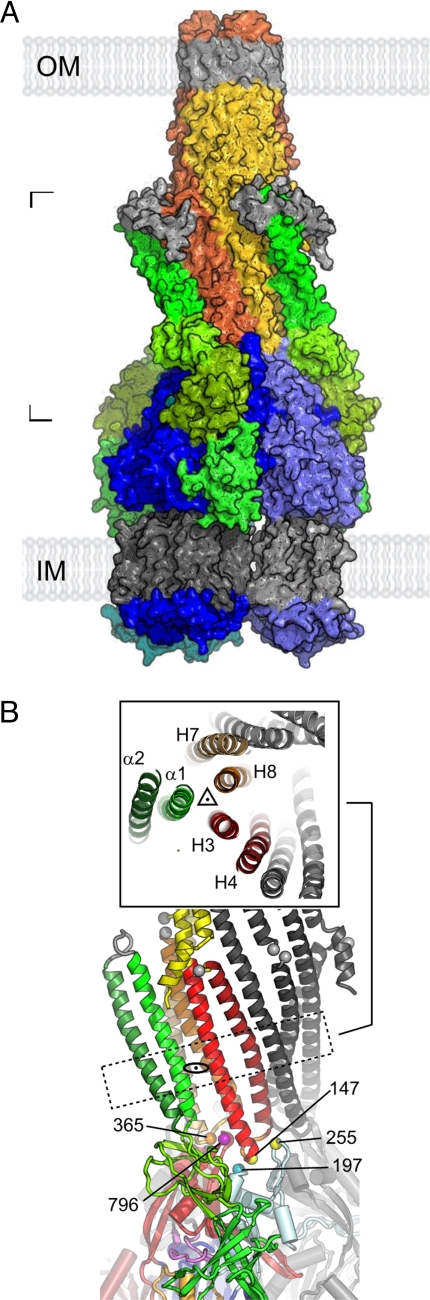
The assembled structure of a complete tripartite bacterial multidrug efflux pump (original) (raw)
Abstract
Bacteria like Escherichia coli and Pseudomonas aeruginosa expel drugs via tripartite multidrug efflux pumps spanning both inner and outer membranes and the intervening periplasm. In these pumps a periplasmic adaptor protein connects a substrate-binding inner membrane transporter to an outer membrane-anchored TolC-type exit duct. High-resolution structures of all 3 components are available, but a pump model has been precluded by the incomplete adaptor structure, because of the apparent disorder of its N and C termini. We reveal that the adaptor termini assemble a β-roll structure forming the final domain adjacent to the inner membrane. The completed structure enabled in vivo cross-linking to map intermolecular contacts between the adaptor AcrA and the transporter AcrB, defining a periplasmic interface between several transporter subdomains and the contiguous β-roll, β-barrel, and lipoyl domains of the adaptor. With short and long cross-links expressed as distance restraints, the flexible linear topology of the adaptor allowed a multidomain docking approach to model the transporter–adaptor complex, revealing that the adaptor docks to a transporter region of comparative stability distinct from those key to the proposed rotatory pump mechanism, putative drug-binding pockets, and the binding site of inhibitory DARPins. Finally, we combined this docking with our previous resolution of the AcrA hairpin–TolC interaction to develop a model of the assembled tripartite complex, satisfying all of the experimentally-derived distance constraints. This AcrA3-AcrB3-TolC3 model presents a 610,000-Da, 270-Å-long efflux pump crossing the entire bacterial cell envelope.
Keywords: antibiotic resistance, docked model, membrane complex, TolC exit duct
Tripartite efflux pumps enable Gram-negative bacteria such as Escherichia coli and Pseudomonas aeruginosa to expel antibiotics and other noxious agents, while closely-related machineries export virulence proteins (1–5). They are therefore key to bacterial survival and the problem of multidrug resistance. Drug efflux pumps span the entire cell envelope, i.e., the inner and outer membranes and intervening periplasmic space (Fig. 1). They comprise an inner membrane transporter that provides energy and binds efflux substrates, an outer membrane-anchored exit duct of the TolC family, and a periplasmic adaptor protein linked to the inner membrane (6, 7). To understand how these pumps function and how they might be counteracted, it is essential to derive a model of the complete tripartite assembly that is based on high-resolution structures of the 3 components and biochemical characterization of their interactions. The trimeric structures of the E. coli TolC (1,284 residues) and the AcrB transporter (a proton antiporter of 3,096 residues) are known (8–10). TolC is anchored in the outer membrane by a β-barrel and projects across the periplasm a 100-Å α-helical tunnel, a single pore (8), that is opened by realignment of periplasmic entrance helices (8, 11–13). It has been proposed that drugs destined for the exit duct bind in defined pockets of the AcrB transporter (shown in Fig. 1), and that variation in the structural states of the 3 transporter subunits reflects a cyclical rotatory pump mechanism (14–16). Whereas the inner membrane AcrB has a 70-Å-high periplasmic extension that makes limited contact with TolC (6, 17, 18), assembly of the active transenvelope pump requires the periplasmic adaptor component. The adaptor forms a complex with the transporter and recruits TolC, inducing and/or stabilizing its open state (2, 6, 7).
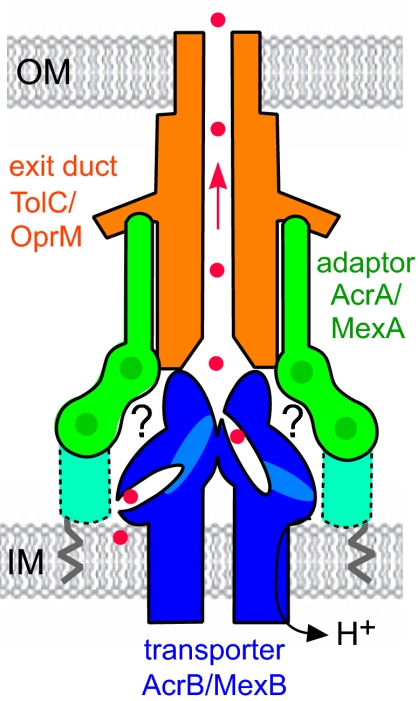
Fig. 1.
Schematic of the tripartite multidrug efflux pump. The pumps comprise an exit duct (shown in orange; TolC in E. coli, OprM in P. aeruginosa) anchored in the outer membrane (OM), an integral inner membrane (IM) transporter (in blue; AcrB and MexB) and a periplasmic adaptor protein (in green; AcrA and MexA) linked to the inner membrane by a fatty acid (zigzag line). The adaptor binds the exit duct α-helical domain via its α-hairpin (23) and the transporter via unknown interactions (indicated by ?). The adaptor linear multidomain structure is characterized by interdomain flexibility, but it is incomplete, missing the MP domain indicated by the dotted outline. Red dots indicate antibacterial drugs bound to putative pockets in the transporter, passing through TolC (arrowed), and out of the cell.
We previously crystallized the 360-residue monomer of the P. aeruginosa adaptor MexA and revealed 3 linearly-arranged domains spanning ≈65% of the protein: a β-barrel domain, a lipoyl domain, and a 47-Å-long α-helical hairpin (17–21). This partial structure, confirmed in the MexA homologue AcrA of E. coli (22), allowed in vivo site-specific cross-linking to map intermolecular contacts between TolC and AcrA (23). In the resulting lowest-energy docked model, the N-terminal α-helix of the adaptor hairpin packs against the outer coil of the TolC α-helical barrel close to the entrance aperture, allowing space for the outward movement of the TolC inner coil during transition to the open state (8, 11, 12, 23). Although this interaction would be compatible with a trimeric arrangement for the adaptor, matching that of the other 2 components, the stoichiometry of interacting components is not yet clear.
Further progress toward a data-driven model of the entire trimeric efflux pump has been precluded by the lack of a complete adaptor monomer structure. The N and C termini, comprising 130 residues, could not be located in the electron density (19, 20), apparently because of their disorder (22, 24). The N-terminal residue of the mature adaptor is acylated (25) and membrane-anchored, so it is likely that the missing domain is closest to the inner membrane (dotted area in Fig. 1) and involved in interaction with the transporter (17, 26). Establishing the complete adaptor structure is therefore key to gaining a data-based view of the adaptor–transporter complex (the translocase) and, from this, the assembled tripartite pump.
Results and Discussion
Structure of the Final Pump Domain: Completing the Periplasmic Adaptor.
To uncover the final adaptor domain (Fig. 1) and complete the structures of the pump components, we re-examined the existing crystallographic data from the MexA monoclinic crystals (19) and developed an improved analysis and averaging of the rigid body interdomain movements, as summarized in Table S1. This analysis established that the apparent disorder actually reflects an unrecognized subtlety in the noncrystallographic symmetry (NCS) of the crystal, which has 13 distinct copies of the adaptor (19). One of the well-ordered copies, chain F, could be refined as an all-atom model at 3.2-Å resolution. From this copy and the analysis of the NCS we were able to establish positions of a further 9 copies in the crystal, giving a final re-refined model with _R_work 23.9% (_R_free 26.4%) (Table S1).
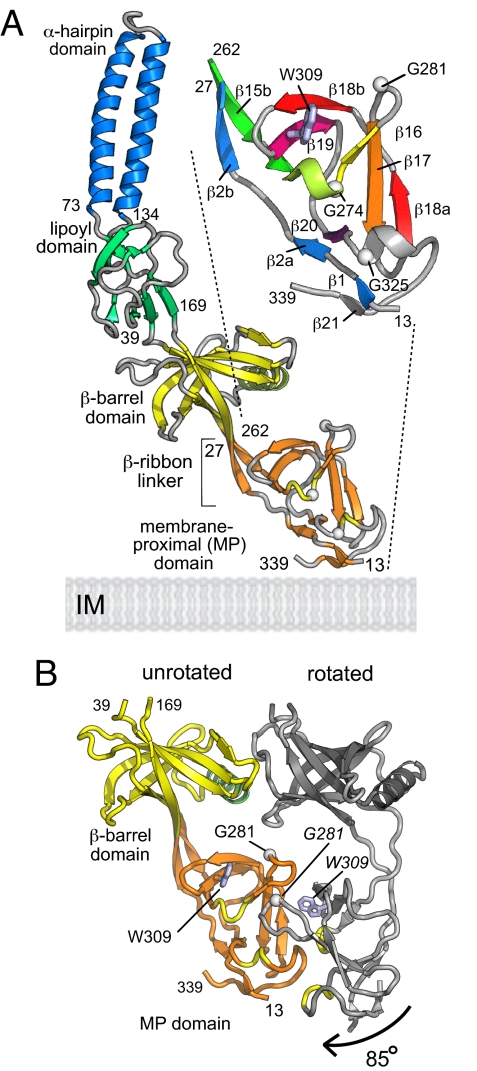
Fig. 2A presents this re-refined model of the MexA adaptor, showing the 4 domains including (in orange) the well-ordered membrane proximal (MP) domain, in which the N and C termini (residues 13–27 and 262–339) together form a compact β-roll extending contiguously from the β-barrel domain on a β-ribbon linker. The N-terminal strand (blue elements in the enlarged inset of Fig. 2A) passes directly through the domain whereas the C terminus contributes an up–down–up β-sheet (β16–β17–β18a as yellow–orange–red, with Gly-281 as a white Cα) and a β-hairpin (β18b–β19 in red–purple, with Trp-309 as a gray side chain) that forms the top of the domain, connected to the base by an extended S-shape linker (gray, β19 to β20). The resulting β-roll arrangement produces a domain with a distinctive concave face characterized by the Gly-281 β-turn and 2 exposed helical turns each of which contains a conserved Gly residue (yellow coil and white Cα atoms in Fig. 2A). Sequence alignments to other efflux pump adaptors (e.g., in Fig. S1 to E. coli AcrA) show that the MP domain is conserved, as are the key features noted above. The equivalence of secondary structure elements in the β-roll and the contiguous β-barrel domain (Fig. S2) indicates that this region of the adaptor structure may have evolved by domain duplication.
Fig. 2.
The completed structure of the periplasmic adaptor. (A) Structure of MexA including the MP domain. The 4 adaptor domains are: α-hairpin (blue), lipoyl (green), β-barrel (yellow), and MP β-roll (orange). Turns are gray except for 2 MP domain helical turns (yellow) that include Gly residues (white Cα atoms) on the concave surface effecting crystal contacts. The enlarged inset gives a smoothed representation of the MP domain topology, with elements numbered according to the adaptor family sequence alignment (Fig. S1) and colored from blue to red. Trp-309 is shown in gray. (B) The MP domain: conformational variation, rotation, and crystal contacts. The MexA adaptor barrel (yellow) and MP (orange) domain, shown in the unrotated MP domain conformation, establish crystal contacts with a neighboring copy (gray), with the MP domain in its rotated conformation. Helical turns on the MP domain concave face are in yellow, and Gly-281 and Trp-309 are shown as white Cα atoms and gray side-chains, respectively (labeled in italics on the rotated domain).
Analysis of the MP domain copies in the crystal (shown in Fig. S3) revealed that apparently favorable interactions between neighboring adaptor molecules result in 2 distinct orientations of the MP domains relative to the rest of the adaptor (Fig. 2B). One orientation, which we designate “unrotated”, is that shown in Fig. 2A, with an ordered β-ribbon linker. The β-barrel and MP domain of this form are shown colored in Fig. 2B, contrasting with the conformation of the adjacent “rotated” conformation of the adaptor, shown in gray. The MP domain of the rotated form is essentially unaltered but is twisted through ≈85 ° clockwise because of a change in the conformation of the β-linker. The conformational change in the linker region results from the close crystal contact that brings the concave faces of the MP domains of the adjacent unrotated and rotated forms into apposition (orange and gray forms, Fig. 2B), interlocking the β-turns (G281, white Cα, Fig. 2B) and helical turns (yellow, Fig. 2B) at the interface. The unrotated orientation of the MP domain was also fortuitously evident in 1 MexA copy that had no close neighbor in the crystal (chain I, Fig. S3), suggesting that this form may represent the preferred resting state of the linker i.e., in the absence of a stabilizing domain contact. The MP domain crystal contacts suggest a propensity of the concave surface to form domain interactions.
Completing the β-roll MP domain, which interacts via its N terminus with the inner membrane, adds ≈45 Å to the height of the linear flexible adaptor, which allows the distal α-helical domain to reach and pack against the TolC entrance helices (23). Initial biochemical and genetic data have implicated each of the MP, β-barrel, and lipoyl domains in complex formation with the inner membrane transporter (17, 26–28), so we set out to determine their contributions to this key interaction.
Mapping the Adaptor–Transporter Contact Surfaces by in Vivo Cross-Linking.
The adaptor–transporter contact surfaces were elucidated by extensive in vivo cross-linking experiments in the E. coli AcrA–AcrB–TolC pump, as used to define the adaptor α-hairpin interaction with TolC (23). The AcrA and MexA sequences are closely homologous, including the MP domain (Fig. S1), and so an AcrA structural model was completed straightforwardly. The single native N-terminal cysteine of AcrA is palmitylated (25) and unreactive, and the 2 AcrB native cysteines were changed to serines. Selected surface-exposed residues were then converted to cysteines on the AcrA adaptor and the AcrB transporter, generating from each a series of specific single-cysteine variants to probe both periplasmic surfaces.
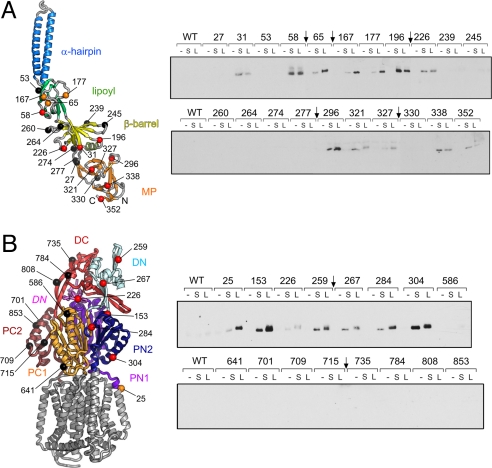
Fig. 3A Left shows the 21 cysteines introduced into AcrA, throughout the contiguous MP, β-barrel, and lipoyl domains. E. coli cultures expressing each cysteine AcrA variant and His-tagged AcrB were treated with each of 2 flexible, membrane-permeable, hetero-bifunctional cross-linkers that react at one end with a sulfhydryl-group and at the other with a primary amine-group. _N_-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) has a short (S) cleavable spacer arm of 6.8 Å, whereas the equivalent arm of sulfosuccinimidyl 6-(3′(2-pyridyldithio)-propionamido) hexanoate (LC-SPDP) is 15.6 Å long (L). Cross-linked AcrA–AcrB complexes, formed between the AcrA-engineered cysteine and an accessible AcrB lysine side chain, were affinity-purified by using the C-terminal His tag on AcrB and proteins immunoblotted for AcrA (Fig. 3A Right). Twelve of the 21 AcrA Cys variants cross-linked, the majority by both S and L cross-linkers (red residues in Fig. 3A Left), but in 4 cases (those substituted at Ala-65, Glu-167, Thr-177, and Glu-327, colored orange in Fig. 3A Left) the S cross-link was reproducibly weaker. Residues colored black in Fig. 3A Left gave no cross-linking, equivalent to WT AcrA, i.e., with no engineered cysteine.
Fig. 3.
In vivo cross-linking of AcrA adaptor and AcrB transporter. (Left) Cysteine substitutions in AcrA (A) and AcrB (B) are shown on backbone ribbons. (Right) The cross-linking results from the AcrA (A) and AcrB (B) variants are presented showing immunoblots of AcrA protein. The AcrA adaptor (A) has 21 Cys substitutions on the lipoyl (green), β-barrel (yellow), and MP β-barrel (orange) domains. On the AcrB transporter periplasmic surface (B), 16 Cys substitutions span the PN1, DN, PN2, PC1, DC, and PC2 subdomains; the integral membrane part of the transporter is shown in gray for reference. In vivo cross-linking was performed by using either the short reagent SPDP (S) or longer LC-SPDP (L). Negative controls without cross-linker (−) or with protein (WT) having no Cys substitution were included in every assay series (duplicate WT controls were removed, indicated by arrows). The residues on the protein structures (Left) are colored red if cross-linked by both S and L linkers, orange if linked only by L linker, or black if negative, i.e., giving no cross-link.
Reciprocal experiments were performed to determine the periplasmic regions of the AcrB transporter interacting with the adaptor. The 16 AcrB cysteine substitutions shown in Fig. 3B Left are exposed on the surface of the periplasmic porter (P) and TolC-docking (D) domains, subdivided into the respective N-terminal (PN1, PN2, DN) and C-terminal (PC1, PC2, DC) subdomains (15), and in one case (Lys-226) on DN intruding from the adjacent subunit (labeled in italics in Fig. 3B Left). Cross-linking reactions between each of the (His-tagged) AcrB cysteine variants and WT AcrA were performed as above and are presented in Fig. 3B Right. Nine of the 16 substituted residues were cross-linked, 7 with both S and L linkers (red in Fig. 3B Left), whereas Leu-25 and Lys-226 were reproducibly weaker with the S linker (orange in Fig. 3B Left). The 7 negative (unlinked) residues are colored black in Fig. 3B Left.
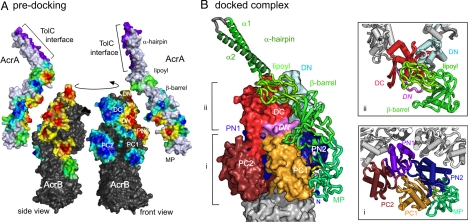
A summary of all 74 (S and L) cross-linking assays is displayed predocking on the AcrA adaptor and AcrB transporter monomers by coloring their surfaces on a simple gradient (Fig. 4A, front and side views) from red, indicating proximity to residues cross-linked, through to blue around residues that did not cross-link. The red cross-linking hot-spots on the transporter subunit form a well-defined area on the half of the periplasmic surface formed from N-terminal subdomains (PN1, PN2, and DN), and span the entire periplasmic height of the transporter (Leu-25, the lowest positive at the inner membrane interface, showed bias to the L linker). The negative residues (indicated by the large blue areas in Fig. 4A) are generally on the C-terminal subdomains. Placed in juxtaposition to the AcrB hot-spots, the AcrA adaptor presents hot-spots on a curved face spanning the lower part of the lipoyl domain and both β-barrel and MP domains. Residues exposed on the opposite face of the adaptor either did not form cross-links or did so primarily with the L cross-linker.
Fig. 4.
Docking of AcrA and AcrB directed by the cross-linking data. (A) AcrA and AcrB interaction hotspots predocking. Cross-linking hotspots on the AcrA and AcrB surfaces predocking, colored on a gradient reflecting all cross-linking data: from dark blue near negative Cys-substituted residues (no cross-link), through green near residues with a bias to the L linker, to yellow/orange/red with increasing proximity to positives linked by both S and L linker. The TolC interface of the adaptor hairpin (23) is in magenta. The arrow indicates rotation from side to front view. (B) Docked complex of AcrA on an AcrB subunit. The 4 AcrA adaptor domains are shown in green shades, with 2 helical turns of the MP concave surface in yellow, β-turn Gly residues in white and the N-terminal residue in blue. The transporter periplasmic subdomain colors are as in Fig. 3B. Cross-sections at 2 levels of the complex (indicated by the brackets) are boxed on the right and illustrate the domain–domain contacts of the adaptor MP domain (i) and β-barrel and lipoyl domains (ii).
Data-Driven Model of the Adaptor–Transporter Complex.
To derive a molecular model of the AcrA–AcrB complex consistent with the constraints defined by the cross-linking distances, we applied a data-driven docking approach comparable to that that established the TolC–AcrA adaptor interaction (23). For the AcrB transporter the subunit chosen for refinement was that closest to the conformation observed in symmetrical trimeric AcrB structures (16), but an additional estimate of average backbone variation allowed for conformational changes in the periplasmic region (14, 15) (see Methods). It was, nevertheless, immediately clear that no docking based on rigid AcrA adaptor structure could satisfy all of the cross-linking data, and so the methods were adapted to explicitly treat the adaptor as a series of rigid bodies with interdomain flexibility as has been observed in the crystal structures (19, 20, 22).
Starting with initially random positions of the AcrA adaptor around the AcrB trimer, the distance constraints were applied as pseudobonds to pull the domains into positions that attempt to optimize the network of cross-linking distances (details in SI Text). The number of restraints would initially seem small for a large complex, but the well-located cysteines and the naturally sparse and random distribution of target lysines limit the range of possible orientations for each rigid domain subject to the requirement for structurally-reasonable interdomain linkers. Indeed, the great majority of docked complexes were dead-end solutions, lacking some required cross-links, and only a unique orientation of the adaptor completely satisfied the data. This final model is presented in Fig. 4B, where AcrA (green), is shown docked on the surface of an AcrB subunit colored to indicate its subdomains (15). The underlying distances between all of the 19 positive cross-linked cysteines and available reactive lysine side-chains are set out in Table S2. The model is also supported by the majority of the negative results, i.e., where there were no cross-links (detailed in Table S3). Fourteen of the 18 negative AcrA and AcrB cysteine residues lie as predicted by the model outside of the cross-linking distance, and the 4 apparent misfits, negative yet apparently within cross-linking distance, appear to be buried at the interface in the docked model or directed away from available lysine partners by side-chain packing.
Once side-chain packing was optimized in the docked model, we removed the cross-link restraints. The proteins remained docked together during further energy minimization, confirming that the docked solution has no unfavorable contacts masked by the cross-linking forces. Comparing Fig. 4 A with B side by side emphasizes how closely it reflects the juxtaposition of mapping hotspots.
The docked AcrA adaptor in Fig. 4B is positioned at a slight angle to the vertical, interacting closely with AcrB at the interface between the exposed periplasmic subdomains of the N-terminal (PN1, DN, and PN2, as blues) and C-terminal (PC1, DC, and PC2, in orange or red) halves of the transporter. Adjacent to the inner membrane, the adaptor MP domain contacts the surfaces of the porter subdomains PN2 and PC1. This is highlighted by the lower cross-section (Fig. 4Bi). Here, the adaptor MP domain is seen to establish a close fit with the transporter via the same concave surface that makes homotypic contacts in the crystal (Fig. 2B). The contiguous adaptor β-barrel domain makes contact with the upper regions of the PN2–PC1 junction (Fig. 4B_ii_) and bridges to the docking domain of the transporter, principally via DN, but also DC and crucially with the projecting loop of the DN subdomain from the adjacent subunit (Fig. 4B_ii_, violet, labeled in italics). The adaptor lipoyl domain also binds at the interface between the N- and C-terminal halves of the transporter TolC-docking domain (DN and DC; Fig. 4B_ii_) while the α-hairpin extends upward from the lipoyl domain.
Each AcrB subunit undergoes conformational changes that are proposed to underpin a rotatory pumping mechanism of the transporter (14, 15), and at the periplasmic surface (e.g., Fig. S4) these localize to the region around the PC2 subdomain. This region is well away from adaptor binding on the relatively stable surface areas of the PN2, PC1, DN, and DC subdomains, suggesting that the docked adaptor would not be required to undergo large conformational changes during the transporter pumping mechanism. Our results are therefore consistent with a single conformation of the docked adaptor on all 3 subunits in the trimeric transporter. Docking 3 adaptors onto the trimeric transporter was therefore performed directly without further refinement, in so doing presenting each of the 3 α-hairpins with its N-terminal helix directed inward (Fig. S4) to form an interface with the entrance coils of a TolC subunit, as required by our previous cross-linking results (23).
Assembling the Complete Drug Efflux Pump.
Finally, we assembled the entire pump by docking the trimeric TolC onto the modeled AcrA–AcrB trimeric complex (Fig. 5A). The adaptor α-helical hairpins were rotated to bind on the TolC entrance coiled coils (the outer helices H3/H4; refs. 8 and 11) at the interaction angle previously determined (23) and this interface was reoptimized (29). The result satisfied all of the previous restraints (Table S4) and retained the structural features and register of the previous TolC–AcrA docking while lifting the adaptor α-hairpin slightly upward. Detail of the refined TolC–adaptor interface is shown in Fig. 5B. It reveals a remarkable pseudodimeric interaction between the AcrA α-hairpin and helices H3 and H4 of the TolC entrance coils (shown as a pseudodyad symbol between green and red elements in Fig. 5B), and a pseudo 3-fold arrangement in the center of the interaction with both the outer coil H3/H4 and the inner coil H7/H8 (shown in the cross-section of Fig. 5B). A similar structural arrangement of antiparallel α-hairpins was observed at the center of the MexA asymmetric unit, where adaptor chains F and G meet (Fig. S3).
Fig. 5.
Assembly of the tripartite efflux pump. (A) Completed pump model. The surface rendering of the TolC3–AcrA3–AcrB3 complex is colored by its components. The TolC trimer (orange, red, and yellow subunits with gray equatorial domains and membrane regions) is docked onto the AcrA (green)-docked AcrB trimer (blue/light blue subunits with gray membrane regions) that was generated in Fig. 4. The location of the inner and outer membrane is indicated. (B) Detail of the TolC–AcrA–AcrB docking region. For clarity the TolC equatorial domains have been omitted between the gray Cα atoms. The boxed cross-section (corresponding to the dotted rectangle at the level of the adaptor lipoyl and α-hairpin domains) shows the TolC helical coils (8) H3 (light red) and H4 (dark red), H7 (light orange) and H8 (dark orange), and the AcrA α-hairpin with α1 (bright green) and α2 (dark green). Pseudodimeric arrangement is indicated by the dyad symbol while a pseudotrimeric arrangement is indicated by the 3-fold symbol in the cross-section. Colored loops and marked Cα positions show TolC–AcrB subunit register (with AcrB colored as for Fig. 4B).
The register of the AcrB–TolC assembly enforced by the cross-linking data allows a close tip-to-tip fit between them (Fig. 5A, blue/dark blue to orange/yellow subunits). Fig. 5B shows the loop from TolC helices H3 to H4 (indicated by Gly-147) nestling between the 2 β-hairpins of the AcrB docking subdomains DN and DC (indicated by residues 255 and 796, respectively) and accommodated above the shorter turn that contains the marked AcrB Gln-197. The positions of the yellow Cα atoms in Fig. 5B illustrate that this register is equally compatible with the reported experimental disulphide bond formation between residues on the TolC loop and AcrB β-turn even though it differs from previous models deriving from these data (18, 30). Significantly, establishing the close fit between the apex of AcrB and the tip of TolC was facilitated by implementing the partially open state of TolC (ref. 13 and Fig. S6). This finding indicates that the fit could be optimal in the fully open form in vivo.
A Central Structural Role for the Adaptor in Pump Assembly.
The assembled structure of the complete tripartite pump presented in Fig. 5 differs substantially from previous purely computational models (13, 16, 30), contains complete structures of the 3 components, and is driven, and validated by a substantial number of site-specific in vivo cross-links. It illustrates the key role of the periplasmic adaptor in establishing an active pump from the weakly interacting inner and outer membrane components, one consequence of which is that each adaptor buries a calculated combined surface area of 7,750 Å2: 4,440 Å2 in binding the AcrB transporter and 3,310 Å2 in recruiting the TolC exit duct. This role is facilitated by the adaptor's linear, multidomain structure in which there is high intrinsic flexibility (22, 24, 31), reflected in hinge-like movements between the lipoyl and both the α-hairpin and the β-barrel domains (refs. 19 and 22, and Fig. S5_A_), and rotation of the MP domain (Fig. 2B and Fig. S5_B_). The resulting close fit between TolC and AcrB is stabilized by the single adaptor on each subunit so there is no need for additional adaptor molecules to generate a continuous seal around the junction of the inner and outer membrane components.
In the pump the adaptor α-hairpin interaction with the TolC entrance was most compatible with coiled coils in an “open-state” arrangement (Fig. S6), generating a coiled-coil interface at the core with extensive contacts and pseudosymmetry deriving from the high structural similarity between the TolC coiled-coil helical pairs and the α-hairpin of the adaptor (Fig. 5B). As we proposed (23), it is likely that this adaptor interaction stabilizes the TolC open state in the assembled pump. The possibility exists that structural changes of the transporter could be transmitted via the adaptor to induce an asymmetric opening of TolC, but this idea is not supported by our data as the adaptor binds to an area of low asymmetry. The evidence for an asymmetric opening of TolC is based largely on a mutated form (13) that lacks key residues interlinking the coiled-coils in the closed state (11, 12). Although TolC recruitment and TolC opening are substrate-dependent, distinct steps in the export of large virulence proteins directed by homologous systems (4), the high throughput of the drug efflux system suggests a constitutively open state for TolC might be more realistic. The mechanism suggested by our modeling is that TolC symmetrically opens when assembled onto AcrB by the adaptor and remains open. There is no requirement for a closed TolC conformation in the assembled efflux pump, because substrate movement will occur entirely within the bodies of AcrB and TolC without leakage to the periplasm. Given both its flexibility and its position away from areas showing large conformational change, the linear adaptor is likely to retain its contacts with the transporter in the pumping cycle. Recently, artificial peptides called DARPins have been produced that bind to the transporter (32) and effectively inhibit the pump. One obvious mechanism for the inhibitory activity would be the disruption of the adaptor–transporter contacts. However, when the DARPin binding sites are compared with the adaptor interface there is no overlap (Fig. S4).
The assembly presented here highlights the structural importance of the periplasmic adaptor protein. However, further involvement of the adaptor in the efflux mechanism is not precluded by our results. It has been suggested that the adaptor might aid access of drugs from the periplasm into the pump (7). Our model shows that the adaptor makes no contacts in the cleft between PC1 and PC2 subdomains thought to be key for such access (refs. 14 and 15 and Fig. 4Bi). However, drug binding in the interior of the transporter has been localized to a hydrophobic binding pocket at the junction of the PN2 and PC1 subdomains (14, 15, 33) that is immediately adjacent to the site of MP domain binding (Fig. 4B_i_). In the transporter crystal structure, overlying residues prevent MP domain access. Nevertheless, one can speculate that the flexible MP domain, localized as it is immediately at the inner membrane, might have a role in facilitating access of hydrophobic substrates to this binding pocket (32, 33).
As a full crystallographic solution of the assembled membrane pump is not likely to be available in the near future, the complete assembly of TolC, AcrB, and AcrA presented here affords a data-driven view of the entire active pump. It represents a total molecular mass of >610,000 Da determined at the resolution of the cross-linking constraints that we estimate to be ≈8 Å, comparable to the resolution of protein complexes in electron microscopy. This remarkable machinery is 270 Å long and establishes a multipurpose gated efflux route spanning the entire bacterial cell envelope, i.e., both membranes and the periplasmic space. The assembled pump is an obvious target for the design of novel chemotherapeutic agents against multidrug-resistant bacteria.
Methods
Re-Refinement of the MexA Structure.
Model building of the complete MexA adaptor structure was based on re-refinement of the crystallographic data collected from the previously-described MexA monoclinic crystal form (ref. 19 and Table S1). The monoclinic crystal asymmetric unit contains 13 NCS copies of MexA (Fig. S3 and refs. 19 and 20). A continuous Cα tracing of the unknown region of one of these copies (chain F) was possible, and a polyalanine model was produced from this. Subsequent refinement allowed modeling of an additional 96 residues (Table S1), and poly(Ala) models of the unknown region in a further 9 chains were produced (A and D copies could not be modeled). Rebuilding was with COOT (34) and CCP4 utilities (35). After minimization in CNS 1.1 (36) with tight NCS restraints, the final step was restrained _B_-factor refinement in PHENIX 1.3 (37). Figures were made with PyMOL (38).
Site-Specific in Vivo Cross-Linking.
Cross-linking was carried out in the double knockout strain E. coli MCΔAcrAB (17), a (DE3) lysogen of MC1061. The native AcrB was extended with 4 C-terminal histidines, and the 2 native cysteine residues (Cys-493 and Cys-887) were converted to serines. This cysteineless AcrB was expressed from vector pACYC184 (39), and WT and variant AcrA were expressed from vector pNIC-28-Bsa4 (Novagen, with modifications by Sune Lobedanz), all at basal levels without T7 promotor induction. Site-directed Cys substitutions (QuikChange; Stratagene), using double-stranded plasmid template and long mutagenic primers, were confirmed by DNA sequencing (Geneservice). All Cys variants assembled functional pumps, conferring resistance to the substrate novobiocin. Doubly-transformed E.coli MCΔAcrAB cultures were grown to an A600 of 0.8 in 50 mL of LB medium with antibiotic selection. Three aliquots of cells from each culture were washed, and cross-linking reactions were carried out as described (23) except that all immunoblotting was with polyclonal rabbit antisera against AcrA. Each variant was assayed, with negative controls, at least 3 times. The cross-linking results were displayed (Fig. 4A) by using PyMOL (ref. 38; details in SI Text).
Data-Driven Docking of AcrA onto AcrB and AcrA–TolC Reoptimization.
Docking of AcrA to AcrB was performed essentially as described (23, 36) modified for a multidomain adaptor and a multisubunit transporter. Cross-links distance restraints had a 1/_R_6 weighting scheme to select partners at optimal distances (40). AcrB backbone and buried side chains were constrained as a single rigid body. The AcrA backbone was constrained as 4 rigid domains, with intervening linker sections given limited flexibility (details in SI Text). Cross-link distances were from the Cβ of variant positions, with 2 Å added for the Cys side chain, increasing the short to 8.8 Å and the long to 17.6 Å. A range of ±2.5 Å for AcrB was added to this, based on an rmsd analysis (Fig. S4), and an additional range of ±3.5 Å for the Nζ atom radius of target Lys rotamers. This gives 8.8 ± 6.0 Å and 17.6 ±6.0 Å for short and long cross-links, respectively (Tables S2–S4). The TolC–AcrA docking (23) was re-refined with HEX 4.8 (29) using a partial open-state structure (13).
Supplementary Material
Supporting Information
Acknowledgments.
We thank Dave Ritchie, Sune Lobedanz, and Hemma Brandstätter for technical help and discussions and Graham Eliff and Jay Pema for computing support. The work was supported by a Wellcome Trust Program Grant (to V.K. and C.H.) and the Cambridge Oppenheimer Research Fund (M.F.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2V4D).
See Commentary on page 6893.
References
- 1.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: The bacterial exit duct for proteins and drugs. Annu Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 2.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three's company: Component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struct Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H, Zgurskaya HI. AcrAB and related multidrug efflux pumps of Escherichia coli. J Mol Microbiol Biotechnol. 2001;3:215–218. [PubMed] [Google Scholar]
- 4.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from Escherichia coli: Reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole K. Multidrug resistance in Gram-negative bacteria. Curr Opin Microbiol. 2001;4:500–508. doi: 10.1016/s1369-5274(00)00242-3. [DOI] [PubMed] [Google Scholar]
- 6.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 7.Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta. 2009 doi: 10.1061/j.bbapap.2008.010. [DOI] [PubMed] [Google Scholar]
- 8.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 9.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 10.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 11.Andersen C, et al. Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci USA. 2002;99:11103–11108. doi: 10.1073/pnas.162039399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eswaran J, Hughes C, Koronakis V. Locking TolC entrance helices to prevent protein translocation by the bacterial type I export apparatus. J Mol Biol. 2003;327:309–315. doi: 10.1016/s0022-2836(03)00116-5. [DOI] [PubMed] [Google Scholar]
- 13.Bavro VN, et al. Assembly and channel opening in a bacterial drug efflux machine. Mol Cell. 2008;30:114–121. doi: 10.1016/j.molcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 15.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 16.Murakami S. Multidrug efflux transporter, AcrB: The pumping mechanism. Curr Opin Struct Biol. 2008;18:459–465. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Touzé T, et al. Interactions underlying assembly of the Escherichia coli AcrAB–TolC multidrug efflux system. Mol Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- 18.Tamura N, Murakami S, Oyama Y, Ishiguro M, Yamaguchi A. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry. 2005;44:11115–11121. doi: 10.1021/bi050452u. [DOI] [PubMed] [Google Scholar]
- 19.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akama H, et al. Crystal structure of the membrane fusion protein MexA of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: Periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 22.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure (London) 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobedanz S, et al. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc Natl Acad Sci USA. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ip H, Stratton K, Zgurskaya H, Liu J. pH-induced conformational changes of AcrA the membrane fusion protein of Escherichia coli multidrug efflux system. J Biol Chem. 2003;278:50474–50482. doi: 10.1074/jbc.M305152200. [DOI] [PubMed] [Google Scholar]
- 25.Yoneyama H, Maseda H, Kamiguchi H, Nakae T. Function of the membrane fusion protein MexA of the MexAB–OprM efflux pump in Pseudomonas aeruginosa without an anchoring membrane. J Biol Chem. 2000;275:4628–4634. doi: 10.1074/jbc.275.7.4628. [DOI] [PubMed] [Google Scholar]
- 26.Elkins CA, Nikaido H. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J Bacteriol. 2003;185:5349–5356. doi: 10.1128/JB.185.18.5349-5356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric analysis of the multicomponent multidrug efflux transporters from Gram-negative bacteria. J Bacteriol. 2002;184:6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J Bacteriol. 2008;190:691–698. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie DW, Kemp GJ. Protein docking using spherical polar Fourier correlations. Proteins. 2000;39:178–194. [PubMed] [Google Scholar]
- 30.Fernandez-Recio J, et al. A model of a transmembrane drug-efflux pump from Gram-negative bacteria. FEBS Lett. 2004;578:5–9. doi: 10.1016/j.febslet.2004.10.097. [DOI] [PubMed] [Google Scholar]
- 31.Vaccaro L, Koronakis V, Sansom MS. Flexibility in a drug transport accessory protein: Molecular dynamics simulations of MexA. Biophys J. 2006;91:558–564. doi: 10.1529/biophysj.105.080010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MGG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2006;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohnert JA, et al. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, et al. Crystallography, NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 39.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz DM, Ihling C, Clore GM, Sinz A. Mapping the topology and determination of a low-resolution 3-dimensional structure of the calmodulin–melittin complex by chemical cross-linking. Biochemistry. 2004;43:4703–4715. doi: 10.1021/bi036149f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information