Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry (original) (raw)
Abstract
Bipolar disorder (BP) is a disabling and often life-threatening disorder that affects ≈1% of the population worldwide. To identify genetic variants that increase the risk of BP, we genotyped on the Illumina HumanHap550 Beadchip 2,076 bipolar cases and 1,676 controls of European ancestry from the National Institute of Mental Health Human Genetics Initiative Repository, and the Prechter Repository and samples collected in London, Toronto, and Dundee. We imputed SNP genotypes and tested for SNP-BP association in each sample and then performed meta-analysis across samples. The strongest association P value for this 2-study meta-analysis was 2.4 × 10−6. We next imputed SNP genotypes and tested for SNP-BP association based on the publicly available Affymetrix 500K genotype data from the Wellcome Trust Case Control Consortium for 1,868 BP cases and a reference set of 12,831 individuals. A 3-study meta-analysis of 3,683 nonoverlapping cases and 14,507 extended controls on >2.3 M genotyped and imputed SNPs resulted in 3 chromosomal regions with association P ≈ 10−7: 1p31.1 (no known genes), 3p21 (>25 known genes), and 5q15 (MCTP1). The most strongly associated nonsynonymous SNP rs1042779 (OR = 1.19, P = 1.8 × 10−7) is in the ITIH1 gene on chromosome 3, with other strongly associated nonsynonymous SNPs in GNL3, NEK4, and ITIH3. Thus, these chromosomal regions harbor genes implicated in cell cycle, neurogenesis, neuroplasticity, and neurosignaling. In addition, we replicated the reported ANK3 association results for SNP rs10994336 in the nonoverlapping GSK sample (OR = 1.37, P = 0.042). Although these results are promising, analysis of additional samples will be required to confirm that variant(s) in these regions influence BP risk.
Keywords: genetics, genome-wide association study
Bipolar disorder (BP) is characterized by dramatic mood changes, with individuals experiencing alternating episodes of depression and mania interspersed with periods of normal function. BP is chronic, severely disabling, and life-threatening, with increased risk of suicide and estimated lifetime prevalence of ≈1% (1).
BP has a substantial genetic component. Monozygotic twin concordance rate estimates range from 45 to 70% and sibling recurrence risk estimates from 5 to 10 (2). Nonetheless, the underlying genetic and neurobiological triggers of BP remain elusive. Numerous linkage and candidate gene studies have sought to identify BP linked regions and associated genes, but no loci have been convincingly identified. Several groups have recently reported results of BP genome-wide association studies (GWAS), using pooled (3) or individually genotyped (4–6) samples; these studies identified SNPs in CACNA1C (alpha 1C subunit of the L-type voltage-gated calcium channel), ANK3 (ankyrin 3), and DGKH (diacylglycerol kinase, eta) as potentially associated with BP.
To test for SNP-BP association in additional well-characterized samples, we analyzed data for >2.3 million genotyped and imputed SNPs on >3,700 individuals in 2 sample sets: (i) 1,177 bipolar I (BP I) cases and 772 controls from the National Institutes of Mental Health Genetic Initiative Repository and the Prechter Repository [National Institute of Mental Health (NIMH)/Pritzker], and (ii) 899 BP cases and 904 controls from London and Dundee, U.K., and Toronto, Canada whose collection was sponsored by GlaxoSmithKline Research and Development (GSK). We analyzed the NIMH/Pritzker and GSK GWAS samples separately and performed a 2-study meta-analysis. We then imputed and analyzed the publicly available Affymetrix 500K genotype data from the Wellcome Trust Case Control Consortium (WTCCC) (4) for 1,868 BP cases and an expanded reference set of 12,831 individuals. The expanded reference set comprised a blood donor sample and 6 non-BP disease case groups. After removing from the GSK London sample 261 cases that overlapped with the WTCCC sample, we performed a 3-study meta-analysis of 3,683 cases and 14,507 extended reference set individuals. We identified 3 regions that harbored SNPs with association P ≈ 10−7.
Results
For the NIMH/Pritzker GWAS, 1,177 BP I cases (473 sibling pairs and 231 unrelated individuals) from the NIMH and Prechter Repositories and 772 controls from the NIMH Repository (Table 1) were genotyped on the Illumina HumanHap 550K chip; 512,844 autosomal SNPs passed QC (see Methods) and had minor allele frequency (MAF)≥1%. For the GSK GWAS, 899 BP cases and 904 controls were genotyped (Table 1); 512,508 autosomal SNPs passed QC and had MAF ≥1%. We carried out genotype imputation, using these data together with estimated haplotypes for the HapMap CEU (Utah residents with ancestry from northern and western Europe) samples, resulting in 2,473,048 (NIMH/Pritzker) or 2,465,069 (GSK) genotyped or imputed autosomal SNPs with MAF ≥1%. Previous work has shown good concordance of imputed and experimental genotypes (7–9).
Table 1.
NIMH/Pritzker, GSK (complete and reduced sample), and WTCCC bipolar case and control characteristics
| Study sample | n | Age at recruitment (yrs) | Female, % | Diagnosis, % BP I | |
|---|---|---|---|---|---|
| Mean (SD) | Range | ||||
| NIMH/Pritzker | |||||
| Cases | 1,177 | 42.2 (12.6) | 14–88 | 62.8 | 100 |
| Controls | 772 | 42.2 (13.4) | 20–69 | 50.3 | — |
| GSK | |||||
| Cases | |||||
| Complete sample | 899 | 47.1 (12.2) | 18–84 | 64.2 | 90.6 |
| Reduced sample | 638 | 46.8 (12.3) | 18–84 | 64.9 | 88.6 |
| Controls | 904 | 39.5 (16.3) | 18–89 | 58.6 | — |
| WTCCC | |||||
| Cases | 1,868 | 40–49 * | <40 to >70 | 63 | 71 |
| Controls | 12,831 | 51 | — |
We tested for SNP-BP association under an additive genetic model, using the observed allele count for genotyped SNPs and estimated allele dosage for imputed SNPs. For both studies, we included as covariates principal components (PCs) based on the genotype data to help correct for potential population stratification; for GSK, we also included study site as a covariate. Genomic control values (10) for genotyped and imputed SNPs were 1.03 and 1.03 in the NIMH/Pritzker sample and 1.02 and 1.03 in the GSK sample (Fig. S1 A–D). After applying genomic control to each set of results, we combined results between samples, using a fixed effects meta-analysis, with resulting genomic control value 1.01 (Fig. S2_A_). No SNP in either study or in the 2-study meta-analysis attained P < 5 × 10−8, corresponding to genome-wide significance of 0.05 assuming the equivalent of 1 million independent tests (11) (Fig. S3_A_). We identified 2 regions with SNP association P < 10−6 (Table S1A): rs12998006 (P = 7.6 × 10−7, OR = 1.34) on chromosome 2 near CPS1 (carbamoyl-phosphate synthetase 1 isoform a) (Fig. S4_A_) and rs2813164 (P = 8.3 × 10−7, OR = 1.31) on chromosome 1 between NEK7 (never in mitosis-related gene 7) and ATP6V1G3 (ATPase, H+ transporting, lysosomal, V1 subunit G3) (Fig. S4_B_).
To increase power to detect BP-associated loci, we obtained genotype data from the WTCCC (4). These data included 1,868 BP cases and an extended reference set of 12,831 individuals comprised of the National Blood Service (NBS) controls and individuals with coronary artery disease, Crohn's disease, hypertension, rheumatoid arthritis, type 1 diabetes, or type 2 diabetes (Table 1). Given the low population prevalence of BP (1%), use of an unscreened expanded reference set should result in little loss of power to detect BP association. These samples had been genotyped on the Affymetrix 500K chip; 397,653 autosomal SNPs passed WTCCC QC criteria and had MAF ≥1%. We carried out imputation as before, resulting in 2,431,899 genotyped or imputed SNPs that passed QC criteria and had MAF ≥1%.
To assess whether the non-BP disease cases could reasonably serve as part of an extended reference set for our BP association study, we first compared the non-BP disease cases to the NBS controls. We observed no strong SNP associations except in the HLA region. After removal of genotypes for autoimmune disease cases (Crohn's disease, rheumatoid arthritis, type 1 diabetes) from the analysis of the HLA region, the genomic control value for non-BP cases vs. NBS controls was 1.03.
We tested for SNP-BP association in the WTCCC BP case and extended reference set under an additive model with the genotype-based PCs as covariates. Genomic control values for genotyped and imputed SNPs were both 1.15 (Fig. S1 G and H). Analysis without PCs resulted in genomic control values of 1.18 and 1.17, suggesting that differences between cases and controls were only partially captured by the PCs, consistent with observations by WTCCC (4) investigators.
We dropped 261 cases present in both the WTCCC and GSK samples from the GSK sample (Table 1 and Fig. S1 E and F), reanalyzed the remaining GSK samples, and applied genomic control to results from each of the 3 studies. We then performed a 3-study meta-analysis of 3,683 cases and 14,507 controls genotyped or imputed for 2,366,197 autosomal SNPs. The meta-analysis genomic control value was 1.04 (Figs. S2_B_ and S3_B_). No SNP in the 3-study meta-analysis reached genome-wide significance at P < 5 × 10−8.
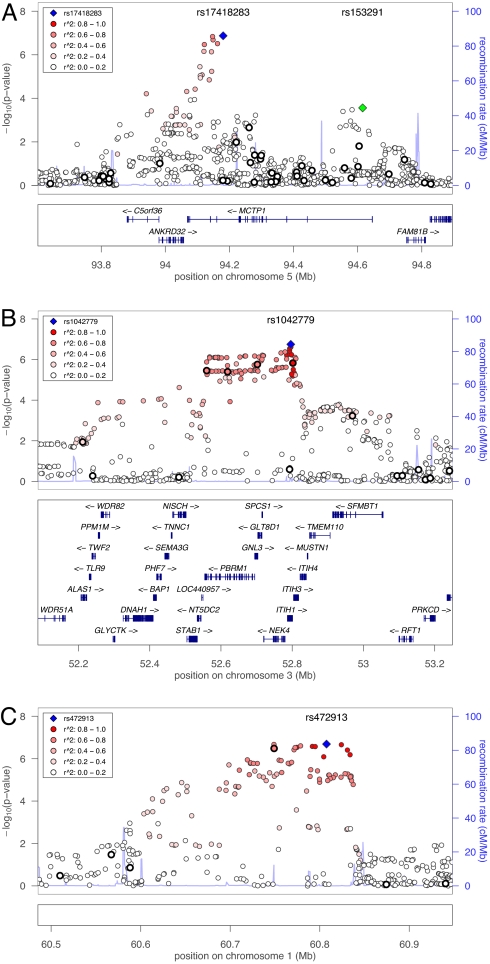
In the 3-study meta-analysis, 53 SNPs from 3 independent regions had P < 10−6 [Table 2(top regional SNP) and Tables S2 and S3]. On chromosome 5q15, we observed strongest association evidence in the genome with SNP rs17418283 (OR = 1.21, _P_ = 1.3 × 10−7) located in an intron of _MCTP1_ (multiple C2 domains, transmembrane 1 isoform) (Fig. 1_A_). We observed a second signal ≈400 kb away in the first intron of _MCTP1_ at rs153291 (OR = 1.13, _P_ = 2.7 × 10−4). Inclusion of rs17418283 as a covariate in the analysis did not change the evidence for association for rs153291 (OR = 1.12, _P_ = 2.8 × 10−4) (Fig. S5). On chromosome 3p21, we observed strongest association evidence (OR = 1.19, _P_ = 1.8 × 10−7) with rs1042779 in a large LD block from 52.2 to 53.2 Mb (Fig. 1_B_). rs1042779 (Arg595Gln) and the nearby rs678 (Glu585Val) (OR = 1.19, _P_ = 2.5 × 10−7) are nonsynonymous SNPs in _ITIH1_ (inter-alpha trypsin inhibitor, heavy chain 1). 33 SNPs in this region showed strong evidence for association (_P_ < 10−6). On chromosome 1p32.1, we observed strongest association evidence with rs472913 (OR = 1.18, _P_ = 2.0 × 10−7) ≈500 kb from the closest known gene, _NF1A_ (nuclear factor 1 A-type) (Fig. 1_C_). In all 3 regions, the large WTCCC case/extended reference set showed the strongest association evidence, but there was no significant evidence of heterogeneity among the 3 studies (Table S2). On chromosomes 1 and 3, inclusion of the most strongly associated SNP as a covariate in the analysis diminished the evidence for association to _P_ > 10−2 for other SNPs in the region; these conditional analysis results are consistent with chromosome 1 and 3 association signals that reflect 1 BP-predisposing variant, or multiple BP-predisposing variants in high LD (Fig. S5).
Table 2.
NIMH/Pritzker, GSK (reduced sample), and WTCCC bipolar meta-analysis association results: loci with P < 10−6
| SNP | Chr | Position* bp | Nearest gene(s) | Risk/nonrisk allele | Control risk allele freq | NIMH/Pritzker | GSK | WTCCC | Meta-analysis | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR 95% CI | P | OR 95% CI | P | OR (95% CI) | P | ||||||
| rs472913 | 1 | 60,807,579 | — | C/G | 0.50 | 1.12 (0.97–1.29) | 0.11 | 1.17 (1.00–1.36) | 0.051 | 1.20 (1.11–1.28) | 6.3 × _10_−7 | 1.18 (1.11–1.25) | 2.0 × 10−7 |
| rs1042779 | 3 | 52,796,051 | NEK4; ITIH1 | A/G | 0.63 | 1.20 (1.04–1.38) | 0.015 | 1.31 (1.11–1.54) | 0.0012 | 1.16 (1.07–1.25) | 0.00012 | 1.19 (1.11–1.27) | 1.8 × 10−7 |
| rs17418283 | 5 | 94,180,344 | MCTP1 | C/T | 0.28 | 1.09 (0.93–1.28) | 0.31 | 1.19 (1.01–1.41) | 0.038 | 1.25 (1.15–1.36) | 9.7 × _10_−8 | 1.21 (1.13–1.30) | 1.3 × 10−7 |
Fig. 1.
Plot of −log10 P values for NIMH/Pritzker, GSK, and WTCCC BP association meta-analysis for chromosomal regions with P < 10−6. refFLAT annotated genes are shown in A–C Lower. SNPs genotyped in all 3 samples are denoted by a thick black circle. Stronger red intersity indicates higher _r_2 with the most significant SNP (purple diamond).
To assess the sensitivity of our results to adjustment for PCs, we reanalyzed data from each study without PCs (Table S4). Meta-analysis P values for the top SNPs with and without PC adjustment varied by up to approximately 1 order of magnitude (Table S5). The most notable change was that evidence for our most strongly associated nonsynonymous SNP, rs1042779 on chromosome 3, became stronger (OR = 1.20, P = 2.7 × 10−8 vs. OR = 1.19, P = 1.8 × 10−7), indicating there may have been some population stratification at this locus.
We next considered the impact on our top results of data imputation. On chromosome 3, the most strongly associated SNP was genotyped in NIMH/Pritzker and GSK; on chromosomes 5 and 1 the most strongly associated SNP was imputed in all 3 studies. Imputation quality of the most strongly associated SNP from each region was high (estimated _r_2 > 0.8) (Table S4); previous work demonstrates that results based on imputed data of this quality generally agree well with corresponding results based on direct genotyping (refs. 7 and 8 and Y. Li, C. J. Willer, J. Ding, P. Scheet, and G. R. Abecasis, personal communication). Further, in the chromosome 1 region, SNP rs1461356 (OR = 1.79, P = 3.3 × 10−7) was directly genotyped in all 3 studies, as was rs2289247 (OR = 1.16, P = 1.7 × 10−6) in the chromosome 3 region. On chromosome 5, no completely genotyped SNP had P < 10−4.
Because of the strong WTCCC contribution to the 3 top results, we further evaluated the use of the WTCCC extended reference set in our analysis. We tested for association between the WTCCC BP cases and the much smaller NBS-only control sample. With this much-reduced sample, we saw more modest evidence of association for each SNP, although similar effect sizes for the chromosome 1 and 3 SNPs: rs472913 (OR = 1.17, P = 0.0020 vs. OR = 1.20, P = 6.3 × 10−7 in the BP cases vs. extended reference set), rs1042779 (OR = 1.17, P = 0.0024 vs. OR = 1.16, P = 0.00012), and rs17418283 (OR = 1.11, P = 0.076 vs. OR = 1.25, P = 9.7 × 10−8) (Table S4_C_). We also tested for allele frequency differences between the NBS controls and the non-BP cases. SNPs rs472913 (P = 0.75) and rs1042779 (P = 0.56) on chromosomes 1 and 3 showed no evidence of a difference. For rs17418283, the allele frequencies in the BP cases, NBS controls, and non-BP cases were 0.31, 0.29, and 0.27, and the allele frequencies in the NBS and non-BP cases were significantly different (P = 0.0067), but this difference was considerably less significant than that observed in the BP/extended reference set analysis (P = 9.7 × 10−8).
These data quality and robustness analyses suggest that the evidence for BP association remains strong for the chromosome 1 and 3 regions and less so for the chromosome 5 region, although the presence of a second signal in the chromosome 5 region strengthens our interest in that region.
Discussion
We carried out genome-wide association analyses of 2 new and 1 previously published (WTCCC) BP GWAS, and then performed metaanalyses of the 2 new studies with and without the WTCCC study. The sample size of the 3-study meta-analysis was 3,683 cases and 14,507 controls. No SNP reached genome-wide significance. For the most strongly associated SNP from each of the 7 regions with P < 10−5 in the 2-study meta-analysis, none had P < 0.1 in the WTCCC data or <0.0001 in the 3-study meta-analysis (Table S1_B_). However, 6 of these 7 associations went in the same direction in the WTCCC data.
In the 3-study meta-analysis, we identified 3 regions with P ≈ 10−7: 1p31.1, 3p21, and 5q15. The most strongly associated SNP genome-wide is rs17418283, located in an intron of MCTP1 on chromosome 5. MCTP1 is an extensively spliced, highly conserved membrane protein that binds Ca2+ with high affinity in the absence of phospholipids (12) and is highly expressed in the brain (http://symatlas.gnf.org/SymAtlas/). This gene may have 2 independent association signals, because SNPs in the first intron of MCTP1 are also moderately and independently associated with BP. Our finding is intriguing because Ferreira et al. (6) recently implicated another Ca2+-related gene, the L-type calcium channel subunit gene, CACNA1C, in BP (P = 7.0 × 10−8). The chromosome 5 region also contains ANKRD32 (ankyrin repeat domain 32), which encodes an uncharacterized ankyrin-repeat-containing protein. There is no known functional relationship between ANKRD32 and the ANK3 gene implicated by Ferreira et al. (6).
We observed the strongest nonsynonymous SNP-BP association genome-wide for rs1042779 on chromosome 3 at 52.8 Mb. There are 10 genes in the most strongly associated 243 kb region, and >25 genes in the ≈1 Mb larger region bounded by flanking recombination hot spots (Fig. 1B). rs1042779 is an Arg595Gln SNP in ITIH1. We also observed association with the nonsynonymous Glu585Val SNP rs678 in the same exon of ITIH1 (r_2 = 0.96 for rs678 and rs1042779 in HapMap CEU). Four additional nonsynonymous SNPs in this region had P < 10−5: rs2289247 (Val367Met) and rs11177 Arg27Gln in GNL3 (guanine nucleotide binding like-3)_, rs1029871 (Pro225Ala) in NEK4, and rs3617 (Gln315Lys) in ITIH3. For the Glu585Val and Pro225Ala variants, Glu and Pro are the conserved alleles in mammals, and the Val and Ala alleles are functionally nonconservative changes. ITIH1 encodes a serine protease inhibitor highly expressed in liver (13). The family of inter-alpha trypsin inhibitors is thought to have anti-proteolytic activities and to play an anti-inflammatory role (14). GNL3 encodes nucleostemin, which was isolated from rat CNS, is expressed in the nucleolus of stem cells, and is thought to be a critical regulator of the cell cycle. Expression of GNL3 rapidly declines upon neuronal cell differentiation, and both over- and under-expression lead to decreased stem cell proliferation in the CNS (15). Aberrant regulation of nucleostemin would be consistent with the neurotrophic hypothesis of mood disorders, which posits that stem-cell proliferative potential in the brain modulates BP risk (16). This association signal is 11 Mb proximal to a BP linkage peak (LOD = 2.01) reported for the families that are the source of the NIMH/Pritzker BP cases analyzed here (17).
On chromosome 1 at 60.8 Mb, there is no well annotated gene within 500 kb of the most strongly associated SNP, rs472913. rs472913 is in moderately high LD with rs2989476 (_r_2 = 0.74), the most strongly associated BP SNP in the WTCCC BP case/extended reference set analysis (4). This SNP is in an intron of a possible transcript annotated by Unigene as P3 NTera2D1 teratocarcinoma, and defined by multiple expressed sequence tags, including one expressed in brain.
After the 3 loci with the strongest associations on chromosomes 1, 3, and 5, there were additional notable observations. On chromosome 2, we identified rs13409348 (OR = 1.20, P = 2.7 × 10−6) (Table S2) located in intron 4 of the gene CTNNA2 (encoding alpha N catenin 2). CTNNA2 is expressed almost exclusively in distinct neuronal populations in primates (18). alpha N-catenin is thought to be a key regulator of the stability of synaptic contacts and the motility of dendritic spines, a key aspect of neuronal plasticity (19). Its deficiency in mice causes axon migration defects (20) and an abnormal startle response (21), a murine behavior indicative of dysregulation of sensorimotor gating that is often considered an endophenotype of psychosis in humans (22). The gene LRRTM1 (Leucine-rich repeat transmembrane neuronal 1) is located in an intron of CTNNA2. We observed associations of a haplotype near LRRTM1 with schizophrenia (P = 0.0014 in 1,002 families) and handedness (P = 0.00002) (23). This risk haplotype is best tagged by the rs1446109 A allele, which is ≈1 Mb from rs13409348 and shows modest evidence of association in our meta-analysis (A risk allele, OR = 1.08, P = 0.08). We also identified rs2537859 (OR = 1.16, P = 4.2 × 10−6) (Table S2) on chromosome 4, 42 kb upstream of KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral). KIT encodes a cytokine receptor of the tyrosine kinase family, expressed in multiple cells including neurons (24). KIT binds stem cell factor and plays a role in cell survival, proliferation, and differentiation (25). Nonsynonymous mutations and deletions in KIT are associated with multiple diseases and KIT overexpression in the brain can induce gliomas (26).
In the 2- and 3-study meta-analyses we identified a region on chromosome 1 at ≈195 Mb with 2 strong but distinct association signals ≈300 kb apart and separated by a strong recombination hotspot. rs2813164 (Table S1) emerged in the 2-study meta-analysis and is located between NEK7 http://www.pnas.org/cgi/data/0813386106/DCSupplemental/Supplemental_PDF#nameddest=SF4and ATP6V1G3 (Fig. S4_B_). rs12568099 emerged in the 3-study meta-analysis (Table S2) and is located 25 kb downstream of PTPRC (Fig. S4_C_); there also was evidence for association around NEK7. Within the region, ATP6V1G3 is of particular interest as it encodes a component of vacuolar proton-pumping ATPase involved in organelle and synaptic vesicle acidification (27).
Previous genome-wide association studies and subsequent metaanalyses of BP have identified SNPs that reached genome-wide significance or had support across multiple studies; for a description of the overlap between our sample and those of other studies, see SI Materials and Methods. The Ferreira et al. (6) meta-analysis identified SNPs with strong evidence of association in the regions of CACNA1C (rs1006737, OR = 1.18, P = 7.0 × 10−8) and ANK3 (rs10994336, OR = 1.45, P = 9.1 × 10−9). Schulze et al. (28) reported support for association with ANK3 in a German sample (rs10994336, OR = 1.70, P = 0.0001) and stronger evidence from meta-analysis with NIMH case and control samples (OR = 1.70, P = 1.7 × 10−5). In the GSK samples that did not overlap the sample of Ferreira et al. (6), we observed modest replication of rs10994336 (OR = 1.37, P = 0.042) and slightly stronger evidence when we included the NIMH samples overlapping those of Shultze et al., but not Ferreria et al. (OR = 1.39, P = 0.012); a fixed-effects meta-analysis of the results of Ferreria et al., the German Schulze et al. sample, and the nonoverlapping GSK and NIMH samples, yielded compelling evidence for association (OR = 1.47, P = 1.1 × 10−10). The _ANK3_-encoded protein, Ankyrin G, is a cytoskeleton-binding molecule located at the Nodes of Ranvier and involved in the assembly of a variety of membrane proteins, including voltage-gated sodium (29) and potassium (30) channels and cell adhesion molecules (reviewed in ref. 31). We did not observe association with rs1006737 (OR = 1.01, P = 0.81). The Baum et al. (3) meta-analysis identified a strongly associated SNP in DGKH (rs1012053, OR = 1.59, P = 1.5 × 10−8); we did not observe association in the independent GSK sample (OR = 1.01, P = 0.76). The lack of confirmation of association in our independent NIMH/Pritzker and GSK samples for these loci may reflect the need for larger sample sizes to detect small effects, differences in sample inclusion criteria including heterogeneity in BP cases, or false positive results in the original studies.
Our GWAS results and others reported to date suggest there are few, if any, common SNP variants with large effects on BP risk. BP differs from complex diseases such as macular degeneration (32), Crohn's disease (4), and type 1 diabetes (4), which are influenced by one or more loci of large effect. Type 2 diabetes appears intermediate in that it has a single locus (TCF7L2) of moderate effect together with loci of substantially smaller effects requiring large samples for detection (33). BP may be due to a combination of many common variants with weak effects, potentially including those identified above, and rarer variants with a range of effect sizes. For common variants, detection and verification will require larger GWA and follow-up samples. The strong familial component of BP and the lack of common variants of large effect suggest that rarer SNP variants along with other types of genetic differences such as copy number variations (CNVs) or epigenetic modifications may play a substantial role. Sequencing of BP cases and controls will help unravel the role of rare SNPs and copy number variants. Detection of BP risk variants anywhere in the allele frequency spectrum has the potential to lead to greater understanding of the biology that underlies BP, the sources of heterogeneity among BP cases, and to the subsequent development of new BP treatments.
In summary, we have identified chromosomal regions and specific genes that may harbor variants that contribute to BP risk and merit further study. Several of the genes encoded in these chromosomal regions are consistent with the notion that subtle differences in neural development and/or neuroplasticity may underlie mood disorders. Further work is required to determine whether these loci contain causal variants that could be more strongly associated with BP risk. If so, it would be of great interest to study the neural localization, expression, and regulation of these genes in human and animal brain, particularly in circuits relevant to mood disorders.
Methods
Sample Ascertainment and Selection.
Subjects for all participating studies gave informed consent and study protocols were approved by the relevant institutional review board or ethics committee.
NIMH/Pritzker.
We selected samples for previously-studied individuals from 2 sources: 1,130 Bipolar I (BP I) cases and 772 controls from the NIMH Human Genetics Initiative Repository (http://zork.wustl.edu/nimh/) and 47 BP I cases from the University of Michigan Prechter Repository. NIMH Repository cases were collected as part of a multicenter study of individuals with BP and their families (34–35). Cases were diagnosed according to DMS-III or DSM-IV criteria, using the Diagnostic Interview for Genetic Studies (DIGS) (36) (n = 1,081) or Family Interview for Genetic Studies (FIGS) (37) and/or medical record review (n = 67); we excluded cases with low confidence diagnoses. From each available non-Ashkenazi European-origin family, when possible we selected 2 BP I siblings including the proband if available (n = 946 individuals in 473 sibling pairs); otherwise we selected a single BP I case (n = 184).
We selected an additional 47 non-Ashkenazi European-origin BP I cases who met DSM-IV criteria from the University of Michigan Prechter Bipolar Genetics Repository. Subjects were recruited using flyers distributed at the University of Michigan Depression Center and on the Center website and interviewed using the DIGS.
We selected as controls 772 non-Ashkenazi European-origin individuals aged 20–70 years from the NIMH Human Genetics Initiative Repository who reported they had not been diagnosed with or treated for BP or schizophrenia, and had not heard voices that others could not hear. We excluded individuals with suspected major depression based on answers to questions related to depressive mood.
To minimize population stratification, we selected individuals reporting primarily European ancestry and matched NIMH controls to NIMH cases based on self-reported ancestry in the DIGS.
GSK.
We selected 899 BP cases (814 BP I and 85 BP II) and 904 controls from subjects recruited at 3 study sites: the Institute of Psychiatry (IOP) in London, U.K. (483 cases and 462 controls); the Centre for Addiction and Mental Health in Toronto, Canada (334 cases and 257 controls); and the University of Dundee, U.K. (82 cases and 185 controls). Cases were recruited through advertisements in hospitals, clinics, primary care physician offices, and patient support groups, were ≥18 years of age at interview, and reported Caucasian ethnicity. They were interviewed using the Schedules for Clinical Assessment in Neuropsychiatry (SCAN). BP diagnoses were established according to DSM-IV or ICD-10 criteria, using the computerized algorithm (CATEGO) for the SCAN2.1 interview (WHO) (38). Cases were excluded if they received a diagnosis of i.v. drug dependency or reported i.v. drug use or if they had mood incongruent psychotic symptoms or if manic episodes only occurred in conjunction with or as a result of alcohol, substance abuse, substance dependence, medical illnesses, or medications. 261 cases recruited at the IOP were present in the WTCCC BP sample. When we performed a 3-study meta-analysis, we excluded these overlapping cases from the GSK sample and used 638 cases and 904 controls as the GSK analysis sample (Table 1 and Table S1). Controls were ≥18 years of age, reported Caucasian ethnicity, and denied the presence of any psychiatric disorders in a questionnaire.
WTCCC.
We accessed phenotype and GWAS genotype data on 1,868 BP cases and 12,831 additional individuals provided by the WTCCC (www.wtccc.org.uk/info/access_to_data_samples.shtml). All samples were ascertained in the U.K., reported European ancestry, and clustered with the WTCCC in multidimensional scaling of the WTCCC and the 270 HapMap samples. BP and related phenotypes were diagnosed using Research Diagnostic Criteria (4). 71% of cases were diagnosed as BP I, the remainder as schizoaffective disorder bipolar type (15%), BP II (9%), or manic disorder (5%). For our primary WTCCC control group, we included 12,831 individuals unscreened for BP: 1,458 National Blood Service controls together with a total of 11,373 individuals from the coronary artery disease (n = 1,926), Crohn's disease (n = 1,748), hypertension (n = 1,952), rheumatoid arthritis (n = 1,860), type 1 diabetes (n = 1,963), and type 2 diabetes (n = 1,924) groups. We could not include the WTCCC 1958 Birth Cohort data owing to access constraints.
Sample Genotyping and Quality Control (QC).
NIMH/Pritzker.
NIMH and Prechter samples were genotyped on the Illumina HumanHap550 Beadchip at Stanford University and the University of Michigan following manufacturer recommendations. Phase1 (phase 2) comprised 999 (1,122) samples, including 29 (19) duplicate samples and 15 parent-offspring trios (1 parent-offspring duo,1 parent-offspring trio, 8 parent-offspring quartets). In each phase, we first called genotypes using the Illumina default cluster file based on the HapMap CEU samples and excluded samples with success rate <98.0%. Genotypes were then reclustered based on our own genotyped samples; for each SNP, we retained the genotypes from the clustering method yielding higher genotyping success rate. Nine individuals with inconsistent reported and genotype-based sex were dropped as were 6 samples to eliminate cryptic relatedness. We removed 6 individuals with evidence for ancestry that differed from the rest of the sample (see below).
Of the 541,327 genotyped autosomal SNPs, we excluded 12,592 because of (i) total non-Mendelian and duplicate errors >2; (ii) data inconsistent with Hardy-Weinberg equilibrium (P < 10−5) in unrelated cases and controls; or (iii) SNP call rate <95% in either phase. Genotyping success rate (99.9%), duplicate genotype concordance (99.992%), and parent-child Mendelian consistency rates (99.93%) were high. There was no evidence for differential quality parameters between phases 1 and 2. Among the 528,735 SNPs that passed QC, 512,844 had MAF ≥1% in the combined sample and were used in association analysis.
GSK.
DNAs for 1,896 GSK individuals, including duplicated samples for 109 individuals, were genotyped on the Illumina HumanHap550 Beadchip at Illumina following manufacturer recommendations. Genotype calls were generated as described in ref. 39 with minor modifications. Initial calls were made using the Illumina CEU cluster file. 61 samples from 43 individuals with call rate <95% were dropped as were 11 individuals with inconsistent reported and genotype-based gender, 13 individuals to eliminate cryptic relatedness, and 26 outlier individuals in the stratification analysis (see below). SNPs with call frequency <99% were reclustered based on the GSK samples. 20,172 SNPs with (a) call frequency <95%; (b) call frequency 95–98% and cluster separation score <0.3, or heterozygote excess frequency >0.1 or less than −0.1; or (c) call frequency >98% and cluster separation <0.25 or heterozygote excess frequency >0.3 or less than −0.3 were dropped. Of the 521,990 SNPs that passed QC, 512,508 (full sample) and 512,668 (reduced sample) had MAF≥0.01 and were used in association analysis. Duplicate genotype concordance was 99.99%.
WTCCC.
Samples were genotyped on the Affymetrix GeneChip 500K Mapping Array Set as described in ref. 4. 397,653 SNPs passed WTCCC QC and had MAF≥0.01.
Genotype Imputation.
For each study, we imputed genotypes for up to ≈2 million autosomal SNPs with MAF≥0.01, using the genotypes from the Illumina HumanHap550 Beadchip (NIMH/Pritzker, GSK) or the Affymetrix GeneChip 500K Mapping Array Set (WTCCC) and phased chromosomes for the 60 HapMap CEU founders. Genotypes were imputed using a Hidden Markov model as programmed in MACH (9) for SNPs not present in the genotyping platform or whose genotype data failed QC. We imputed WTCCC and Pritzker to Build 35 and GSK to Build 36. We retained imputed SNPs with estimated _r_2 > 0.3 and imputed MAF≥0.01. The numbers of imputed SNPs used in analysis were 1,960,204, 1,952,561 (1,952,849), and 2,034,246 for NIMH/Pritzker, GSK full sample (reduced sample), and WTCCC, respectively.
The SI Materials Materials and Methods contains an expanded version of the following sections.
Assessment of Stratification.
In each set of study samples we performed principal components (PC) analysis based on a subset of the sample genotypes (40) in unrelated individuals. We excluded individuals in the NIMH/Pritzker and GSK samples with PC >6 SD from the mean of one or more of the top 10 PCs; to mirror the analysis used by the WTCCC (4), we did not exclude WTCCC samples.
GWA Analysis.
We eliminated 694 SNPs with allele frequency differences >0.2 for any pair of studies. We analyzed the observed allele counts or imputed allele dosages, using logistic regression assuming an additive genetic model, with genotype-based PCs and study site (GSK only) as covariates, and then repeated the analysis without PCs. In the NIMH/Pritzker sample, we used a sandwich estimator (41) to adjust the estimated variances. For the WTCCC sample, we compared the BP cases to the extended reference set as our primary analysis, except in the HLA region on chromosome 6 from 27.2 to 34.0 Mb where we excluded the 5,571 autoimmune disease cases (type 1 diabetes, Crohn's disease, rheumatoid arthritis) from the GWA analysis.
Meta-Analysis of GWA Samples.
We performed a fixed effects meta-analysis, using the OR and 95% confidence intervals to combine the association evidence from the study-specific GWA analyses. We used association results for experimentally derived genotypes when available, and for imputed genotypes otherwise. 2,366,197 autosomal SNPs passed QC and had MAF≥0.01 in all 3 samples. We adjusted for the genomic control values in each study separately for genotyped and imputed SNPs by increasing the standard error of the OR estimate to correspond to the genomic control P value. Evidence for heterogeneity between ORs was assessed using Cochrans's Q statistic and _I_2 (9).
Supplementary Material
Supporting Information
Acknowledgments.
We thank the participants who donated their time and DNA to make this study possible; Elzbieta Sliwerska for assistance with genotyping; Terry Gliedt, Peggy White, and Randy Pruim for assistance with data management and manuscript preparation; members of the National Institutes of Mental Health Human Genetics Initiative (see SI Acknowledgments) and the University of Michigan Prechter Bipolar DNA Repository for generously providing phenotype data and DNA samples; the staff at the recruiting sites in London, Toronto, and Dundee, and at GlaxoSmithKline for contributions to recruitment and study management; and the other members of the Pritzker Neuropsychiatric Disorders Research Consortium (PNDRC) for their contribution to the research. This study makes use of data generated by the Wellcome Trust Case Control Consortium (WTCCC) (see SI Acknowledgments). Many of the authors (L.J.S., W.G., M.F., J.Z.L., M. Burmeister, D.A., R.C.T., F.M., A.F.S., W.E. B., J.D.B., E.G.J., S.J.W, R.M.M., H.A., M. Boehnke) are members of the PNDRC. This work was supported by the Pritzker Neuropsychiatric Disorders Research Fund, L.L.C.
Footnotes
Conflict of interest statement: P. Muglia, X.Q.K., F.T., C.F., A.A., and D.K.B. are, or were, full-time employees of GlaxoSmithKline when this article was written. GlaxoSmithKline sponsored the bipolar collection at the London, Toronto, and Dundee sites. R.D., K.M., P. McGuffin, J.S.S., J.L.K., L.M., J.B.V., and A.E.F. worked at those sites
References
- 1.Merikangas KR, et al. Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Arch Gen Psychiatry. 2007;64:543–552. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craddock N, Forty L. Genetics of affective (mood) disorders. Eur J Hum Genet. 2006;14:660–668. doi: 10.1038/sj.ejhg.5201549. [DOI] [PubMed] [Google Scholar]
- 3.Baum AE, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sklar P, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira MA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 8.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoggart J, Clark TG, DeIorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 12.Shin OH, Han W, Wang Y, Südhof TC. Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J Biol Chem. 2005;280:1641–1651. doi: 10.1074/jbc.M407305200. [DOI] [PubMed] [Google Scholar]
- 13.Bost F, et al. Isolation and characterization of the human inter-alpha-trypsin inhibitor heavy-chain H1 gene. Eur J Biochem. 1993;218:283–291. doi: 10.1111/j.1432-1033.1993.tb18376.x. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H. Endogenous anti-inflammatory substances, inter-alpha-inhibitor and bikunin. Biol Chem. 2006;387:1545–1549. doi: 10.1515/BC.2006.192. [DOI] [PubMed] [Google Scholar]
- 15.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Kelsoe JR, et al. A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA. 2001;98:585–590. doi: 10.1073/pnas.011358498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith A, Bourdeau I, Wang J, Bondy CA. Expression of Catenin family members CTNNA1, CTNNA2, CTNNB1 and JUP in the primate prefrontal cortex and hippocampus. Brain Res Mol Brain Res. 2005;135:225–231. doi: 10.1016/j.molbrainres.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 19.Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- 20.Uemura M, Takeichi M. Alpha N-catenin deficiency causes defects in axon migration and nuclear organization in restricted regions of the mouse brain. Dev Dyn. 2006;235:2559–2566. doi: 10.1002/dvdy.20841. [DOI] [PubMed] [Google Scholar]
- 21.Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat Genet. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- 22.Seong E, Seasholtz AF, Burmeister M. Mouse models for psychiatric disorders. Trends Genet. 2002;18:643–650. doi: 10.1016/s0168-9525(02)02807-x. [DOI] [PubMed] [Google Scholar]
- 23.Francks C, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang SC, Fedoroff S. Cellular localization of stem cell factor and c-kit receptor in the mouse nervous system. J Neurosci Res. 1997;47:1–15. [PubMed] [Google Scholar]
- 25.Jensen BM, Akin C, Gilfillan AM. Pharmacological targeting of the KIT growth factor receptor: A therapeutic consideration for mast cell disorders. Br J Pharmacol. 2008;154:1572–1582. doi: 10.1038/bjp.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blom T, et al. KIT overexpression induces proliferation in astrocytes in an imatinib-responsive manner and associates with proliferation index in gliomas. Int J Cancer. 2008;123:793–800. doi: 10.1002/ijc.23576. [DOI] [PubMed] [Google Scholar]
- 27.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: Molecular structure andfunction, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 28.Schulze TG, et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malhotra JD, Kazen-Gillespie K, Hortsch M, Isom LL. Sodium channel beta subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell–cell contact. J Biol Chem. 2000;275:11383–11388. doi: 10.1074/jbc.275.15.11383. [DOI] [PubMed] [Google Scholar]
- 30.Pan Z, et al. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett V, Chen L. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr Opin Cell Biol. 2001;13:61–67. doi: 10.1016/s0955-0674(00)00175-7. [DOI] [PubMed] [Google Scholar]
- 32.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeggini, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nurnberger JI, Jr, et al. Genomic survey of bipolar illness in the NIMH genetics initiative pedigrees: A preliminary report. Am J Med Genet. 1997;74:227–237. doi: 10.1002/(sici)1096-8628(19970531)74:3<227::aid-ajmg1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 35.Dick DM, et al. Genomewide linkage analyses of bipolar disorder: A new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nurnberger JI, Jr, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Gen Initiative Arch Gen Psych. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 37.Maxwell ME. Family Interview for Genetic Studies (FIGS): A Manual for FIGS, Clinical Neurogenetics Branch, Intramural Research Program, National Institute of Mental Health, Bethesda, Maryland. Bethesda, MD: NIMH; 1992. [Google Scholar]
- 38.Celik C. Geneva: WHO; 2003. Computer assisted personal interviewing application for the Schedules for Clinical Assessment in Neuropsychiatry Version 2.1 and diagnostic algorithms for WHO ICD 10 chapter V DCR and for statistical manual IV. Release I. Ed. 1.0.3.5. [Google Scholar]
- 39.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.White H. Maximum likelihood estimation of misspecified model. Econometrica. 1982;50:1–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information