adeno-squamous NSCLC cell line, PDGFRA expression was associated with focal PFGRA gene amplification, which was similarly detected in a small fraction of squamous cell NSCLC primary tumor specimens. Moreover, in this NSCLC cell line, focal amplification of the gene encoding the PDGFR ligand PDGFC was also detected, and silencing PDGFRA or PDGFC expression by RNA interference inhibited proliferation. A similar co-dependency on PDGFRA and PDGFC was observed in the sunitinib-sensitive rhabdomyosarcoma cell line. These findings suggest that, in addition to GISTs, rare tumors that demonstrate PDGFC-mediated PDGFRA activation may also be clinically responsive to pharmacologic PDGFRA or PDGFC inhibition.
Keywords: Platelet-derived growth factor receptor, PDGF-C, NSCLC, sarcoma, sunitinib
INTRODUCTION
Sunitinib is a multi-targeted tyrosine kinase inhibitor that potently inhibits VEGF, PDGF, and c-KIT receptor kinases (1). In renal cell carcinoma (RCC) sunitinib demonstrated superiority over standard interferon-alpha therapy (2), sunitinib is now recommended for previously untreated patients with advanced RCC. Sunitinib is also approved for treatment of imatinib-refractory GISTs, many of which harbor activating c-KIT or PDGFR kinase domain mutations (3). A recent phase II clinical study has revealed efficacy of single-agent sunitinib in advanced NSCLC patients (4). Accumulating evidence indicates that inhibition of VEGF signaling using various anti-angiogenic agents can suppress tumor growth and improve patient survival (2, 5, 6); however, it is unclear from studies involving multi-kinase inhibitors, such as sunitinib, as to the relative contribution of VEGF receptor inhibition in suppressing tumor growth.
The PDGFR/PDGF system includes two receptors (PDGFRA and PDGFRB) and four ligands (PDGF-A, B, C and D) (7). Ligand binding induces receptor dimerization, enabling autophosphorylation of specific tyrosine residues and subsequent recruitment of a variety of signal transduction molecules (8). PDGFR regulates normal cellular growth and differentiation (9), and expression of activated PDGFR promotes oncogenic transformation (10), suggesting that activating mutations or gene rearrangements could play a role in human tumorigenesis. Numerous in vitro and in vivo studies demonstrated that inhibition of PDGFRA signaling disrupts cancer cell survival in the subset of GISTs with activating PDGFRA mutations (11, 12). In a recent study of 150 NSCLC patient samples, activated PDGFRA was detected in 13% of cases (13), suggesting that a subset of these patients might benefit from therapies directed against PDGFRA. Moreover, PDGFRA overexpression has been observed in metastatic versus non-metastatic medulloblastoma patient samples, and disrupting PDGFRA function inhibited the metastatic potential of medulloblastoma cells in vitro (14).
We recently reported the development of a high-throughput platform for profiling a large panel of human cancer cell lines with molecularly targeted inhibitors to identify subsets with significant sensitivity (15). That analysis revealed several examples of genotype-associated sensitivities to selective kinase inhibitors, demonstrating the utility of this strategy to reveal cell autonomous tumor cell responses to anticancer agents. Here, we describe the profiling of 637 cancer cell lines for sensitivity to single-agent sunitinib, using a monoculture format that precludes any contribution of drug effects on angiogenesis. Our studies revealed that the majority of tested cell lines are highly refractory to sunitinib. Of the two cell lines demonstrating sunitinib sensitivity, both were found to express high levels of PDGFRA and PDGFC mRNA and phosphorylated PDGFRA protein. shRNA knockdown of PDGFRA was as effective as sunitinib in decreasing cell proliferation in both cell lines, and targeting the PDGFC ligand alone was similarly effective.
Our findings suggest that while anti-angiogenesis activity probably accounts for the majority of the clinical benefit associated with sunitinib treatment in solid tumors, in rare cases, beyond _PDGFRA_-mutant GISTs, activated PDGFRA may be the critical target, and that selective PDGFRA inhibitors may be useful in the clinical management of a subset of tumors that exhibit PDGFRA activation. Moreover, in tumors with evidence of PDGFC ligand overexpression, neutralizing antibodies may be an equally effective therapeutic modality.
MATERIALS AND METHODS
Human cancer cell lines and cell viability assays
Human cancer cell lines were obtained from commercial vendors and were maintained and tested for viability using an automated platform, as previously described (15). Cells were treated for 72 hours with 1μM sunitinib and then assayed for cytostatic or cytotoxic responses. We elected to use this concentration based on steady state plasma concentrations of approximately 0.2μM at clinically recommended doses of sunitinib in patients, and based on the experimental time-points addressed in the studies.
Protein detection
Immunodetection of proteins following SDS-PAGE was performed using standard protocols. Equal protein loading was assessed using a β-tubulin antibody (Sigma). Akt, Erk1/2, phospho-Erk1/2 (T202/Y204), PDGFRA, phospho-PDGFRA (Y720), phospho-PDGFRA (Y754), STAT3 and phospho-STAT3 (S727) antibodies were from Cell Signaling Technology (Beverly, MA). The phospho-Akt (S473) antibody was from BioSource International (Camarillo, CA). All antibodies were used at a 1:1,000 dilution, except β-tubulin (1:10,000).
Kinase inhibitors
Sunitinib was obtained from MGH pharmacy. Sorafenib and Imatinib were purchased from American Custom Chemicals Corporation (ACC, San Diego, CA). The in vitro kinase specificity profile of all three compounds is listed in Supp. Table 1.
Table 1.
A. Elevated PDGFRA copy number in a subset of NSCLC cell lines. Cell lines with copy number >3 are highlighted for each gene. Data was derived from Affymetrix Nsp 250K SNP array data from 88 NSCLC cell lines. B, The most highly up- and down-regulated mRNAs in the NCI-H1703 cell line compared to all of the NSCLC cell lines. Gene expression data were available for 90 of the NSCLC cell lines screened with sunitinib. Genes were included if the fold change was greater than or less than 1.2. LBFC, the lower bound of the 90% confidence intervals of fold change; UBFC, the upper bound of the 90% confidence intervals of fold change. All data were analyzed using the dChip software.
| Chromosome | Gene | NCI-H1693 | NCI-H1703 | NCI-H2085 | NCI-H23 | NCI-H661 |
|---|---|---|---|---|---|---|
| 4 | PDGFRA | 4.88 | 4.36 | 3.72 | 3.28 | 3.61 |
| 4 | KIT | 4.88 | 1.98 | 3.77 | 2.98 | 3.62 |
| 4 | PDGFC | 1.75 | 5.93 | 1.94 | 1.07 | 1.74 |
| 5 | PDGFRB | 2.56 | 2.29 | 2.03 | 1.63 | 1.91 |
| 11 | PDGFD | 2.33 | 1.68 | 2.33 | 1.57 | 2.15 |
| 22 | PDGFB | 1.54 | 2.34 | 1.87 | 1.73 | 1.99 |
| Gene | Chromosome | Fold change | LBFC | UBFC |
|---|---|---|---|---|
| PDGFRA | 4q11 | 213.13 | 148.75 | 340.59 |
| PDGFRA | 4q11 | 39.88 | 33.38 | 49.42 |
| FLT4: fms-related tyrosine kinase 4 | 5q35 | 8.24 | 6.70 | 10.52 |
| FGFR1: fibroblast growth factor receptor 1 | 8p11 | 4.85 | 4.01 | 6.11 |
| SHC1: SHC transforming protein 1 | 1q21 | 2.89 | 2.57 | 3.27 |
| PLCE1: phospholipase C, epsilon 1 | 10q23 | 2.45 | 2.08 | 2.95 |
| SEMA3C | 7q21 | 1.71 | 1.52 | 1.94 |
| HMGA1: high mobility group AT-hook 1 | 6p21 | 1.55 | 1.37 | 1.77 |
| HMGA1: high mobility group AT-hook 1 | 6p21 | 1.45 | 1.28 | 1.67 |
| EGFR: Epidermal growth factor receptor | 7p12 | −1.59 | −1.30 | −1.91 |
| EGFR: Epidermal growth factor receptor | 7p12 | −1.63 | −1.38 | −1.88 |
| VEGF: vascular endothelial growth factor | 6p12 | −1.89 | −1.66 | −2.14 |
| MET: met proto-oncogene | 7q31 | −2.55 | −2.20 | −2.94 |
| DDR1: discoidin domain receptor family, member 1 | 6p21 | −2.69 | −2.33 | −3.07 |
| RGS2: regulator of G-protein signalling 2, 24kDa | 1q31 | −3.32 | −2.32 | −4.42 |
| MET: met proto-oncogene | 7q31 | −3.49 | −2.76 | −4.24 |
| EGFR: epidermal growth factor receptor | 7p12 | −3.70 | −2.93 | −4.53 |
| IRS1: Insulin receptor substrate 1 | 2q36 | −3.93 | −3.27 | −4.78 |
| EPS8: epidermal growth factor receptor pathway substrate 8 | 12q13 | −4.91 | −3.76 | −6.15 |
| IRS1: insulin receptor substrate 1 | 2q36 | −6.85 | −5.87 | −7.94 |
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) was performed as described previously (16). Probes for PDGFRA and c-KIT were derived from BAC clones RP11-58C6 (PDGFRA) and RP11-977G3 (c-KIT) and purchased from Invitrogen (Carlsbad, CA).
DNA sequencing
Genomic DNA was isolated using the Gentra purification system. PDGFRA, PDGFRB and c-KIT coding sequences were amplified from genomic DNA by PCR. PCR products were purified and subjected to bidirectional sequencing by using BigDye v1.1 (Applied Biosystems) in combination with an ABI3100 sequencer (Applied Biosystems). Primers used for sequencing are listed in Supp. Table 2. Electropherograms were analyzed by using Sequence Navigator software (Applied Biosystems). All mutations were confirmed by at least two independent PCR amplifications.
Cell cycle analysis
Cells were pulsed with 10μM BrdU for 1–2 hours prior to collection, centrifuged and fixed in ice-cold 70% ethanol. Cells were washed with PBS/0.5% BSA and incubated in denaturing solution (2M HCL) for 20 min at RT. After a further wash with PBS/0.5% BSA the cells were resuspended in 0.1M sodium borate for 2 min at RT. After an additional wash cells were suspended in anti-BrdU monoclonal antibody for 20 min (1:500, Becton Dickinson, San Jose, CA). Cells were washed in PBS/0.5% BSA and the pellet was resuspended in FITC-conjugated anti-mouse IgG (1:50, Vector Laboratories, Burlingame, CA) for 20 min. After an additional wash in PBS/0.5% BSA cells were stained with 10μg/mL propidium iodide (Sigma) and treated with RNAse A (Sigma) prior to two-dimensional FACS analysis using CELLQUEST software (Becton Dickinson).
SNP and gene expression analyses
Gene copy numbers were determined as previously described using the GeneChip® Human Mapping 250K. The array was then scanned on the GeneChip® Scanner 3000 7G and analyzed using GTYPE version 4.0 with the Dynamic Model Mapping Algorithm and the GeneChip® Human Mapping 500K Set library files (Mapping 250K_Nsp).
For gene expression studies, RNA was extracted using the Qiagen RNA easy kit (P/N 74106), and amplified and biotin-labeled with the Arcturus RiboAmp® RNA Amplification Kit using biotinylated ribonucleotides (Perkin Elmer PN Biotin-11-UTP, NEL543001EA/Biotin-11-CTP, NEL542001EA) during in vitro transcription. Labeled aRNA was hybridized to Affymetric GeneChip Human X3P (GPL1352) using protocols described within Affymetrix’s GeneChip Expression Analysis Technical Manual (PN701021 Rev. 3) Data was acquired using the Affymetrix GeneChip 3000 Scanner with autoloader and 7G upgrade. GCOS ver 1.4 software was used to run the scanner and analyze the data. The expression value each gene was calculated using Affymetrix GeneChip software and data were analyzed using dChip software (http://biosun1.harvard.edu/complab/dchip/) (17). Probe sets were filtered using two criteria: (1) coefficient of variation between 0.5 and 1000, and (2) P call rate in arrays ≥20%.
Quantatative PCR
Total RNA was isolated and purified from cells using STAT-60 (Tel-Test Inc., Friendswood, TX, USA). cDNA was transcribed from 2μg of total RNA using the AffinityScript Multi Temperature cDNA Synthesis kit (Stratagene, La Jolla, CA). Q-PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA) and with an ABI PRISM 7000 real-time cycler (Applied Biosystems, Foster City, CA). Quantification was based on standard curves for each primer set from a serial dilution of the NCI-H1703 cell line cDNA. All samples were analyzed in triplicate. Primers sequences were: GAPDH F, GAGTCAACGGATTTGGTCGT; GAPDH R, TTGATTTTGGAGGGATCTCG; PDGFRA F, AAATTGTGTCCACCGTGATCT; PDGFRA R, AGGCCAAAGTCACAGATCTTC; PDGFC F, AACGGAGTACAAGATCCTCAGC; PDGFC R, CCATCACTGGGTTCCTCAAC.
RNAi studies
shRNAs targeting sequences within the genes encoding either PDGFRA (n=10) or its ligand PDGFC (n=5) were expressed from the pLKO.1 lentiviral vector (Supp. Table 3). NCI-H1703 and A-204 cells were infected in the presence of polybrene (8 μg/ml). A cell line demonstrating sunitinib-insensitivity (A549) was used to determine infection efficiency based on puromycin resistance, and to confirm specificity. Protein lysates and RNA were collected 48 hours post-infection, and cell numbers were determined 72 hours post-infection.
PDGFC neutralizing antibody experiments
Cells were seeded in 1% FBS medium and treated the following day with 5–20 ng/ml of an anti-PDGFC neutralizing antibody (R&D Systems Inc, Minneapolis). Normal goat IgG at 20ng/ml concentration was used as a control. Cells were fixed and stained 5 days following treatment and cell viability measured as previously described (15).
RESULTS
Rare human cancer cell lines are sensitive to single-agent sunitinib treatment
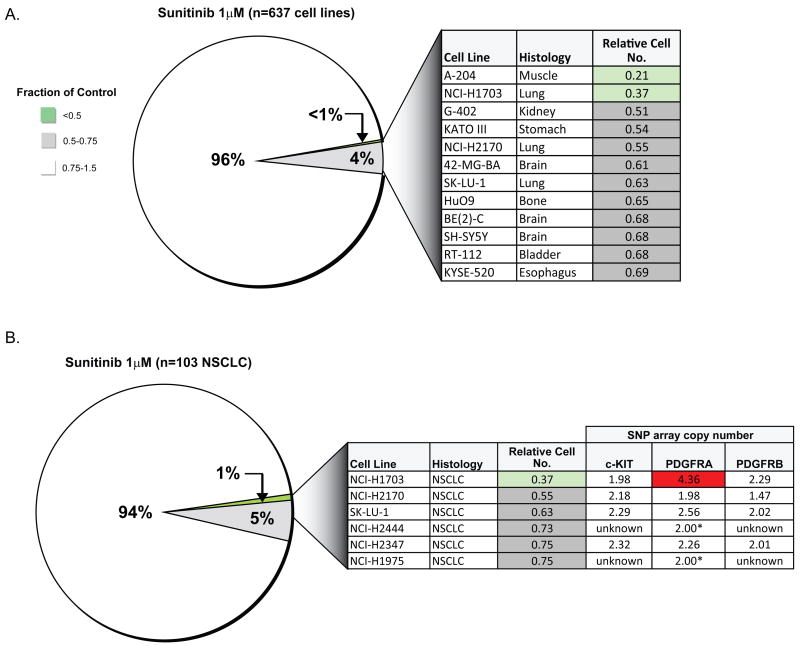
Using an automated platform to examine drug sensitivity in cancer cell lines (15), we tested the sunitinib sensitivity of 637 established human cancer cell lines derived from a wide variety of solid tumor types (Supp. Figure 1) (1). Cells were treated for 72 hours with 1μM sunitinib and then assayed for cytostatic or cytotoxic responses. While the vast majority of tested cell lines were largely refractory to treatment, two cell lines (A-204 rhabdomyosarcoma and NCI-H1703 NSCLC) displayed significant sunitinib sensitivity, as indicated by a greater than 50% reduction in cell number (Figure 1A). We note that cell lines derived from GISTs, which demonstrate clinical sunitinib sensitivity, reflecting inhibition of mutationally-activated PDGFR or c-KIT kinases, were absent from the panel of tested lines. A few additional lines demonstrated a relatively weaker response to sunitinib.
Figure 1.
A, Pie chart representation of the sensitivity of 637 human cancer cell lines to treatment with 1μM sunitinib. The drug effect was calculated as the fraction of untreated cells present after 72 hours of treatment. The color scheme corresponds to the relative inhibitory effect of treatment, with ratios reflecting the number of cells remaining following exposure to inhibitor. Details regarding the most sensitive cell lines identified are shown in the chart, and the cell lines are shown in order of decreasing sensitivity (top to bottom). B, Pie chart representation of the sensitivity of 103 NSCLC cell lines to 1μM sunitinib. Copy number data were generated from 250K Nsp SNP array profiles (or FISH, as indicated by asterisk).
The sunitinib-sensitive NSCLC-derived cell line harbors focal PDGFRA gene amplification
Among 103 NSCLC cell lines tested, significant sunitinib sensitivity was observed only in the adeno-squamous NCI-H1703 line (Figure 1B). SNP array data available for 88 of these lines revealed that NCI-H1703 cells harbor focal PDGFRA gene amplification (Figure 1B). This was confirmed by interphase FISH analysis (Supp. Figure 2A). There was no evidence of either c-KIT or PDGFRB genomic amplification or protein expression in these cells (data not shown). Sequence analysis of the entire coding sequence of PDGFRA, PDGFRB and c-KIT in this cell line revealed a single mutation in exon 9 of PDGFRA (S478P) – within the extracellular domain, which would not be expected to result in PDGFR activation.
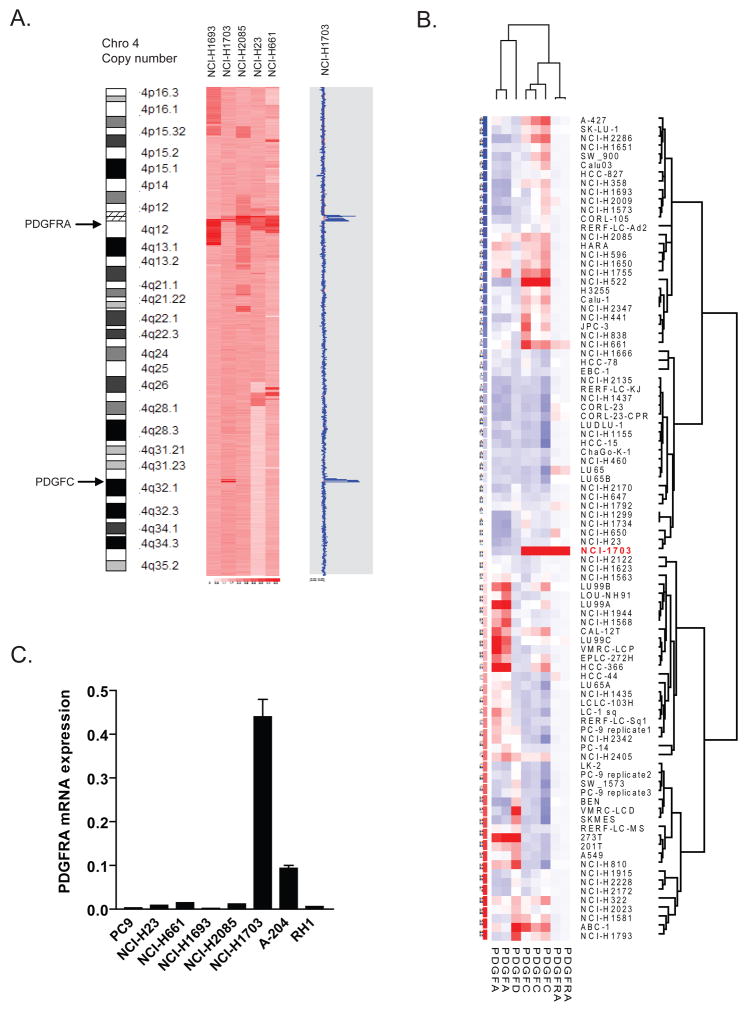
Figure 2.
A, SNP array analysis of chromosome 4 for the five NSCLC cell lines exhibiting elevated PDGFRA copy number demonstrates increased PDGFC ligand copy number (5.93) in NCI-H1703 cells. The blue tracing indicates the degree of amplification of each SNP in the array. The red line underlying the blue tracing indicates copy number of 2. B, PDGFRA and PDGF ligand mRNA expression in 90 NSCLC cell lines. NCI-H1703 is indicated by red lettering. Probe sets for ligand PDGFB are excluded following a filter based on P call rate in arrays <20%. C, Relative PDGFRA mRNA expression levels in NSCLC and rhabdomyosarcoma cell lines as determined by quantitative RT-PCR.
The SNP array data revealed similarly elevated PDGFRA gene copy number in four other NSCLC cell lines (NCI-H1693, NCI-H2085, NCI-H23 and NCI-H661), however, these lines were sunitinib-insensitive (Table 1A, Figure 2A). FISH analyses of these cell lines confirmed PDGFRA amplification (Supp. Figure 3). However, analysis of the transcriptional expression profile of tyrosine kinase signaling pathway-associated genes in the 90 NSCLC cell line panel revealed that only NCI-H1703 showed significant expression of PDGFRA mRNA (Supp. Figure 4). Furthermore, when the gene expression profile of NCI-H1703 cells was compared with the other 89 NSCLC cell lines for the most significant up-regulated and down-regulated mRNA transcripts, the most highly expressed mRNA in NCI-H1703 cells corresponded to PDGFRA (fold change of 213) (Table 1B). When we focused on those probes involved in PDGFR signaling, none of the other four NSCLC cell lines with increased PDGFRA copy number displayed increased PDGFRA mRNA expression (Figure 2B). The observed increase in PDGFRA mRNA expression in the NCI-H1703 cells was confirmed by Q-PCR (Figure 2C).
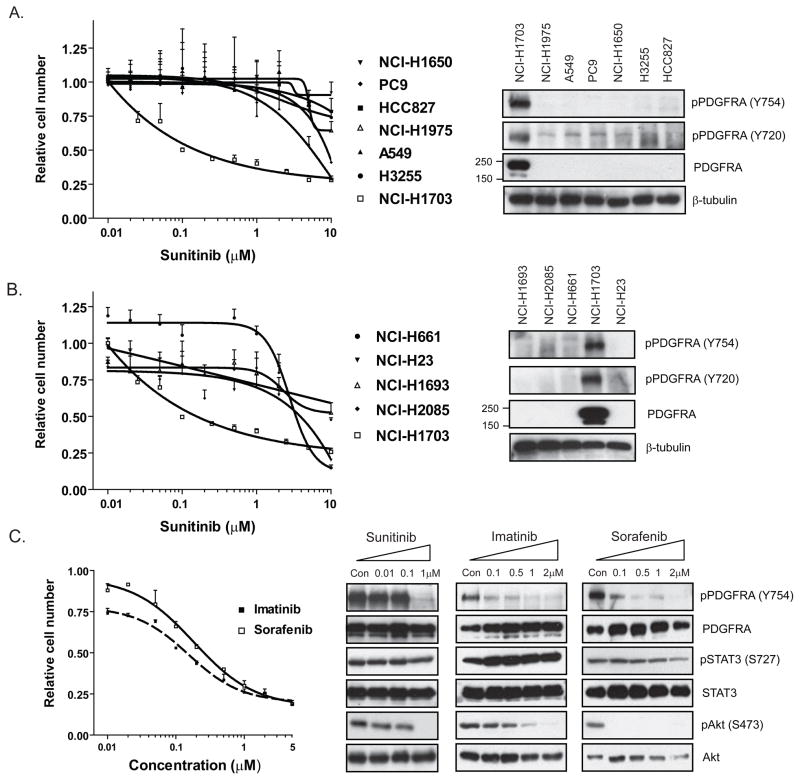
Figure 3.
Dose-response curves demonstrating the effect of sunitinib on cell numbers 72 hours after treatment for NCI-H1703 and a panel of NSCLC cell lines with either normal (A) or increased (B) PDGFRA copy number, and immunoblots corresponding to these same cell lines demonstrating total PDGFRA and phospho-PDGFRA levels. C, Dose response curves demonstrating the effect of the additional PDGFR inhibitors imatinib and sorafenib on cell numbers 72 hours after treatment in the NCI-H1703 cell line. Immunoblots demonstrating the effect of treating the NCI-H1703 cell line for 6 hours with the indicated concentrations of sunitinib, imatinib and sorafenib on phosphorylation of PDGFRA and the downstream effectors, STAT3 and Akt. Note that p-STAT3 levels are largely unaffected by drug treatment, whereas p-Akt levels are reduced.
Suntinib dose-response curves for the NCI-H1703 cell line versus a panel of NSCLC cell lines with normal (Figure 3A) or increased PDGFRA gene copy number (Figure 3B) confirmed the unique sensitivity in NCI-H1703 cells. Moreover, expression of phosphorylated and total PDGFRA protein was only detected in NCI-H1703 cells (Figure 3A, B), and PDGFRA protein was not detected in any of the sunitinib-insensitive cell lines. In fact, when we extended this panel to include an additional 26 NSCLC sunitinib-insensitive cell lines, we were unable to detect expression of PDGFRA in any other lines (Supp. Figure 5). Therefore, the increased transcriptional expression of PDGFRA in NCI-H1703 results in increased PDGFRA protein and is associated with elevated phospho-PDGFRA, which potentially mediates sensitivity to sunitinib.
To assess PDGFRA amplification in clinical NSCLC cases, we analyzed 143 NSCLC primary tumor specimens by FISH and detected 3/81(3.7%) cases of focal PDGFRA amplification in squamous cell carcinomas (Supp. Figure 2B). PDGFRA amplification was not detected in any of 62 adenocarcinoma cases analyzed. Thus, focal PDGFRA gene amplification arises at relatively low frequency in NSCLC, and may be more common in the squamous cell setting.
Inhibition of PDGFRA activation in NCI-H1703 cells disrupts downstream signaling
Treatment of NCI-H1703 cells with sunitinib for 6 hrs resulted in complete inhibition of PDGFRA protein phosphorylation as well as that of Akt, a PDGFR effector (Figure 3C). Sunitinib had no effect on such signaling in the sunitinib-insensitive cell lines (not shown). To verify that PDGFRA-dependent signaling was indeed the basis for the observed sunitinib sensitivity of NCI-H1703 cells, we treated the cells with two additional PDGFRA kinase inhibitors– sorafenib and imatinib. Both compounds exhibited a similar activity to that of sunitinib (Figure 3C), whereas none of the sunitinib-insensitive NSCLC cell lines displayed sensitivity to either agent (not shown). Furthermore, like sunitinib, both compounds suppressed Akt signaling in NCI-H1703 cells (Figure 3C). Together, these findings suggest that the NCI-H1703 NSCLC cells are dependent on activated PDGFRA signaling.
To investigate the underlying mechanism for the ability of sunitinib to reduce cell number in NCI-H1703 cells, we examined PARP cleavage, an indicator of apoptosis, and cell cycle profile. There was no evidence of PARP cleavage following treatment with a 1μM concentration of sunitinib at either 24, 48 or 72 hrs in this cell line (data not shown), whereas cell cycle analysis confirmed a significant S-phase arrest at each of these time-points (Supp. Figure 6), consistent with a cytostatic response to drug exposure.
PDGFRA activation is associated with sensitivity to sunitinib in a rhabdomyosarcoma cell line
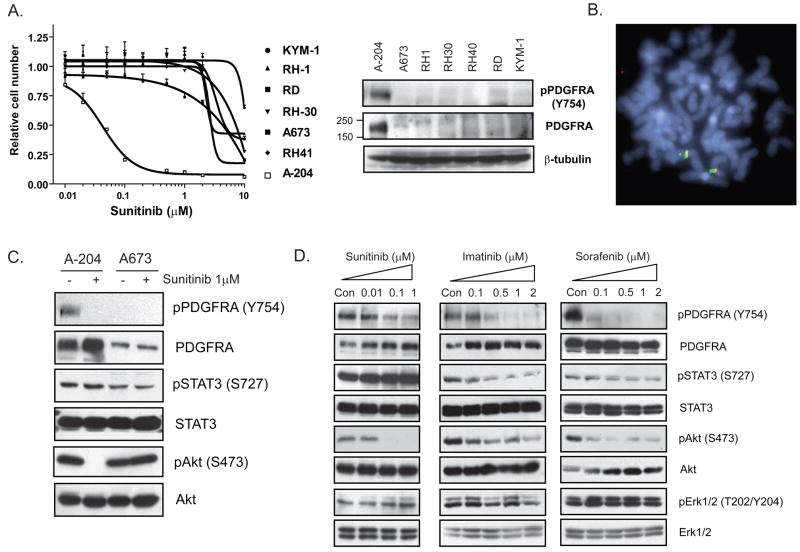
As described above, in the initial screen of 637 cell lines for sunitinib sensitivity, a rhabdomyosarcoma cell line, A-204, was the most highly drug-sensitive line detected (Figure 1A). To determine whether the observed sensitivity could be extended to other rhabdomyosarcoma cell lines, a panel of 6 additional rhabdomyosarcoma lines was tested for sunitinib sensitivity (Figure 4A). Of the tested lines, only A-204 demonstrated sunitinib sensitivity and in only this cell line was PDGFRA protein detectable (Figure 4A, lane 1). Unlike in NCI-H1703 cells, FISH analysis did not reveal PDGFRA gene amplification in any of these lines (Figure 4B), and DNA sequence analysis of PDGFRA, PDGFRB and c-KIT in A-204 cells did not reveal any mutations. However, as in NCI-H1703 cells, we detected a substantial (15-fold) increase in PDGFRA mRNA expression in A-204 cells (Figure 2C). Moreover, sunitinib treatment completely abolished Akt signaling in this line compared to a sunitinib-insensitive rhabdomyosarcoma line (Figure 4C). In addition, treatment A-204 cells with sorafenib and imatinib also disrupted Akt signaling (Figure 4D) and similarly inhibited proliferation (not shown). These results suggest that rare rhabdomyosarcoma cells are dependent on activated PDGFRA signaling, associated with increased expression of PDGFRA mRNA.
Figure 4.
A, Dose-response curves demonstrating the effect of sunitinib on cell numbers 72 hours post-treatment for several rhabdomyosarcoma cell lines. Immunoblots demonstrating expression of phospho-PDGFRA and total PDGFRA in these lines are shown to the right, with β-tubulin as a loading control. B, FISH analysis of sunitinib-sensitive A-204 cells using PDGFRA (RP11-58C6 - red) and c-KIT probes (RP11-977G3 - green). C, Immunoblots demonstrating the effect of treating the A-204 (sunitinib-sensitive) and A673 (sunitinib-resistant) cell lines for 6 hours with 1μM sunitinib on phosphorylation of PDGFRA and the downstream effectors, STAT3 and Akt. D, Effect of treating A-204 for 6 hours with the PDGFR inhibitors sunitinib, imatinib and sorafenib on phosphorylation of PDGFRA and the downstream effectors, STAT3, Akt and Erk1/2.
Amplification of the gene encoding the PDGFRA ligand PDGFC mediates PDGFRA activation
Gene expression profiles of 90 NSCLC cell lines using a filtered list of genes involved in PDGFR signaling revealed that NCI-H1703 was the only line displaying significant transcriptional up-regulation of PDGFRA together with the gene encoding one of its ligands PDGFC (Figure 2B). The increased PDGFC mRNA in NCI-H1703 cells (and in A-204 cells) was confirmed by Q-PCR (Supp. Figure 7A). Only the NSCLC cell line NCI-H661 (sunitinib-insensitive) demonstrated similarly elevated PDGFC mRNA, but in the absence of expression of PDGFRA mRNA or protein. Further analysis of SNP array data from 88 NSCLC lines revealed a unique co-amplification of the PDGFRA (4q12) and PDGFC (4q32) genes on chromosome 4 in NCI-H1703 cells, which was not observed in any of the other cell lines (Figure 2A).
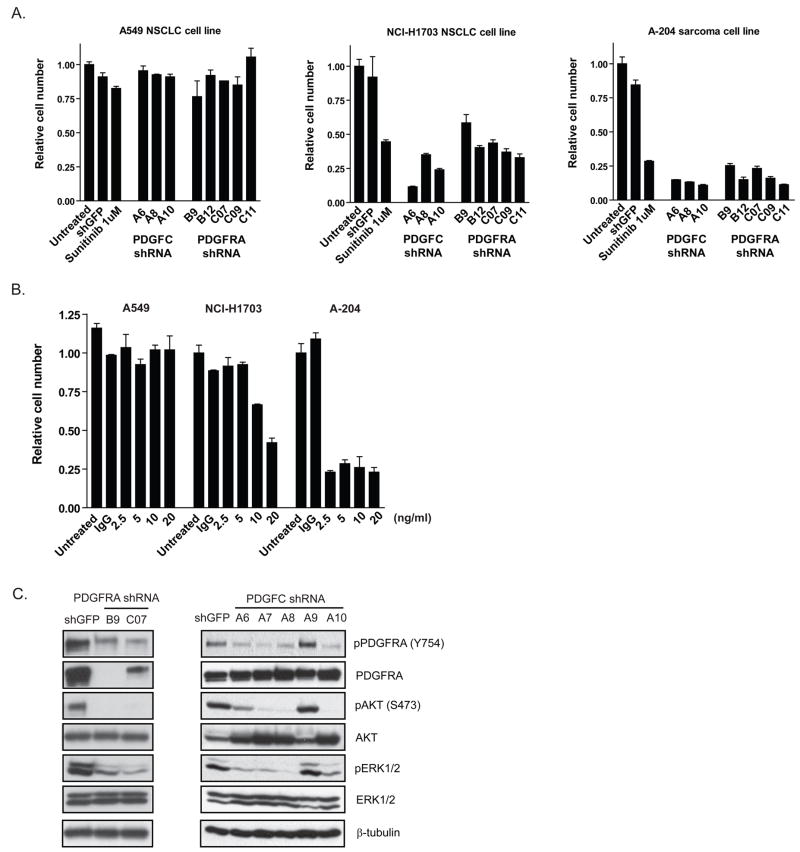
ShRNA-mediated knockdown of PDGFRA and PDGFC was used to directly assess their functional requirement in both the NCI-H1703 and A-204 cell lines (Supp. Figure 7B). There was no effect of these shRNAs on a sunitinib-insensitive cell line (A549) that lacks PDGFRA expression. In contrast, knockdown of PDGFRA in NCI-H1703 and A-204 cells significantly reduced proliferation to a similar extent as sunitinib treatment (Figure 5A). Furthermore, knockdown of PDGFC expression also reduced proliferation in both lines, and the observed decrease was of the same magnitude seen following sunitinib treatment (Figure 5A).
Figure 5.
A, Lentiviral-delivered shRNAs were used to target GFP (negative control), PDGFRA and PDGFC in A549, NCI-H1703 and A-204 cells, and cell numbers were measured 72 hours post-infection. Treatment with 1μM sunitinib served as positive control. B, A549, NCI-H1703 and A-204 cells were treated with a neutralizing anti-PDGFC antibody (2.5–20ng/ml) or normal IgG control antibody (at 20ng/ml). Cell numbers were measured 5 days post-treatment. C, Effect of shRNA-mediated depletion of PDGFRA and PDGFC in NCI-H1703 cells on PDGFRA phosphorylation and downstream signaling 24 (PDGFRA) and 48 (PDGFC) hours post-infection.
We also examined the activity of a neutralizing anti-PDGFC antibody to confirm the ligand knockdown findings and to assess the potential therapeutic value of anti-PDGFC antibodies in such tumor cells. We treated three cell lines (A549, NCI-H1703 and A-204) with a concentration range of the anti-PDGFC antibody. While there was no detectable effect on proliferation of A549 cells, the antibody reduced proliferation in the NCI-H1703 and A-204 cell lines to a similar extent to that seen following sunitinib treatment (Figure 5B). Notably, the effect was observed in A-204 cells even at the lowest antibody concentration, potentially reflecting relatively higher PDGFC expression in the NCI-H1703 cells (Supp. Figure 7A). Combining sunitinib and the anti-PDGFC antibody did not result in any additive inhibitory effects on these cells (not shown). ShRNA-mediated depletion of PDGFRA and PDGFC was used to determine the effect on PDGFRA activation and downstream signaling in the NCI-H1703 cells (Figure 5C). ShRNA-mediated depletion of both receptor and ligand resulted in decreased PDGFRA phosphorylation, and inhibition of Akt and Erk1/2 phosphorylation. Together, these results indicate that both of the sunitinib-sensitive cell lines demonstrate a similar dependency on increased PDGFRA and PDGFC expression for sustained proliferation.
DISCUSSION
Our cancer cell line profiling analysis with the multi-kinase inhibitor sunitinib have revealed that drug sensitivity in a monoculture context is restricted to a small number of lines exhibiting activated PDGFRA signaling. Moreover, in these cells, PDGFRA activation is coupled to critical downstream effectors such as Akt, and disrupting these pathways appears to mediate the inhibitory effects of sunitinib on proliferation. Previous reports of PDGFRA activation in cancer have been largely confined to GISTs (activating PDGFRA mutations) and rare cases of idiopathic hypereosinophilic syndrome (FIP1L1-PDGFRA fusion transcripts) (18, 19). Our findings suggest that in additional rare cases of NSCLC and sarcoma PDGFRA activation may be important in maintaining the malignant phenotype.
The clinical success of sunitinib in renal cancer has been suggested to reflect its role as a VEGFR inhibitor, and consequent effects on angiogenesis. Notably, renal cancers are highly vascularized tumors, suggesting a potential critical requirement for angiogenesis in that disease setting. However, it sunitinib’s ability to target additional kinases, such as PDGFR, might contribute to its clinical activity in renal cancer. We note that our cell line panel included 19 renal cancer cell lines, none of which demonstrated significant sunitinib sensitivity. This suggests that PDGFR is not likely to provide a critical dependency signal in renal cancer; however a contributing role of PDGFR inhibition in sunitinib’s clinical activity cannot be excluded. Whereas in a conventional xenograft model, any observed consequence of drug treatment on tumor growth could potentially reflect direct effects on tumor cells as well as effects on the stroma and vasculature, our monoculture-based platform provides a means to isolate the tumor cell-autonomous drug response.
In both of the sunitinib-sensitive cancer cell lines identified, PDGFRA activation appears to be mediated by increased expression of the receptor as well as one of its ligands, PDGFC. This is in contrast to other models of receptor tyrosine kinase activation associated with gene amplification, where ligand-independent activation is typically postulated (20, 21). In NCI-H1703 cells, activation of PDGFRA signaling pathways is a consequence of focal PDGFRA and PDGFC gene co-amplification. To our knowledge, this is the first report in NSCLCs of over-expression of both an oncogenic receptor tyrosine kinase and its ligand; although, co-expression of the PDGFRA or PDGFRB receptors with their cognate ligands at higher levels than seen in adjacent normal tissue has been reported for some gliomas and osteosarcomas (22, 23). Intriguingly, targeting PDGFC had a greater effect on proliferation in both sunitinib-sensitive cell lines than targeting PDGFRA. This raises the possibility that PDGFRA is not the sole critical target of PDGFC in these cells.
Our findings also suggest that antibodies directed against PDGFR ligands may have therapeutic potential in PDGFRA-dependent cancer. Traditionally, therapeutic antibodies have been targeted to cell surface receptors implicated in tumor cell proliferation or maintenance, rather than against their cognate ligands (24). Such antibodies typically demonstrate a more favorable toxicity profile than small molecule kinase inhibitors, and when considered in the context of significant toxicities associated with sunitinib, our findings suggest potential clinical advantages of antibody-mediated targeting of the PDGFC ligand in some cancers.
Our observation that the PDGFRA gene amplification in the NCI-H1703 adeno-squamous NSCLC cell line was also seen in a subset of squamous cell NSCLC clinical samples but in none of the adenocarcinoma samples screened by FISH, raises the possibility that this represents an oncogenic mechanism unique to this histological sub-type. In agreement with our findings, Rikova et al detected PDGFRA activation using a phospho-proteomic screen in 8 NSCLC patient samples as well as in the NCI-H1703 cell line (13). Whereas NSCLC adenocarcinoma patients are being actively recruited into clinical trials of EGFR tyrosine kinase inhibitors (in the setting of activating EGFR mutations) and anaplastic lymphoma kinase inhibitors (ALK translocations), to date, no drug-sensitizing genotypes have been identified for squamous cell NSCLC patients (25, 26). It remains to be seen whether retrospective analyses of sunitinib-responsive NSCLC patients will reveal enrichment for PDGFRA gene amplification or expression, and whether such patients’ tumors demonstrate squamous histology.
Curiously, PDGFRA expression was only detected in the NCI-H1703 NSCLC cells, despite the fact that four other cell lines demonstrated increased PDGFRA gene copy number. Thus, focal amplification of the PDGFRA gene may uniquely yield high level PDGFR expression, potentially reflecting an additional genomic alteration within this locus that influences the regulatory regions of PDGFRA gene transcription. Similarly, it remains unclear as to the molecular mechanism underlying increased PDGFRA or PDGFC mRNA expression in the A-204 cells. Sarcomas often harbor chromosomal translocations giving rise to oncogenic activation, and these can affect PDGFR signaling. For example, dermatofibrosarcoma protuberans and giant cell fibroblastomas harbor chromosomal rearrangements involving chromosome 17 and 22, in which the collagen type Ialpha1 (COLIA1) gene undergoes fusion with the gene encoding PDGFB (27). In one study of 42 cases of uterine sarcoma, 70% of tumors displayed increased PDGFRA expression compared to that seen in adjacent normal tissue (28). Likewise, in a study of 54 osteosarcoma patients, increased PDGFRA and PDGFRB expression was observed in tumors in more than 75% of cases (29). Notably, most Ewing sarcomas are associated with a gene fusion that produces a transcription factor (EWS/FLI-1) that promotes PDGFC mRNA expression (30). However, imatinib therapy in this setting demonstrates minimal clinical activity (31). In these tumors, which are notoriously refractory to chemotherapy, targeting PDGFR signaling pathways may provide a useful alternative therapy.
In summary, our findings demonstrate that ligand-mediated activation of PDGFRA signaling may be a critical mediator of cell proliferation in a small subset of NSCLCs and rhabdomyosarcomas, and may sensitize these cancer cells to either selective small molecule PDGFR kinase inhibitors or ligand-neutralizing antibodies. Our findings suggest that sunitinib as well as other PDGFR kinase inhibitors may provide genotype-associated clinical benefit beyond the setting of _PDGFR_-mutant or _c-KIT_-mutant GISTs.
Acknowledgments
We are grateful to members of the Settleman laboratory for helpful discussion throughout the course of these studies. We also thank Michelle Longworth for assistance with the cell cycle analysis. This work was supported by the NCI SPORE in Lung Cancer award P20 CA090578-06.
References
- 1.Mendel DB, Laird AD, Xin X, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9(1):327–37. [PubMed] [Google Scholar]
- 2.Motzer RJ, Hutson TE, Tomczak P, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356(2):115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 3.Demetri GD, van Oosterom AT, Garrett CR, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368(9544):1329–38. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 4.Socinski MA, Novello S, Brahmer JR, et al. Multicenter, phase II trial of sunitinib in previously treated, advanced non-small-cell lung cancer. J Clin Oncol. 2008;26(4):650–6. doi: 10.1200/JCO.2007.13.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350(23):2335–42. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 6.Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355(24):2542–50. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 7.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22(10):1276–312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valius M, Kazlauskas A. Phospholipase C-gamma 1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor’s mitogenic signal. Cell. 1993;73(2):321–34. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- 9.Ross R, Raines EW, Bowen-Pope DF. The biology of platelet-derived growth factor. Cell. 1986;46(2):155–69. doi: 10.1016/0092-8674(86)90733-6. [DOI] [PubMed] [Google Scholar]
- 10.Gazit A, Igarashi H, Chiu IM, et al. Expression of the normal human sis/PDGF-2 coding sequence induces cellular transformation. Cell. 1984;39(1):89–97. doi: 10.1016/0092-8674(84)90194-6. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–9. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 12.Corless CL, Schroeder A, Griffith D, et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23(23):5357–64. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 13.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald TJ, Brown KM, LaFleur B, et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2001;29(2):143–52. doi: 10.1038/ng731. [DOI] [PubMed] [Google Scholar]
- 15.McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104(50):19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68(9):3389–95. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 17.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20(8):1233–40. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 18.Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299(5607):708–10. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 19.Cools J, DeAngelo DJ, Gotlib J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348(13):1201–14. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 20.Smolen GA, Sordella R, Muir B, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A. 2006;103(7):2316–21. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennan PJ, Kumagai T, Berezov A, Murali R, Greene MI. HER2/neu: mechanisms of dimerization/oligomerization. Oncogene. 2000;19(53):6093–101. doi: 10.1038/sj.onc.1203967. [DOI] [PubMed] [Google Scholar]
- 22.Sulzbacher I, Traxler M, Mosberger I, Lang S, Chott A. Platelet-derived growth factor-AA and -alpha receptor expression suggests an autocrine and/or paracrine loop in osteosarcoma. Mod Pathol. 2000;13(6):632–7. doi: 10.1038/modpathol.3880109. [DOI] [PubMed] [Google Scholar]
- 23.Guha A, Dashner K, Black PM, Wagner JA, Stiles CD. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. Int J Cancer. 1995;60(2):168–73. doi: 10.1002/ijc.2910600206. [DOI] [PubMed] [Google Scholar]
- 24.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol. 2005;23(9):1147–57. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 25.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26(15):2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 26.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 27.Patel KU, Szabo SS, Hernandez VS, et al. Dermatofibrosarcoma protuberans COL1A1-PDGFB fusion is identified in virtually all dermatofibrosarcoma protuberans cases when investigated by newly developed multiplex reverse transcription polymerase chain reaction and fluorescence in situ hybridization assays. Hum Pathol. 2008;39(2):184–93. doi: 10.1016/j.humpath.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Adams SF, Hickson JA, Hutto JY, Montag AG, Lengyel E, Yamada SD. PDGFR-alpha as a potential therapeutic target in uterine sarcomas. Gynecol Oncol. 2007;104(3):524–8. doi: 10.1016/j.ygyno.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Kubo T, Piperdi S, Rosenblum J, et al. Platelet-derived growth factor receptor as a prognostic marker and a therapeutic target for imatinib mesylate therapy in osteosarcoma. Cancer. 2008;112(10):2119–29. doi: 10.1002/cncr.23437. [DOI] [PubMed] [Google Scholar]
- 30.Zwerner JP, May WA. PDGF-C is an EWS/FLI induced transforming growth factor in Ewing family tumors. Oncogene. 2001;20(5):626–33. doi: 10.1038/sj.onc.1204133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bond M, Bernstein ML, Pappo A, et al. A phase II study of imatinib mesylate in children with refractory or relapsed solid tumors: a Children’s Oncology Group study. Pediatr Blood Cancer. 2008;50(2):254–8. doi: 10.1002/pbc.21132. [DOI] [PubMed] [Google Scholar]