Cdc42 and Rac Family GTPases Regulate Mode and Speed but Not Direction of Primary Fibroblast Migration during Platelet-Derived Growth Factor-Dependent Chemotaxis (original) (raw)
Abstract
Cdc42 and Rac family GTPases are important regulators of morphology, motility, and polarity in a variety of mammalian cell types. However, comprehensive analysis of their roles in the morphological and behavioral aspects of chemotaxis within a single experimental system is still lacking. Here we demonstrate using a direct viewing chemotaxis assay that of all of the Cdc42/Rac1-related GTPases expressed in primary fibroblasts, Cdc42, Rac1, and RhoG are required for efficient migration towards platelet-derived growth factor (PDGF). During migration, Cdc42-, Rac1-, and RhoG-deficient cells show aberrant morphology characterized as cell elongation and cell body rounding, loss of lamellipodia, and formation of thick membrane extensions, respectively. Analysis of individual cell trajectories reveals that cell speed is significantly reduced, as well as persistence, but to a smaller degree, while the directional response to the gradient of PDGF is not affected. Combined knockdown of Cdc42, Rac1, and RhoG results in greater inhibition of cell speed than when each protein is knocked down alone, but the cells are still capable of migrating toward PDGF. We conclude that, Cdc42, Rac1, and RhoG function cooperatively during cell migration and that, while each GTPase is implicated in the control of morphology and cell speed, these and other Cdc42/Rac-related GTPases are not essential for the directional response toward PDGF.
The migration of cells toward or away from the source of a diffusible signaling factor is known as chemotaxis, a fundamental form of cell behavior implicated in a wide range of physiological and pathological processes, including wound repair, immune response, and cancer metastasis. Many different mammalian cell types exhibit chemotaxis, from “professional” migratory cells such as neutrophils, which exhibit rapid amoebalike motility, to larger cells such as fibroblasts, which exhibit slow and complex movements. A wide variety of signaling molecules serve as putative chemoattractants for various mammalian cell types, ranging from metal ions such as calcium (7) to short bacterial peptides such as _N_-formyl-methionyl-leucyl-phenylalanine (fMLP) (19, 26), growth factors, and chemokines such as platelet-derived growth factor (PDGF) (21) and SDF-1α (4), and even vast extracellular matrix proteins including collagens (16), fibronectin (15), and hyaluronan (25). Consequently, the underlying cellular mechanisms that regulate chemotaxis are likely to vary between different cell types and in response to the activation of different receptor classes.
Ultimately, however, for a cell to chemotax it must first polarize, orienting itself along the direction of the chemotactic gradient. The polarization of a cell in response to a chemotactic signal requires the processing of spatiotemporal gradient cues within the cell's surrounding environment and the transmission of this information to the cytoskeleton to enable appropriate morphological responses. Rho family GTPases are believed to play a central role in this process, translating cell surface signals into the mechanics of cell movement through their ability to regulate and remodel the cytoskeleton (8, 17).
It is now clear that the contribution of individual Rho family GTPases in the regulation of cytoskeletal architecture and cell migration can be cell type dependent. This may reflect functional specialization within a cell, which is likely to be dictated by the relative abundance of related GTPases and the availability of specific downstream effectors. For example, expression of the Rho GTPase Rac2 is limited to cells of hematopoietic origin, as is the Cdc42 effector WASP. Consequently, loss of function of a particular GTPase can result in a more severe phenotype for one cell type than for another. Indeed, while the loss of Cdc42 function in primary mouse fibroblasts has been reported to impair migration speed (31), it has no such effect on fibroblastoid cells (5) and actually enhances the speed of Drosophila hemocytes (24) and macrophages (2). In addition, it has been shown that Rac1 is an important regulator of migration speed in fibroblasts (28) but not in macrophages which express both Rac1 and Rac2 (30). Such examples demonstrate that it is imprudent to generalize about the importance of specific Rho GTPases in certain cellular processes. Also, it is beneficial to study related GTPases within a single, well-defined cell system in order to clarify their individual roles in specific cellular processes and to establish whether functional redundancy exists among different family members. However, while a great number of studies have now examined the roles of various Rho family GTPases in the regulation of cell morphology, migration, and chemotaxis, a comprehensive analysis of these proteins in the regulation of these aspects of cell behavior within a single experimental system is still lacking.
In the present study we perform a detailed analysis of the role of all of the Cdc42 and Rac-related GTPases in the chemotaxis of primary fibroblasts using a Dunn direct-viewing chamber, which enables the long-term observation of cells in a chemotactic gradient. Mouse embryonic fibroblasts (MEFs), which exhibit robust and highly reproducible chemotaxis toward PDGF-BB in vitro, are used as the chemotaxis model. Short interfering RNAs (siRNAs) were used to inhibit the expression of specific GTPases both individually and in combination, and the Dunn chamber was then used in conjunction with fluorescent cell labeling techniques and time-lapse microscopy to directly observe the effects of various siRNAs on the behavior of primary fibroblasts in a chemotactic gradient of PDGF-BB. Our experimental system, which enables direct comparisons to be made between control and test cell populations within the same chemotaxis experiment, provides an extremely powerful method for assessing the significance of differences observed between different treatment groups.
Here we report that Cdc42, Rac1, and RhoG are all important regulators of cell morphology and are required for the efficient chemotaxis of primary fibroblasts in a PDGF gradient. Although the migration of cells in a PDGF-BB gradient is impaired after knockdown of either of these GTPases, the mean direction of cell movement is essentially unaffected. Changes in migration speed, but not the directional response toward PDGF, thus account for the impaired chemotaxis observed. We demonstrate that in the absence of Cdc42, Rac1, or RhoG cells exploit alternative modes of migration, which may reflect the cell's capacity to exploit the functions of the two remaining proteins. Combined knockdown of Cdc42, Rac1, and RhoG results in far greater inhibition of cell migration than when each protein is knocked down alone, demonstrating that these GTPases function cooperatively during fibroblast migration. Finally, we show that the Cdc42/Rac-related GTPases Tc10, Tcl, Wrch1, and Rac3, which are all expressed in primary fibroblasts, are dispensable in the migration and chemotaxis of these cells. We present here one of the first comprehensive studies of the Cdc42- and Rac-related GTPases in the chemotaxis of primary fibroblasts and provide new insights into their cooperative roles in cell migration. In addition, we describe a robust experimental system that can be applied in general to assess and compare the role of signaling molecules in cell migration in vitro.
MATERIALS AND METHODS
Culture of MEFs.
Passage 2 embryonic day 15.5 MEFs were used in all studies. MEFs were cultured in Dulbecco modified Eagle medium supplemented with 0.13% NaHCO3, 1.6% l-glutamine, 0.1% bovine serum albumin, and 100 U/ml penicillin-100 μg/ml streptomycin (MEF culture medium). For routine culture, this medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS); for starvation prior to the study of chemotaxis, the cells were cultured for 16 h in medium supplemented with 0.5% FBS; and for starvation prior to pulldown experiments, the cells were cultured for 16 h in serum free medium.
Cloning of Cdc42 and Rac family GTPases from MEFs.
Total cytoplasmic RNA was isolated from 3 × 106 MEFs by using an RNeasy minikit (Qiagen). One microgram of purified RNA was then used as a template for the synthesis of single-stranded DNA using oligo(dT) primers and the Superscript II reverse transcription-PCR (RT-PCR) system (Invitrogen). PCR was performed on the single-stranded DNA template in order to clone individual Cdc42/Rac family GTPases. The primers used for the amplification of full-length Cdc42/Rac family GTPases are listed in Table S1 in the supplemental material. The following conditions were used for the amplification of all target sequences: an initial 98°C denaturing step; followed by 22 to 28 cycles (Cdc42, Tc10, Tcl, Rac1, Rac3, and RhoG), 35 cycles (Rac2), or 40 cycles (Chp) of a 98°C denaturing step for 15 s; a 60°C annealing step for 30 s; and a 68°C extension step for 1 min. Cycling was followed by a final 10-min extension step at 68°C. PCR products were cloned into a pCR-BLUNT shuttle vector (Invitrogen), sequenced, and finally subcloned into the pEGFP-C2 mammalian expression vector (Clontech).
Transfection of siRNAs.
Frozen passage 2, the MEFs were recovered, resuspended in 10 volumes of OptiMEM medium, and washed by centrifugation before seeding them directly into fibronectin-coated (1 μg/ml) dishes in the presence of transfection reagents. In all cases, the cells transfected with control siRNA (scrambled sequence, medium GC content; Invitrogen catalog no. 12935, lot 1351421) and test siRNA (specific for a given target GTPase) were prepared in parallel from the same frozen stock. For each transfection, 106 cells were seeded in a final volume of 1.2 ml of OptiMEM in the presence of transfection complexes (50 nM siRNA/3 μl of Lipofectamine 2000). After 2.5 h, the transfection medium was replaced with MEF culture medium, and the cells were then cultured for an additional 4 h before being harvested and labeled with fluorescent marker dyes (see Fig. S1 in the supplemental material). Cell labeling was performed by using the PKH cell labeling kit (Sigma) as follows. Cells from control and test transfections were harvested by trypsinization, pelleted by centrifugation, and resuspended in 500 μl of diluent C containing 1 μl of either the PKH67 green or PKH26 red fluorescent marker dye. Cells were incubated in the presence of the dye for 5 min before the addition of an equal volume of FBS to inactivate the labeling reaction. Cells were then washed by the addition of 10 volumes of culture medium and subsequent pelleting by centrifugation. Cell pellets were resuspended in MEF culture medium, and fractions of the control and test transfected cell populations were then mixed and seeded at a density of 0.5 × 105 cells/ml onto coverslips precoated with fibronectin (10 μg/ml). This yielded a mixed population of control and test siRNA-transfected cells, with each subpopulation identifiable by their specific fluorescent membrane marker dye. The remaining, unmixed cells from each treatment group were then reseeded into separate culture dishes for later analysis by either RT-PCR or Western blotting to confirm the inhibition of expression of target mRNAs or proteins. Cell cultures were transferred to medium containing 0.5% serum 32 h after transfection and 16 h prior to the assembly of Dunn chemotaxis chambers. The primers used for the amplification of target sequences during RT-PCRs are listed in Table S2 in the supplemental material. Stealth siRNAs (Invitrogen) for the knockdown of specific Cdc42 and Rac family members were designed using the online BLOCK-iT RNAi Designer software (Invitrogen). The target sequences for the various siRNAs used in the present study are listed in Table S3 in the supplemental material.
Chemotaxis assay.
The Dunn direct viewing chamber (34, 35) in conjunction with time-lapse microscopy was used to record the behavior of MEFs in a stable PDGF-BB gradient over a period of 8 h (Fig. 1A). Throughout the present study modified Dunn chambers were used in which the length of the diffusion gap was approximately 400 μm. An Olympus IX81 inverted epifluorescence microscope equipped with an automated x,y stage (Sigma Koki) was used to capture images of multiple, overlapping observation fields across multiple Dunn chambers (Fig. 1B and C). Dunn chambers were mounted on a transparent thermo-plate (Tokai Hit) positioned at the center of the x,y stage, and accurately maintained at 37°C. Cells were imaged with either an Olympus UPlanFl ×10/0.30 Ph or UPlanFl ×20/0.50 Ph objective lens. The microscope was fitted with tungsten and mercury lamps, separate bright-field and fluorescence shutters (Uniblitz), a shutter controller (Uniblitz), and a charge-coupled device camera (Roper Scientific). All peripheral devices were controlled using Metamorph software (Universal Imaging). Single green and red fluorescence images were acquired at the beginning of the time-lapse sequence in order to identify the initial positions of control and test siRNA-transfected cells (Fig. 1F and see Movie S1 in the supplemental material). All subsequent imagining was conducted under phase contrast. Images were acquired every 10 min for the entire duration of the 8-h observation period. A description of the Dunn chamber and its use can also be found elsewhere (29, 34, 35).
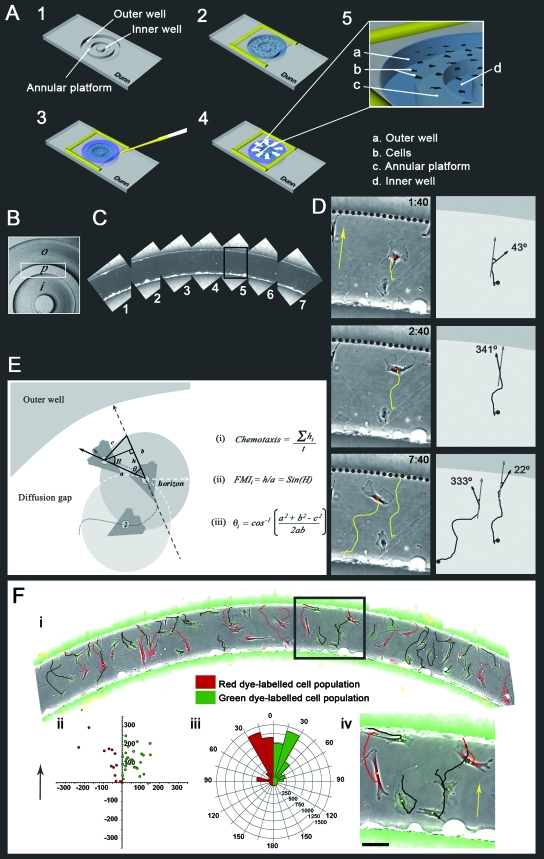
FIG. 1.
(A) The Dunn chamber and its assembly. (Diagram 1) The Dunn chamber is manufactured from glass and consists of an outer and inner well separated by an annular platform that is set 20 μm below the chamber surface. (Diagram 2) A coverslip with adherent cells is inverted and lowered onto the chamber, leaving a small filling slit at one end of the outer well. The coverslip is then sealed to the chamber with wax. (Diagram 3) The chemoattractant is applied to the outer well, and the filling slit is sealed. (Diagram 4) The chemoattractant diffuses radially from the outer to the inner well. (Diagram 5) Cells positioned over the annular bridge lie within a gradient of the chemoattractant. (B) Top-down view of the Dunn chamber illustrating the outer (o) and inner (i) wells and the annular platform (p). (C) Time-lapse imaging of multiple overlapping fields enables the postacquisition reconstruction of a large portion of the annular platform. Fields 1 to 7 correspond to the boxed region in panel B. (D) Enlargement of the boxed region in panel C. The images show selected time frames demonstrating the chemotaxis of MEFs in a PDGF-BB gradient (left column), the subsequent trajectories generated by interactive tracking of individual cells over the course of the film sequence (left and right columns), and the corresponding cell direction, θ, with respect to the direction of the gradient at each respective time point (right column). The yellow arrow indicates the direction of increasing PDGF-BB concentration, and the dotted line indicates the border between the outer well and the annular platform. Scale bar, 50 μm. (E) Cell trajectory data are used to evaluate the chemotaxis, directional response, and speed of individual migrating cells. Equations for the evaluation of chemotaxis (i) and for the derivation of the FMI (ii) for evaluation of the directional response are shown. Calculation of the angle of deviation of cell migration from the direction of the chemotactic gradient (iii) was used for the construction of circular histograms for the graphical representation of chemotaxis and directionality. (F) The coculture of cells labeled with either red or green fluorescent dyes enabled the direct comparison of the two cell populations within the same experiment. (i) Multiple overlapping fields show a large portion of the annular platform of the Dunn chamber containing many red and green fluorescent cells. Images represent composite overlays of red and green fluorescence and phase contrast. Red and green trajectories represent the paths taken by individual cells of the respective label color over an 8-h observation period. (ii) A displacement plot indicating the final position of each cell track, shifted to a common origin for the two dye-labeled populations in panel i. (iii) Circular histogram demonstrating the chemotactic response of the two dye-labeled cell populations. The arrow indicates the direction of increasing PDGF-BB concentration. (iv) Enlargement of the boxed region in panel i. Scale bar, 50 μm.
Analysis of cell behavior.
The subsequent interactive tracking of cells from acquired film sequences was used to generate cell trajectories for the statistical analysis of cell behavior (Fig. 1D and F). Cell trajectories were used to analyze the chemotactic response of individual cells and also to analyze various components of cell migration including speed, directional response to the gradient, and directional persistence. Mean cell speed in the direction of the chemotactic gradient was used for the statistical evaluation of chemotaxis (Fig. 1Ei). The forward migration index (FMI) was used for the statistical evaluation of directionality (6, 33), and the mean angle of deviation from the chemotactic gradient was used for the construction of circular histograms for the graphical representation of both chemotaxis and directionality (Fig. 1D and E; see also Movie S2 in the supplemental material).
Mean angle of deviation and FMI calculations were designed to take into account the annular nature of the Dunn chamber and were performed as follows. As each migrating cell crossed a minimum displacement threshold of 5 μm (Fig. 1E), the cell position upon crossing this threshold was recorded. The angle of deviation, θ, between the newly recorded position (solid arrow) and that of the direction of the chemotactic gradient at the previously recorded position (dashed arrow) was then calculated and stored (Fig. 1Eiii). The FMI value was calculated as the cell displacement in the direction of the chemotactic gradient between the newly recorded and previously recorded cell position, h, divided by actual cell displacement for these two positions, a (Fig. 1Eii). The 5-μm displacement threshold was then reset for the newly recorded cell position, and the process was then repeated until the end of the cell trajectory had been reached. The inclusion of a 5-μm displacement threshold helped ensure that only meaningful cell movements would be processed. Thus, a list of angles and FMI values was generated for each cell trajectory at different time points for the entire duration that the cell could be observed. These values were then used to derive the mean FMI and the mean angle of deviation for each cell within a given experiment. Directional persistence, which is a measure of the propensity for a cell to change its current direction at any given point in time, was calculated as the distance between the initial and final positions of a cell trajectory divided by total distance traveled by that trajectory, incorporating the 5-μm threshold described above.
Circular histograms were used for the graphical representation of chemotaxis and also the directional response toward PDGF. For representation of chemotaxis, the size of each segment represents the percentage of cells whose mean direction of migration lies within the given data bin (given segment arc) multiplied by the mean cell speed for all cells within that data bin (Fig. 1F and see Fig. 5). For representation of the directional response alone, the size of each segment represents the percentage of cells whose mean direction of migration lies within the given data bin (see Fig. 2A and Fig. 8B). Circular histograms were generated using our purpose-written software developed in Microsoft Visual C++.
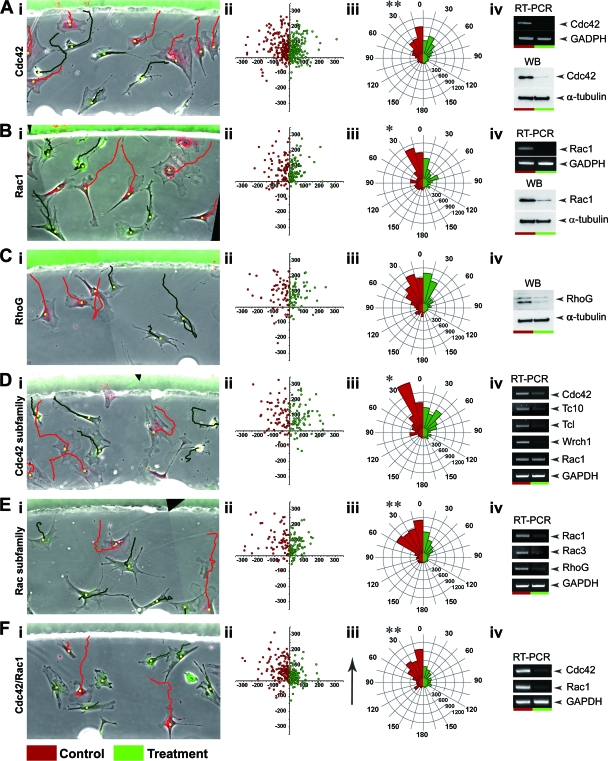
FIG. 5.
Effects of siRNA-mediated knockdown of various Cdc42 and Rac family members, either individually or in combination, on the chemotaxis of MEFs. Images in the left column show the first frames of representative film sequences from Dunn chamber experiments examining the effects of knocking down Cdc42 (Ai), Rac1 (Bi), RhoG (Ci), all Cdc42-related GTPases (Di), all Rac-related GTPases (Ei), and Cdc42 and Rac1 (Fi) on the behavior of MEFs. Images represent composite overlays of red and green fluorescence and phase contrast. In all cases, red fluorescent cells are those transfected with control, scrambled siRNA, while green fluorescent cells are those transfected with siRNA for the relevant GTPase, or a combination of GTPases, under investigation. Red and green trajectories represent the paths taken by individual cells of the respective treatment color over the 8-h observation period. (Aii to Fii) Plots illustrating the final position of cell trajectories from pooled experiments where cells were cells were transfected with either scrambled (red) or GTPase-specific (green) siRNAs. Trajectories have been shifted to a common origin and confined to either the negative (red cells) or positive (green cells) x axis so that control and test data groups can be displayed together. (Aiii to Fiii) Circular histograms summarizing the chemotactic response of MEFs transfected with either scrambled (red) or GTPase-specific (green) siRNAs in a gradient of PDGF-BB. Cells are grouped into data bins ranging from 0 to 180° according to their mean cell direction, with 0° representing perfect chemotaxis. The size of each segment represents the percentage of cells whose mean direction lies within the given segment arc multiplied by the mean cell speed for that data bin. Asterisks represent significant differences in chemotaxis between control and test cell populations, as determined by using a Wilcoxon signed-rank test for paired data. (Aiv to Fiv) Representative results from RT-PCR and Western blotting studies confirming the loss of endogenous mRNA and protein expression, respectively, for the target GTPase in cell cultures used for Dunn chamber experiments.
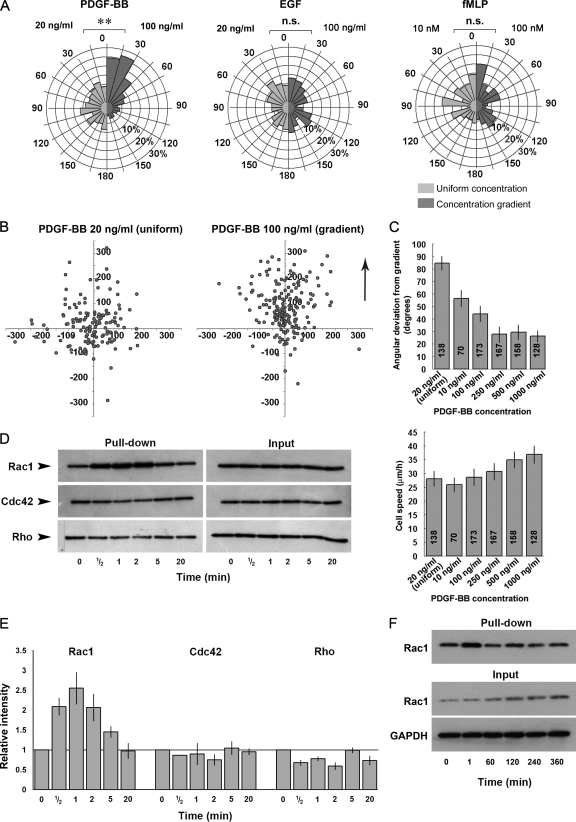
FIG. 2.
(A) Circular histograms summarizing the directional response of MEFs exposed to either a uniform concentration (light gray segments) or a gradient (dark gray segments) of PDGF-BB, EGF, or fMLP. Cells are grouped into data bins ranging from 0 to 180° according to their mean direction of migration, with 0° representing migration directly toward the outer well of the chamber. The size of each segment represents the percentage of cells whose mean direction lies within the given segment arc. The concentrations of chemoattractant used for uniform stimulation and for establishing a gradient are shown on the left and right sides of each histogram, respectively. Note that only PDGF-BB induces a significant directional response to the gradient. Asterisks represent significant differences between the chemotactic responses of the two data groups, as determined by using the Mann-Whitney test for nonparametric data (**, P < 0.01) (B) Plots illustrating the final positions of cell trajectories, shifted to a common origin, from pooled experiments where cells were exposed to either a uniform concentration (left) or gradient (right) of PDGF-BB. The arrow indicates the direction of increasing PDGF-BB concentration with respect to the right hand plot. The axes scales are in microns. (C) Bar charts illustrating the dose-dependent chemotactic (upper) and chemokinetic (lower) response of MEFs to PDGF-BB. (D) Pull-down assays indicating the change in basal activity of Rac1, Cdc42, and RhoA after the uniform stimulation of MEFs with 20 ng of PDGF-BB/ml. (E) Relative change in the activity levels of Rac1, Cdc42, and RhoA after stimulation of MEFs with PDGF, as determined by densitometric analysis. The chart summarizes data from three independent experiments in the case of Rac1 and two independent experiments in the case of Cdc42 and RhoA. (F) Extended time course examining Rac1 activity levels in MEF in response to stimulation with 20 ng of PDGF-BB/ml.
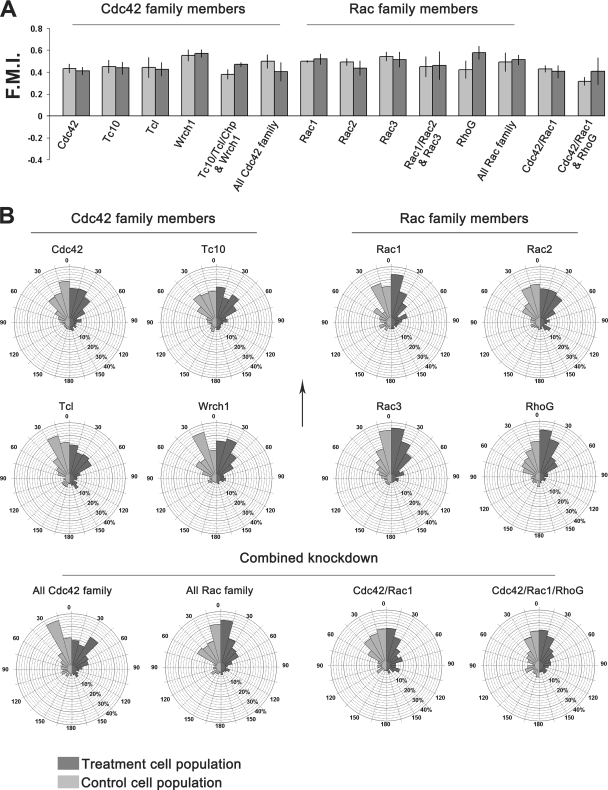
FIG. 8.
(A) The FMI was used for the statistical evaluation of the directional response of cells toward PDGF. Each bar represents the mean of means for the scrambled siRNA transfected cell populations (left bar) and GTPase-specific siRNA transfected cell populations (right bar) within each treatment group. Error bars represent the standard errors of the mean. In all cases, no significant difference exists between control and test cell populations as determined by using the Wilcoxon signed-rank test for paired data. (B) Circular histograms summarizing the direction of the migration of cells transfected with either control (light gray) or test (dark gray) siRNA for each given treatment group. Cells are grouped into data bins ranging from 0 to 180° according to their mean cell direction, with 0° representing migration directly toward the outer well of the chamber. The size of each histogram segment represents the percentage of cells within that treatment population whose mean direction lies within the given segment arc. Each treatment group summarizes data obtained from multiple Dunn chamber experiments. The arrow indicates the direction of increasing PDGF-BB concentration relative to each histogram.
Pulldown assays.
For Cdc42 and Rac1 pulldown assays, cells were harvested on ice in the presence of 50 μg of glutathione _S_-transferase (GST)-CRIB/ml in 500 μl of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 10% glycerol, 1% NP-40, 10 mM NaF, 1 mM Na3VO4, and 1× protease inhibitor cocktail (Amersham). Samples were sonicated on ice four times for 3 s each time and then clarified by centrifugation (15,000 rpm at 4°C for 10 min), and the resulting supernatants then incubated for 30 min at 4°C with constant rotation. GSH-Sepharose 4B beads (Amersham) were added to supernatants, and the samples were then incubated for a further 60 min with constant rotation. The beads were then washed three times with lysis buffer and subjected to Western blotting analysis.
For RhoA pulldowns cells were harvested in the presence of 10 μg of GST-mDia1-RBD-long (amino acids 69 to 451 of mDia1)/ml in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 2 mM MgCl2, 2% glycerol, 1% NP-40, 1 mM EDTA, 10 mM NaF, 1 mM Na3VO4, and 1× protease inhibitor cocktail (Amersham). Samples were then processed as for Cdc42/Rac1 pull-down assays except that the buffer used for washing beads was of lower ionic strength than that used during cell lysis (150 mM NaCl).
Purification of recombinant proteins and microinjection.
One-liter cultures of Escherichia coli BL21 cells transformed with the pGEX-6P-1 expression vector encoding full-length mouse Cdc42 or Rac1 were grown to an optical density at 600 nm of 0.7 at 37°C, cooled to 16°C, and then cultured overnight in the presence of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Bacterial pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, 5 mM dithiothreitol, 10 μM GDP, 1× protease inhibitor cocktail), sonicated three times for 10 s each time, and then clarified by centrifugation. Recombinant protein was captured using GSH-Sepharose 4B beads and washed three times with lysis buffer without protease inhibitors, and the GTPase was then released from GST by overnight cleavage at 4°C using GST-fused precision protease. Supernatant containing the soluble GTPase was dialyzed against microinjection buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM MgCl2, 0.1 mM dithiothreitol, 1 μM GDP) and concentrated by centrifugation (Amicon Ultra), and aliquots were then snap-frozen and stored at −80°C. Recombinant C3 transferase was prepared as described previously (12). Recombinant GTPases were microinjected at a concentration of 1 μg/ml using an Eppendorf transjector and micromanipulator mounted on a Zeiss Axiovert 100 inverted microscope. Recombinant C3 transferase was microinjected at a concentration of 0.1 μg/ml.
Immunocytochemistry.
Cells cultured on coverslips were fixed 4% paraformaldehyde, subsequently permeabilized with 0.1% Triton-X, and then stained for filamentous actin using Oregon Green phalloidin (Molecular Probes). Phalloidin-stained cells were imaged by using epifluorescence microscopy.
Reagents.
Cdc42 antibody was from Cell Signaling (catalog no. 2462). Rac1 (catalog no. 05-389, clone 23A8) and Rac2 (catalog no. 07-604) antibodies were purchased from Upstate. RhoG (C20: sc-1007) and RhoA (26C4: sc-418) antibodies were from Santa Cruz Biotechnology. α-Tubulin (T6199-200UL, clone DM1A) and green fluorescent protein (catalog no. 598) antibodies were obtained from Sigma and MBL, respectively. Recombinant human PDGF-BB and epidermal growth factor (EGF) were purchased from R&D Systems. Wortmannin, bovine fibronectin, fMLP, TRITC (tetramethyl rhodamine isothiocyanate)-dextran (70,000 molecular weight), and the PKH26 and PKH67 fluorescent cell linker kits were obtained from Sigma. Custom-made siRNAs, Lipofectaine 2000 transfection reagent, and OptiMEM were obtained from Invitrogen. Toxin B was a gift from Klaus Aktories. Dunn chambers were purchased from Hawksley Scientific with modifications provided by special request.
RESULTS
Assay for the in vitro analysis of fibroblast migration and chemotaxis.
In this study we utilized a modified Dunn chamber in conjunction with multifield/multichannel time-lapse microscopy to directly observe and follow the long-term behavior of primary mouse fibroblasts in a stable gradient of PDGF-BB (Fig. 1). The subsequent interactive tracking of cells from acquired film sequences was used to generate cell trajectories for the statistical analysis of cell behavior (Fig. 1D and E). Transfection of cells with siRNA was used to specifically inhibit the expression of Cdc42/Rac family GTPases either individually or in combination, while the use of membrane-labeling fluorescent dyes allowed both control and treatment cell populations to be mixed together and subsequently identified within the same culture (Fig. 1F and see Fig. S1 in the supplemental material). The mixing of control and test cell populations enabled comparisons to be undertaken within an identical experimental setting, allowing greater control and thus greater statistical power. Time-lapse imaging of slightly overlapping observation fields enabled the postacquisition reconstruction of a large region of the diffusion gap of the Dunn chamber, allowing greater data sets to be obtained for each treatment group within a given experiment (Fig. 1B, C, and F; see Movie S1 in the supplemental material). For a detailed description of the method used for the culture and subsequent recording and analysis of cell behavior, see Materials and Methods.
Optimization of fibroblast chemotaxis in the Dunn chamber.
In order to establish optimal conditions for the in vitro study of fibroblast chemotaxis, three putative chemoattractants, PDGF-BB (21), EGF (3), and fMLP (26) were initially screened for their ability to induce a chemotactic response in MEF using the modified Dunn chamber. Of these, only PDGF-BB was found to induce significant chemotaxis in this system (Fig. 2A and B; see Movie S1 in the supplemental material). MEFs showed a robust chemotactic response when 100 ng of PDGF-BB/ml was placed in the outer well of the chamber, while control cells exposed to a uniform concentration of 20 ng of PDGF-BB/ml showed no directional preference during migration (Fig. 2A). The addition of either 100 ng of EGF/ml or 50 nM fMLP to the outer well of the chamber did not result in a significant chemotactic response. Subsequent dose-response studies revealed that PDGF-BB could induce robust chemotaxis over a broad range of concentrations, with optimal chemotaxis observed using a PDGF-BB concentration of 250 ng/ml (Fig. 2C, upper chart), while the chemokinetic response induced by PDGF-BB increased with growth factor concentration over all of the concentrations assessed (Fig. 2C, lower chart). In addition, pulldown assays were performed to determine whether the basal activities of Cdc42, Rac1, and RhoA in MEFs were influenced by stimulation with this chemoattractant. PDGF-BB stimulation resulted in the rapid and transient activation of Rac1, with a peak level of activity occurring 30 to 60 s after growth factor addition and returning to basal levels within 20 min (Fig. 2D and E). Cdc42 activity remained relatively unchanged for the entire time course examined, whereas RhoA activity appeared to be slightly suppressed after PDGF-BB stimulation. This latter finding is similar to that previously reported by Sander et al. (18), who demonstrated rapid and transient inactivation of Rho in response to the PDGF-dependent activation of Rac1. An extended time course of Rac1 activation revealed that after the initial burst of activity, the levels of active Rac1 remained comparable to that found for the resting state (Fig. 2F). Of the three archetypal Rho GTPases, therefore, only Rac1 is activated after the stimulation of MEFs with PDGF-BB.
Characterization of the PDGF gradient in the Dunn chamber.
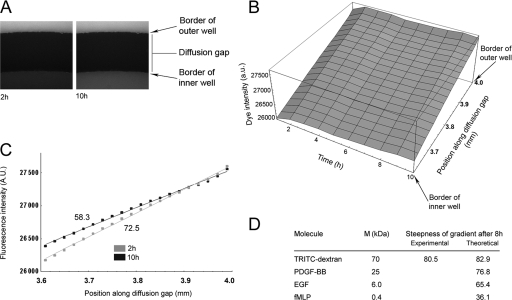
To determine the characteristics of a gradient of a putative chemoattractant within the diffusion gap of the modified Dunn chamber, TRITC-dextran (1 μg/ml, 70,000 molecular weight) was applied to the outer well of the chamber and the formation of a gradient was subsequently monitored over a period of 10 h (Fig. 3A). Analysis of the intensity of the fluorescent dextran across the diffusion gap revealed the rapid formation of a stable linear gradient, the steepness of which slowly reduced with time (Fig. 3B). A least-squares fit of intensity values spanning the diffusion gap was used to determine the gradient of the slope at the 2- and 10-h time points (Fig. 3C), and these values then used to calculate the change in steepness of the gradient over the 8 h period. The change in gradient steepness was similar to that predicted using the theoretical model previously applied by Zicha et al. for the Dunn chamber (35) (Fig. 3D; see also the supplemental material). Validation of the model enabled the subsequent estimation of the change in steepness of a diffusion gradient of the various putative chemoattractants used in the present study over the course of an 8-h Dunn chamber experiment (Fig. 3D). The model predicted that the steepness of the PDGF-BB gradient would fall to ca. 77% of that of its original value over the course of an 8 h Dunn chamber experiment and that the half-life of this gradient would be ∼21 h. Under optimal conditions for chemotaxis (250 ng of PDGF-BB/ml in the outer well), the model predicted that a cell 100 μm in length positioned within the center of the diffusion gap would initially be exposed to a PDGF-BB concentration of 153 ng/ml at its leading edge and a concentration of 91 ng/ml at its tail end, with a change in PDGF concentration the cell is exposed to of ∼0.62 ng/ml/μm. At the end of an 8-h experiment, where the gradient has declined to 77% of that of its initial steepness, the leading edge concentration would have fallen to 118 ng/ml, and the tail end concentration to 70 ng/ml, which corresponds to a change in PDGF concentration of ∼0.48 ng/ml/μm.
FIG. 3.
Characterization of diffusion gradients in the modified Dunn chamber. (A) Fluorescence image stills taken at 2 h (left) and 10 h (right) from a time-lapse sequence monitoring the diffusion of TRITC-dextran (70,000 molecular weight) between the outer and inner wells of the Dunn chamber. (B) Profile of TRITC-dextran fluorescence intensity across the diffusion gap of the Dunn chamber over time. (C) A least-squares fit of intensity data across the diffusion gap at 2 and 10 h was used to derive the steepness of the TRITC-dextran gradient at these time points (expressed in AU/20 μm). The data show that the gradient is essentially linear and remains stable for the entire duration observed and that the steepness of the gradient declines to ca. 80% of that of its original value over an 8-h period. (D) Table summarizing the predicted decline in steepness of a diffusion gradient of the various putative chemoattractants analyzed in the present study. The table also includes the experimentally derived value and the predicted value for the decline in steepness of the TRITC-dextran (70,000 molecular weight) gradient over an 8-h period, which was used to validate the theoretical model. Units represent % of estimated initial gradient steepness. M, molecular mass.
Confirmation of expression of Cdc42/Rac family GTPases in MEFs and evaluation of the efficacy of siRNAs.
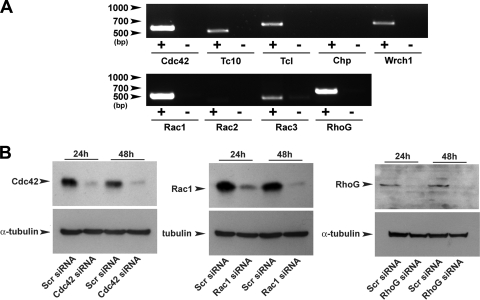
In order to determine the specific contribution of individual Cdc42 and Rac family members in the PDGF-dependent migration of MEFs, siRNAs were used to specifically inhibit the expression of these proteins. Initially, RT-PCR analysis was performed using total cytoplasmic RNA isolated from MEF cultures in order to determine which Cdc42 and Rac family members were expressed in these cells (Fig. 4A). mRNA encoding Cdc42, Rac1, and RhoG could be readily amplified from MEF lysates and appeared to be abundant in these cells. Tc10, Tcl, Wrch1, and Rac3 mRNA could also be amplified, although less efficiently than Cdc42, Rac1, and RhoG mRNA, suggesting that these GTPases are less abundantly expressed in these cells. Chp and Rac2 mRNA were present in only trace amounts and were only detectable when excessive cycling was performed (>35 cycles). Rac2 and Chp are therefore unlikely to represent valid targets for study in the analysis of MEF chemotaxis.
FIG. 4.
Expression analysis and cloning of Cdc42/Rac family members and the evaluation of the efficacy of siRNAs. (A) RT-PCR analysis performed on MEF lysates demonstrating the relative expression levels of different Cdc42 and Rac family members in these cells. Plus and minus columns denote the inclusion and omission of reverse transcriptase during the single-stranded cDNA synthesis reaction. (B) Western blotting analysis demonstrating the efficiency of siRNA-mediated knockdown of endogenous Cdc42 (left), Rac1 (middle), and RhoG (right).
Amplified, full-length cDNAs for Cdc42, Tc10, Tcl, Chp, Rac1, Rac2, Rac3, and RhoG obtained from RT-PCR studies were cloned and used to generate EGFP-fusion constructs, which were subsequently used to evaluate the efficacy of siRNAs for each given GTPase. All siRNAs tested were effective at blocking the expression of their respective target proteins (see Fig. S2 in the supplemental material). In the case of Cdc42, Rac1, and RhoG, Western blotting was also performed to confirm inhibition of expression of the endogenous protein. Although the Cdc42 and Rac1 antibodies both identified a single band of the predicted molecular weight for their respective GTPase, the RhoG antibody identified multiple bands. However, three separate siRNAs targeting nonoverlapping regions of the RhoG sequence all resulted in the loss of the same band of ∼20 kDa when transfected into MEFs (see Fig. S3 in the supplemental material), demonstrating that this band represented RhoG. Western blotting analysis revealed that endogenous levels of Cdc42, Rac1, and RhoG were almost undetectable 48 h after transfection with their respective siRNAs (Fig. 4B).
Rho is dispensable in the PDGF-BB-dependent chemotaxis of fibroblasts.
In initial studies, treatment of MEFs with Clostridium difficile toxin B, which renders all Rho family GTPases inactive through glucosylation (1), abolished the subsequent migration of these cells in a PDGF gradient, demonstrating the fundamental requirement for Rho GTPases in the regulation of cell migration (data not shown). To determine the role of Rho in fibroblast chemotaxis, cells were microinjected with Clostridium botulinum C3 transferase (C3), a specific inhibitor of Rho (20). Four hours after microinjection, cell migration was assessed by using the Dunn chamber. The inclusion of a fluorescent marker dye during microinjection facilitated the relocation of injected cells during Dunn chamber assembly. C3 microinjection had a severe effect on cell morphology (see Fig. S4A and Movie S3 in the supplemental material), inducing cell retraction and the formation of multiple protrusions, morphological changes consistent with those previously reported after microinjection of recombinant C3 into both L-cell fibroblasts (32) and primary rat embryonic fibroblasts (13). Although C3 microinjection did have a profound effect on cell morphology, in the majority of cells observed this treatment had no significant effect on the speed or the directional response of cells to the PDGF gradient (see Fig. S4B and C in the supplemental material). Although in a few cases microinjected cells retracted completely and detached from the substrate, a phenotype previously observed after microinjection of high concentrations of C3 into fibroblasts, the majority of cells were motile and exhibited efficient chemotaxis despite their severely altered phenotype. Microinjection of immunoglobulin G alone had no effect on the morphology of MEFs (see Fig. S5 in the supplemental material). These experiments suggest therefore that, outside its critical role as a regulator of cell adhesion, Rho is not essential for the PDGF-dependent chemotaxis of fibroblasts.
Cdc42 regulates cell morphology and chemotaxis of primary fibroblasts.
Yang et al. have recently reported that Cdc42-null primary fibroblasts are defective in filopodia induction, directed migration, and proliferation, while Pankov et al. reported impaired migration of fibroblasts in response to a gradient of fMLP after siRNA-mediated knockdown of Cdc42 (14, 31). In both of these studies fibroblast chemotaxis was assessed by using a transwell assay. We therefore wanted to determine by direct observation how the loss of Cdc42 function disrupts the chemotaxis process. Depletion of Cdc42 with siRNA resulted in varied cellular phenotypes often characterized by both cell elongation and retraction of the cell body, and in many cases this was also accompanied by the formation of multiple disordered lamellalike extensions (Fig. 5A [see also green cells in Movie S4 in the supplemental material] and Fig. 6C and D) . In some cases, the change in cell morphology after Cdc42 knockdown was profound, leading to extensive cell rounding and the projection of multiple processes from the cell body. This phenotype was similar to that observed for Cdc42-null primary fibroblasts previously reported by others (31). A second siRNA sequence targeting a separate, nonoverlapping region of the Cdc42 sequence resulted in a similar phenotype (data not shown). Since the overexpression of Rho family GTPases often leads to the induction of an exaggerated morphological phenotype by excessively activating downstream signaling, we decided to use microinjection of recombinant protein as a novel approach to attempt to rescue the phenotype observed after Cdc42 knockdown in MEFs. Microinjection of recombinant Cdc42 into Cdc42 knockdown cells resulted in the restoration of a more typical fibroblast phenotype (see Fig. 9I), demonstrating that the morphological change observed was a direct consequence of loss of Cdc42 function.
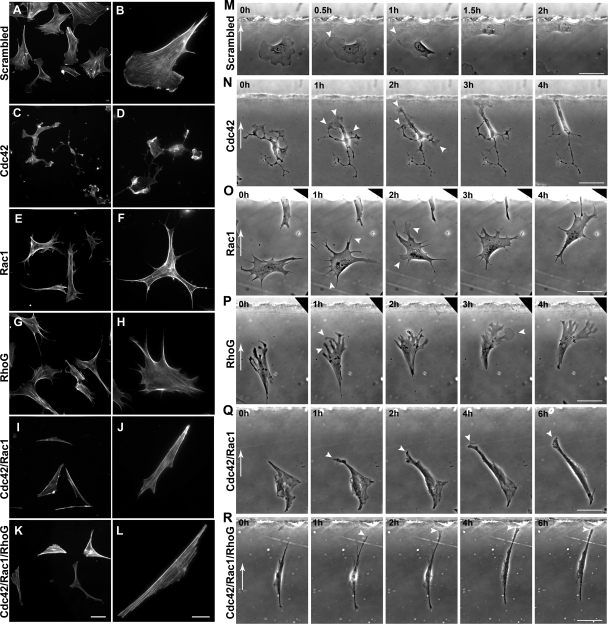
FIG. 6.
Filamentous actin staining in MEFs 48 h after transfection with either scrambled (A and B); Cdc42 (C and D); Rac1 (E and F); RhoG (G and H); Cdc42 and Rac1 (I and J); or Cdc42, Rac1, and RhoG (K and L) siRNAs. Scale bars for the left and right columns represent 50 and 10 μm, respectively, and are valid for all panels within that given column. (M to R) Image sequences from chemotaxis experiments demonstrating the behavior of MEFs in a PDGF-BB gradient 48 h after transfection with either scrambled siRNA (M); Cdc42 siRNA (N); Rac1 siRNA (O); RhoG siRNA (P); Cdc42 and Rac1 siRNA (Q); or Cdc42, Rac1, and RhoG siRNA (R). White arrows in the first frame of each film sequence indicate the direction of increasing PDGF concentration. White arrowheads in panel M mark the growth of a single, broad leading edge of a control, scrambled siRNA-transfected cell toward the PDGF source. The white arrowheads in panel N mark examples of lamellipodia and membrane ruffles emanating from numerous cellular protrusions in a Cdc42 siRNA-transfected cell. Note the migration of the cell toward the outer well over time. White arrowheads in panel O mark the positions of filopodia (1-h time frame), and thicker processes (2-h time frame), in a Rac1-siRNA transfected cell. The cell migrates toward the outer well of the chamber over time. (P) White arrowheads mark the position of fingerlike membrane extensions (1-h time frame), and a transient lamellipodia (3-h time frame) protruding toward the chemoattractant source in a RhoG-siRNA transfected cell. White arrowheads also indicate the extremely slow elongation and migration of Cdc42/Rac1 siRNA (Q) and Cdc42/Rac1/RhoG siRNA (R) transfected cells toward the outer well of the chamber. Scale bars in the far right column are valid for each image within a given row and represent 50 μm. See the corresponding movies in the supplemental material.
FIG. 9.
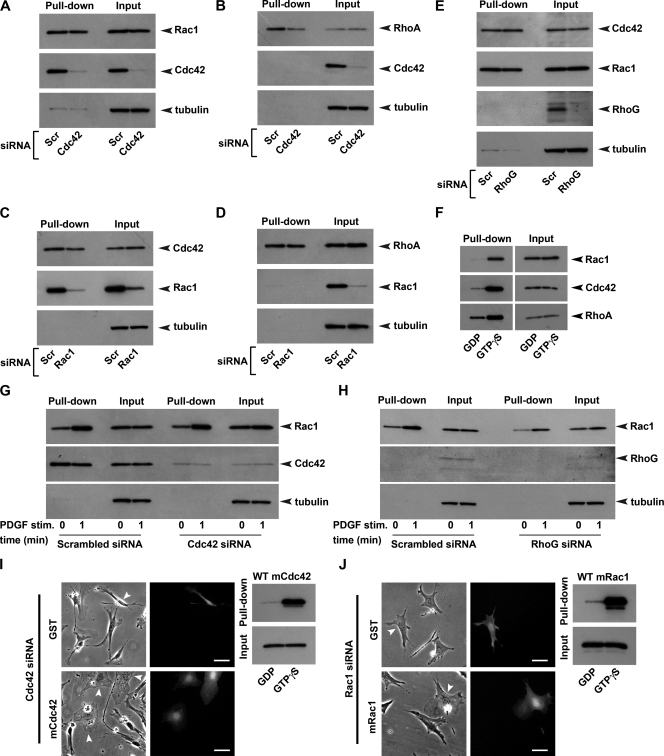
(A to E) Pulldown assays summarizing the effects of the siRNA-dependent knockdown of Cdc42 on the basal activity and expression of Rac1 (A) and RhoA (B), of Rac1 knockdown on the basal activity and expression of Cdc42 (C) and RhoA (D), and of RhoG knockdown on the basal activity and expression of Cdc42 and Rac1 (E) in MEFs. (F) GDP/GTPγS loading experiments demonstrating the specificity of the probes used in pulldown assays for the active form of each respective GTPase. (G and H) Effect of Cdc42 knockdown (G) and RhoG knockdown (H) on the PDGF-BB-dependent activation of Rac1 in MEFs. (I and J) Microinjection of recombinant Cdc42 and Rac1 into Cdc42 knockdown and Rac1 knockdown cells, respectively, rescues fibroblast morphology. (I and J) Phase-contrast (left column) and epifluorescence (right column) images of live cells taken 4 h after microinjection of either GST control (top row) or the recombinant GTPase (bottom row). White arrowheads indicate the position of microinjected cells in phase-contrast images. Comicroinjection of Alexa 488-dextran allowed the subsequent identification of microinjected cells. Scale bars, 50 μm. Purified recombinant mouse Cdc42 and Rac1 were cleaved from GST, and samples were then loaded with either GDP or GTPγS to confirm their activity prior to use in microinjection rescue experiments (adjacent blots).
Analysis of cell trajectory data revealed that Cdc42 knockdown significantly impaired the chemotaxis of fibroblasts within the PDGF-BB gradient (Fig. 5A). In contrast, cells transfected with scrambled siRNA within the same culture exhibited typical fibroblast morphology and were often polarized with a single dominant leading lamella. Cdc42 knockdown cells often retracted and then respread during the course of time-lapse experiments and many cells exhibited sporadic bouts of cell migration (see Movie S4 in the supplemental material).
Impaired chemotaxis of Cdc42 knockdown cells is a consequence of reduced migration speed.
While Cdc42 knockdown significantly impaired the chemotaxis of fibroblasts, it was clear from the observation of these cells in the Dunn chamber that cells were still motile and could still detect and migrate toward the source of the chemoattractant. Observation of Cdc42 knockdown cells at higher magnification revealed that even severely effected cells, which exhibited a highly retracted morphology with multiple, randomly oriented protrusions, were able to orient toward the source of the chemoattractant, often after establishing a dominant leading process, and subsequently migrate toward the outer well of the chamber (Fig. 6N; see Movie S5 in the supplemental material).
We further analyzed cell trajectory data by dissecting the effects of Cdc42 knockdown on various components of cell migration, including speed, the directional response toward the chemoattractant, and directional persistence. Our analysis revealed that Cdc42 knockdown cells exhibited an approximate 25% reduction in the speed of migration compared to control, scrambled-siRNA transfected cells (Fig. 7). However, analysis of the FMI, a sensitive parameter for measuring the directional response of a cell toward a gradient of chemoattractant (6, 33), revealed no significant differences between Cdc42 knockdown and control cell populations (Fig. 8). Analysis of directional persistence, which is a gradient-independent measure of the propensity for a cell to change its direction at any given point in time, revealed that Cdc42 knockdown cells exhibited a significant but small reduction in this value (ca. 6%) compared to controls (control, 0.72 ± 0.019; Cdc42 siRNA, 0.68 ± 0.021).
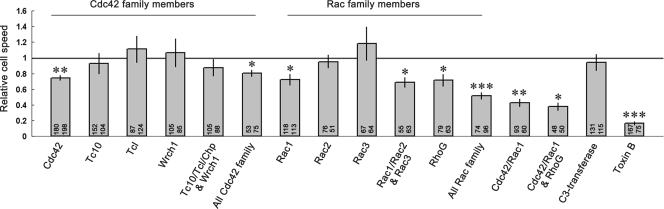
FIG. 7.
Bar charts summarizing the effects of knockdown of various Cdc42- and Rac-related GTPases, either individually or in combination, on the speed of cell migration. Each bar indicates the relative difference in cell speed between the control and test cell populations for each treatment group. The speed of the control cell population for each treatment group is normalized to 1. Values within each bar correspond to the number of control (left) and test (right) cells analyzed for each treatment group. Each treatment group summarizes data obtained from multiple Dunn chamber experiments. Relative differences in speed are calculated by comparing the mean of means for the test and control cell populations within each treatment group. Error bars represent the standard errors of the mean. Asterisks represent the significance of differences, determined by using a two-tailed, paired t test, between control and test cell populations within each treatment group (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To understand why cells exhibited defects in persistence but not the directional response to the gradient (as measured by the FMI), we further analyzed the relationship between these two aspects of migration. The analysis revealed a monotonic relationship, with an increase in directional persistence resulting in an increase in the FMI (see Fig. S6A in the supplemental material). However, the data also revealed that Cdc42-knockdown cells that exhibited lower persistence values tended to exhibit comparatively higher FMI values than their control counterparts. In other words, the alternative mode of migration adopted by Cdc42 siRNA-treated cells was associated with an increased FMI in cells of lower directional persistence compared to cells that exhibited similar persistence values within the control population.
It is possible that the experimental conditions used in our assay that promote a robust chemotactic response in MEFs were potentially masking subtle effects that Cdc42 knockdown confers on the directional response of these cells and that such effects would become apparent if cells were exposed to shallower gradients of the chemoattractant. Given that our mathematical model of diffusion in the Dunn chamber predicted a change in steepness of the PDGF gradient of ca. 23% over the course of an 8-h chemotaxis experiment (Fig. 3D), we examined whether the directional response of Cdc42-knockdown cells differed significantly to that of control cell during the latter stages of the experiment when cells were exposed to a shallower chemotactic gradient. The directional response of cells was assessed during the first and last 2 h of chemotaxis experiments, where the mean changes in PDGF concentration the cells were exposed to were ∼0.6 and 0.5 ng/ml/μg, respectively (∼18% difference). FMI data for Cdc42-knockdown cells was not found to be significantly different from that of control cells during either the early or late stages of chemotaxis experiments (see Fig. S6B in the supplemental material). Therefore, over a change in mean gradient steepness of ca. 18% we were unable to detect any significant difference in the directional response to the PDGF gradient between the Cdc42 knockdown and control cell populations.
Tc10, Tcl, and Wrch1 are all dispensable in the chemotaxis of primary fibroblasts.
While Cdc42 was found to be important for the control of cell morphology and efficient chemotaxis of MEFs, other Cdc42-related GTPases were found to be dispensable in these processes. Knockdown of Tc10, Tcl, and Wrch1, either individually or in combination, had no effect on the speed, directional response, or morphology of MEFs (Fig. 7 and 8). The combined knockdown of all Cdc42 family GTPases resulted in altered cell morphology and a reduced rate of migration similar to that observed after the knockdown of Cdc42 alone (Fig. 5D, 7, and 8). These findings demonstrate that, of all the Cdc42 family GTPases expressed in MEFs, only Cdc42 is required for the efficient chemotaxis of these cells toward PDGF-BB.
Rac1 regulates cell morphology and speed but not the direction of migration during PDGF-BB-dependent chemotaxis.
We also sought to determine the contribution of Rac-related GTPases in the PDGF-dependent migration of MEFs. Knockdown of Rac1 alone resulted in an altered cell phenotype characterized by loss of lamellipodia, the presence of multiple filopodiumlike processes, and also occasionally cell elongation (Fig. 6E and F). Chemotaxis toward PDGF was significantly impaired in these cells (Fig. 5B; see Movie S6 in the supplemental material). However, further analysis of cell trajectory data revealed that defects in chemotaxis were a consequence of reduced speed but not defects in the directional response of cells to the gradient (Fig. 7 and 8). Despite the high level of efficiency with which Rac1 knockdown could be achieved within these cultures (Fig. 5B), these cells exhibited an approximate 30% reduction in the speed of cell migration compared to control cells from the same cultures (Fig. 7). Cell migration occurred despite the absence of lamellipodia and was accompanied by the frequent protrusion of filopodia at the cell periphery, most notably in the direction of increasing PDGF concentration (Fig. 6O; see also Movie S7 in the supplemental material). Cells also frequently extended thicker membrane protrusions toward the chemoattractant source, and occasionally these extensions provided a scaffold for the propagation of small, transient lamellipodiumlike structures.
It has previously been reported that loss of Cdc42 expression results in an approximately twofold reduction in the basal level of Rac1 activity (31). Since Rac1 knockdown results in reduced migration speed in MEFs, it is possible that the reduced speed observed after knockdown of Cdc42 may be an indirect consequence of a reduction in the basal level of active Rac1. Consequently, we performed pull-down assays to compare the endogenous activities of Rac1 in control and Cdc42 knockdown cell cultures. We found that Cdc42 knockdown had no effect on the basal level of Rac1 activity under normal culture conditions (Fig. 9A). Therefore, the loss of speed associated with Cdc42 knockdown cannot be attributed to a reduction in the basal level of Rac1 activity. Also, the transient activation of Rac1 in response to PDGF-BB stimulation, which may be important for establishing a motile state, was not affected in Cdc42 knockdown cells (Fig. 9G).
In contrast to Rac1, however, the basal level of RhoA activity was reduced in Cdc42 knockdown cells (Fig. 9B). This latter finding is intriguing, especially since similarities can be seen between the Cdc42 knockdown phenotype (Fig. 5A) and that of cells microinjected with C3 (see Fig. S4A in the supplemental material). We also note that stress fibers were often less abundant in those cells where morphology had been severely affected by Cdc42 knockdown (Fig. 6C and D). It is therefore tempting to postulate that the retracted cell phenotype frequently observed after knockdown of Cdc42 could, in part, be due to reduced Rho activity in these cells. Pulldown assays revealed that Rac1 knockdown had no detectable effect on the basal level of either RhoA or Cdc42 activity (Fig. 9C and D).
Rac3 is dispensable in the chemotaxis of primary fibroblasts.
Given that loss of Rac1 expression only impairs but does not inhibit cell migration, we wanted to determine whether other Rac-related GTPases either contribute to or compensate for the loss of Rac1 function during cell migration. Dunn chamber studies revealed that siRNA-mediated knockdown of Rac3 had no effect on the morphology or migration of MEFs and that the behavior of these cells was essentially the same as that of control cells throughout the 8-h observation period (Fig. 7 and 8). In addition, combined knockdown of Rac1, Rac2, and Rac3 resulted in morphological change and a reduction in the speed of cell migration similar to that observed following Rac1 knockdown alone (Fig. 7). Transfection of cells with siRNA for Rac2, which is not expressed in these cells at detectable levels, had no effect on cell migration (Fig. 7 and 8). Taken together, these experiments demonstrate that, of these three closely related GTPases, Rac1 is the major regulator of cell morphology and migration in MEFs.
RhoG can regulate morphology and speed of cell migration through mechanisms independent of Rac1.
RhoG, a Rho family GTPase most closely related to the Rac subfamily, has recently been implicated in the migration of both fibroblasts (9) and epitheliumlike cells (10). Since Rac1 has previously been identified as an activation target of the RhoG/Elmar/DOCK180 and DOCK4 complexes, it has been suggested that RhoG may regulate cell motility through the downstream control of Rac1 (9, 10). We therefore wanted to examine the role of RhoG in the migration of MEFs and compare its role to that of Rac1. Although RhoG knockdown did appear to have an inhibitory effect on chemotaxis, these findings were not significant (Fig. 5C; see Movie S8 in the supplemental material). However, RhoG knockdown cells did exhibit a significant reduction (30%) in the speed of cell migration compared to control cells from the same cultures (Fig. 7), while the directional response of these cells to the PDGF gradient was unaffected (Fig. 5C and 8). The mode of migration adopted by RhoG knockdown cells involved the protrusion of both filopodia and, more frequently, other thicker membrane extensions. Although lamellipodia could occasionally be observed in these cells, they were often far smaller (Fig. 6G and H) and more transient than those typically found for control cells (Fig. 6P; see Movies S8 and S9 in the supplemental material). Since these findings were similar to those found after knockdown of Rac1, we sought to determine whether the reduced speed and altered morphology observed was a consequence of low basal Rac1 activity in these cells. However, pulldown assays revealed that the basal Rac1 activity was unaltered in RhoG knockdown cells (Fig. 9E). In HeLa cells, active Rac1 increases rapidly in response to wound generation, and this activation has been shown to be dependent upon RhoG (10). Consequently, the possibility remained that reduced speed of migration may have resulted from an impaired ability of PDGF to stimulate Rac1 activity in RhoG knockdown cells, which in turn may be important for establishing a motile phenotype. However, we found that the PDGF-dependent activation of Rac1 in serum starved MEFs was unaffected by RhoG knockdown (Fig. 9H).
Since the basal level of Rac1 activity and PDGF-dependent activation of Rac1 remained unaltered in RhoG-knockdown cells, we wanted to examine the effects of depleting all of the Rac-related GTPases expressed in MEFs to determine whether Rac1 and RhoG function cooperatively during the migration of these cells and not in a hierarchical manner, as previously suggested for other cell types (10). Cells were cotransected with siRNA specific for Rac1, Rac2, Rac3, and RhoG, and their behaviors were subsequently examined using the Dunn chamber. Inhibition of expression of all Rac family members resulted in significant inhibition of chemotaxis (Fig. 5E). Further analysis of cell trajectory data revealed that the speed of these cells was reduced to ca. 50% of that of controls (Fig. 7), which was greater than that observed after knockdown of Rac1 or RhoG alone. However, these cells were still motile and could still detect and migrate toward PDGF, and specific defects in the directional response of these cells were not observed (Fig. 8). Collectively, these findings demonstrate that while Rac GTPases are required for the efficient chemotaxis of primary fibroblasts, they are not critical for cell migration. These findings also suggest that, in MEFs, RhoG can contribute to cell migration through mechanisms other than those related to the activation of Rac1.
Cdc42, Rac1, and RhoG function cooperatively in the regulation of fibroblast migration.
Given that knockdown of Cdc42, Rac1, and RhoG all impair but do not inhibit the PDGF-dependent chemotaxis of primary fibroblasts, we wanted to determine whether these proteins function cooperatively in the regulation of cell migration. We therefore performed knockdown experiments to determine the effects of loss of combinations of these Rho GTPases. The combined knockdown of both Cdc42 and Rac1 resulted in far greater inhibition of chemotaxis than when either GTPase was knocked down alone (Fig. 5F; see Movie S10 in the supplemental material). Both lamellipodia and filopodia formation were suppressed in these cells (Fig. 6I and J). Inhibition of chemotaxis was a consequence of severe inhibition of cell movement, with mean cell speed reduced to ca. 43% of that of the control cell population (Fig. 7). Although many cells showed limited translocation throughout the 8-h observation period (Fig. 6Q; see Movies S10 and S11 in the supplemental material), application of the 5-μm threshold during the automatic analysis of cell trajectory data, which resulted in the rejection of very small and zero cell displacements, ensured that only translocating cells (86%, n = 99) were included in the analysis of the directional response to the gradient. Analysis of these trajectories (Fig. 8) revealed that the FMI was not significantly different from that found for control cells (94%, n = 124). Finally, the combined knockdown of Cdc42, Rac1, and RhoG resulted in a similar morphology (Fig. 6K and L), dramatic inhibition of cell speed (37% of that of controls), and the same residual response to PDGF as that observed after the combined knockdown of Cdc42 and Rac1 (Fig. 6R, 7, and 8 and see Movies S12 and S13 in the supplemental material).
Chemotaxis of MEF toward PDGF-BB is PI 3-kinase independent.
Since Cdc42 and other related GTPases were found to be dispensable in the PDGF-BB-dependent chemotaxis of MEFs, we wanted to determine whether phosphatidylinositol 3-kinase (PI 3-kinase), which has been implicated as a key regulator of directional sensing and chemotaxis in a wide range of eukaryotic cell systems, was required for this response in MEF. Cells were treated with the PI 3-kinase inhibitor wortmannin (100 nM), and their behavior was then assessed by using a Dunn chamber (see Fig. S7 and Movie S15 in the supplemental material). Recent studies of the cyclic AMP-dependent chemotaxis of the slime mold D. discoideum demonstrate that PI 3-kinase function is only essential for chemotaxis when cells are exposed to suboptimal concentrations of chemoattractant, whereas at higher concentrations other signaling factors can compensate for loss of PI 3-kinase function (27). Consequently, we examined the effects of wortmannin treatment under both suboptimal and optimal conditions for chemotaxis. The chemotaxis of MEFs in the presence of wortmannin was no different than that of control cells when either a low concentration (10 ng/ml) or a high concentration (250 ng/ml) of PDGF-BB was applied to the outer well of the chamber (see Fig. S7A and B in the supplemental material). Further analysis of cell trajectory data did not reveal any defects in either speed or directionality in these cells at either of the growth factor concentrations used. Wortmannin at 100 nM resulted in the potent inhibition of PI 3-kinase activity in MEFs as determined by its ability to block the PI 3-kinase-dependent phosphorylation of Akt in response to PDGF stimulation (see Fig. S7C and D in the supplemental material). These findings demonstrate that the chemotaxis of MEFs toward PDGF-BB is independent of PI 3-kinase function.
DISCUSSION
In this study we have performed a comprehensive comparison of the role of individual members of the entire family of Cdc42/Rac GTPases in PDGF-dependent chemotaxis of primary fibroblasts using a single, well-defined cell assay. In addition, the combined knockdown of multiple family members has provided insights into the cooperative roles of these GTPases during cell migration. Using our experimental system, we have demonstrated roles for Cdc42, Rac1, and RhoG in fibroblasts chemotaxis, while other related GTPases, including Tc10, Tcl, Wrch1, Rac2, and Rac3, were all found to be dispensable for this process. Findings from our study suggest that during fibroblast migration partial redundancy exists among Cdc42 and Rac family GTPases. In the absence of Cdc42, Rac1, or RhoG, cell migration can still occur, albeit less efficiently, as cells exploit alternative modes of movement. In the absence of Cdc42, cells adopt a retracted morphology, and migration is accompanied by the extension of narrow processes, while in the absence of either Rac1 or RhoG cell migration is largely accompanied by the extension of filopodia and other thicker membrane extensions.
Cdc42, Rac1, and RhoG may therefore contribute to different aspects of cell migration, and it may therefore be the case that, following the loss of one of these GTPases, the cell adopts a mode of migration that exploits the functions of the remaining proteins. However, when either Cdc42 and Rac1 or Cdc42, Rac1, and RhoG levels are suppressed simultaneously, cell migration is severely inhibited, since the capacity for cells to form dynamic actin-based protrusive structures is greatly impaired. Interestingly, despite the severe effects that combined depletion of Cdc42, Rac1, and RhoG had on the migration of primary fibroblasts, residual cell movement was directed toward the PDGF source. These findings demonstrate that Cdc42/Rac family GTPases regulate migration speed but not the directional response to the gradient during chemotaxis.
In our study we demonstrate that Cdc42 knockdown in MEFs results in impaired chemotaxis. However, Cdc42 knockdown cells can detect and migrate toward the increasing concentration of PDGF, and the resultant mean direction of cell migration is no different than that of the control cells from the same cultures. We show that reduced speed is the major factor contributing to impaired chemotaxis in Cdc42 knockdown cells and demonstrate that while defects in the persistence of these cells do exist, this does not significantly impair their directional response to the PDGF gradient. Yang et al. report reduced migration of Cdc42-null fibroblasts in a serum gradient using a transwell assay and therefore concluded that directionality is impaired in these cells. The difference in their findings and the findings from the present study may reflect differences in the experimental assays used or the chemotactic stimulus used. The gradient characteristics for a given chemoattractant in terms of stability, steepness, and half-life are likely to differ for the transwell chamber and the Dunn chamber. Therefore, we cannot discount the possibility that Cdc42 knockdown may result in defects in the directional response when the gradient characteristics of PDGF differ from those in our study (for example, in shallow gradients of PDGF). However, given that Yang et al., like us, also report reduced cell migration speed after inhibition of Cdc42 signaling, impaired migration speed alone can account for the reduced transwell migration observed in their study. By direct observation of cell migration and subsequent careful analysis of cell trajectories, we gain detailed insights into the various aspects of migration contributing to the chemotactic response of these cells.
The reduced speed of fibroblast migration after the loss of Cdc42 function reported by us and others contrasts with findings from cells of hematopoietic origin, where deficiencies in Cdc42 signaling have actually been found to enhance migration speed (2, 24). A study using the Bac1 macrophage cell line first ascribed a role for Cdc42 in chemotaxis by demonstrating that microinjection of dominant-negative Cdc42 into macrophages disrupts chemotaxis toward CSF-1, while slightly enhancing migration speed (2). A study of Drosophila hemocyte migration in vivo demonstrates that loss of Cdc42 function in these cells results in reduced directional persistence but also significantly enhanced migration speeds (24). Indeed, the authors of that study demonstrated that while Cdc42-null hemocytes follow a more torturous path to their destination, enhanced migration speed allows them to reach their target as quickly as wild-type cells. It is therefore clear that while Cdc42 plays an important role in cell migration in both fibroblastlike cells and cells of hematopoietic origin, clear differences exist in its regulation of various aspects of this behavior. A phenotype associated with impaired Cdc42 signaling that does, however, appear to be common to many cell types is retraction and elongation of the cell body. It is interesting, therefore, that we have found that basal levels of Rho activity are reduced in Cdc42 knockdown fibroblasts. Given the similarity between the Cdc42 knockdown phenotype and the C3 phenotype observed in our cells, it is tempting to postulate that reduced Rho activity may contribute to the cell retraction observed in cells deficient for Cdc42 signaling.
In the present study we also demonstrate that Rac1 and RhoG, but not Rac3, regulate the morphology and migration of primary fibroblasts. The direct observation of Rac1 and RhoG knockdown cells in a PDGF-BB gradient demonstrates that, in the absence lamellipodia, cells migrate primarily via the extension of filopodia and other thicker membrane processes and that the directional response to the gradient of cells utilizing this mode of migration is as efficient as that observed for cells under control conditions where broad lamellipodia are the dominant protrusive structures. The chemotactic ability of Rac1-deficient cells is consistent with two recent studies characterizing the motility of Rac1-null (28) and Rac1 knockdown (14) fibroblasts, in which transwell assays were used to demonstrate the chemotaxis of Rac1-deficient fibroblasts toward PDGF-BB and fMLP, respectively.
The morphological phenotypes observed after knockdown of Rac1 and Cdc42 in MEF is consistent with the previous report by Srinivasan et al. (23), who examined the roles of these GTPases in neutrophil migration and chemotaxis. These authors demonstrated distinct roles for Rac1 and Cdc42 in the regulation of lamellipodia during neutrophil migration, with the former controlling the formation of these structures and the latter their stability. These authors further demonstrated the loss of chemotaxis toward fMLP in neutrophils expressing a dominant-negative Cdc42 construct. The fact that we still observed migration toward PDGF in MEFs after knockdown of Cdc42 could either reflect differences in the cell types studied or differences in the signaling pathways used during the chemotactic response to these different signaling molecules. Since we were unable to detect significant MEF chemotaxis toward fMLP using our experimental system, this latter possibility could not be addressed.
Given that Rac1 is required for efficient chemotaxis of MEFs in a PDGF gradient, it is interesting that PDGF stimulation only results in a brief, transient burst of Rac1 activity, with active Rac1 returning to a level comparable to that found for the resting state thereafter. It is possible that the initial and transient burst of Rac1 activity functions to mobilize the actin polymerizing machinery in order to enable the cell to make the transition from a quiescent state to a motile state in response to the PDGF stimulus. Once the cell has adopted the motile state, Rac1 would need to be downregulated to prevent excessive activation, which may have deleterious effects for cell motility and polarity, with local changes in the levels of active Rac1 and Cdc42 then maintaining the steady turnover of the actin cytoskeleton during cell migration.
RhoG has previously been implicated in both fibroblast migration and the regulation of Rac1 activity and, consequently, it has been suggested that RhoG regulates cell motility through the downstream regulation of Rac1 (10, 11). However, in the present study we have shown that, although RhoG is also required for efficient cell migration, inhibition of RhoG expression has no effect either on the basal level of Rac1 activity or on the PDGF-dependent activation of Rac1. Furthermore, combined knockdown of both RhoG and Rac1 inhibited the speed of cell migration to a greater extent than that found for knockdown of Rac1 alone. Although RhoG knockdown does not influence the activity of Rac1, these findings do not necessarily exclude a role for RhoG in the regulation of Rac1-mediated cell migration. For example, the loss of RhoG may influence the efficiency of Rac1 function by removing a subset of Rac1 activators critical for migration or by mislocalizing adaptor molecules critical for the efficient recruitment of active Rac1 to specific subcellular compartments during migration. However, findings from the present study do suggest that RhoG can contribute to cell migration through mechanisms other than those involved in the regulation of Rac1.
Finally, our recombinant C3 microinjection studies clearly demonstrate that under conditions where Rho activity is highly attenuated, fibroblasts can chemotax efficiently toward PDGF. Interestingly, Nobes and Hall (13) have previously reported that while microinjection of recombinant, dominant-active RhoA has a severe inhibitory effect on primary rat fibroblast migration during a scratch wound assay, microinjection of C3 has a far less pronounced effect on cell migration despite having profound effects on cell morphology. Clear inhibition of wound closure could only be observed in their study when C3 was microinjected at a concentration well above that required to severely alter cell morphology and abolish stress fibers. Under such conditions, cells often lost attachment to the substrate, indicating the essential role of Rho in maintaining cell adhesion. It has recently been shown that ROCK-1-deficient mice exhibit defective closure of both the eye lids and the ventral body wall during embryonic development (22). In these mice, epithelial cells at the leading margin of the eyelids are unable to organize actomyosin bundles, which appear to be required for the efficient migration of these cells as a sheet across the underlying eye. It may therefore be the case that Rho signaling is more critical for the coordinated migration of a cell population as a single sheet or monolayer, where a “purse string”-based mechanism utilizing actomyosin machinery provides contractile forces that aid the process of forward migration. Our findings suggest, however, that the migration of isolated cells in response to a chemotactic signal is less dependent on Rho function, where the purse-string mechanism of migration does not apply.
In summary, we have performed a comprehensive and detailed analysis of the role of specific Cdc42/Rac family GTPases in the chemotaxis of primary fibroblasts. Our study demonstrates that Cdc42, Rac1, and RhoG function cooperatively during cell migration and reveals new insights into the relationship of these GTPases in the regulation of cell movement. We also demonstrate that while each of these GTPases is implicated in the control of cell morphology and speed, these and other Cdc42/Rac-related GTPases are not required for the directional response toward PDGF during the chemotaxis of primary fibroblasts. Our experimental approach, which combines the Dunn chamber with fluorescent cell labeling techniques, siRNA-mediate protein knockdown, and time-lapse microscopy, provides an extremely powerful method for assessing the significance of differences observed between different treatments, since direct comparisons can be made between control and test cell populations within the same chemotaxis experiment. We believe that this assay will be useful for the future study of various signaling molecules in the migration and chemotaxis of cells in vitro.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Gareth Jones for assistance, Toshimasa Ishizaki and Shingo Yasuda for many valuable comments and technical advice, and also Kimiko Nonomura for preparation of the C3 transferase. We thank Klaus Aktories for kindly providing recombinant toxin B.
This research was supported in part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. J.M. was a postdoctoral Research Fellow of the Japan Society for the Promotion of Science.
Footnotes
▿
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Aktories, K., and J. T. Barbieri. 2005. Bacterial cytotoxins: targeting eukaryotic switches. Nat. Rev. Microbiol. 3397-410. [DOI] [PubMed] [Google Scholar]
- 2.Allen, W. E., D. Zicha, A. J. Ridley, and G. E. Jones. 1998. A role for Cdc42 in macrophage chemotaxis. J. Cell Biol. 1411147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailly, M., J. Wyckoff, B. Bouzahzah, R. Hammerman, V. Sylvestre, M. Cammer, R. Pestell, and J. E. Segall. 2000. Epidermal growth factor receptor distribution during chemotactic responses. Mol. Biol. Cell 113873-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleul, C. C., R. C. Fuhlbrigge, J. M. Casasnovas, A. Aiuti, and T. A. Springer. 1996. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J. Exp. Med. 1841101-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czuchra, A., X. Wu, H. Meyer, J. van Hengel, T. Schroeder, R. Geffers, K. Rottner, and C. Brakebusch. 2005. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol. Biol. Cell 164473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foxman, E. F., E. J. Kunkel, and E. C. Butcher. 1999. Integrating conflicting chemotactic signals. The role of memory in leukocyte navigation. J. Cell Biol. 147577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godwin, S. L., and S. P. Soltoff. 1997. Extracellular calcium and platelet-derived growth factor promote receptor-mediated chemotaxis in osteoblasts through different signaling pathways. J. Biol. Chem. 27211307-11312. [DOI] [PubMed] [Google Scholar]
- 8.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279509-514. [DOI] [PubMed] [Google Scholar]
- 9.Hiramoto, K., M. Negishi, and H. Katoh. 2006. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 3124205-4216. [DOI] [PubMed] [Google Scholar]
- 10.Katoh, H., K. Hiramoto, and M. Negishi. 2006. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 11956-65. [DOI] [PubMed] [Google Scholar]
- 11.Katoh, H., and M. Negishi. 2003. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 424461-464. [DOI] [PubMed] [Google Scholar]
- 12.Morii, N., and S. Narumiya. 1995. Preparation of native and recombinant Clostridium botulinum C3 ADP-ribosyltransferase and identification of Rho proteins by ADP-ribosylation. Methods Enzymol. 256196-206. [DOI] [PubMed] [Google Scholar]
- 13.Nobes, C. D., and A. Hall. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 1441235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pankov, R., Y. Endo, S. Even-Ram, M. Araki, K. Clark, E. Cukierman, K. Matsumoto, and K. M. Yamada. 2005. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 170793-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postlethwaite, A. E., J. Keski-Oja, G. Balian, and A. H. Kang. 1981. Induction of fibroblast chemotaxis by fibronectin. Localization of the chemotactic region to a 140,000-molecular weight non-gelatin-binding fragment. J. Exp. Med. 153494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postlethwaite, A. E., J. M. Seyer, and A. H. Kang. 1978. Chemotactic attraction of human fibroblasts to type I, II, and III collagens and collagen-derived peptides. Proc. Natl. Acad. Sci. USA 75871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridley, A. J. 2001. Rho GTPases and cell migration. J. Cell Sci. 1142713-2722. [DOI] [PubMed] [Google Scholar]
- 18.Sander, E. E., J. P. ten Klooster, S. van Delft, R. A. van der Kammen, and J. G. Collard. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 1471009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiffmann, E., B. A. Corcoran, and S. M. Wahl. 1975. _N_-Formylmethionyl peptides as chemoattractants for leucocytes. Proc. Natl. Acad. Sci. USA 721059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekine, A., M. Fujiwara, and S. Narumiya. 1989. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 2648602-8605. [PubMed] [Google Scholar]
- 21.Seppa, H., G. Grotendorst, S. Seppa, E. Schiffmann, and G. R. Martin. 1982. Platelet-derived growth factor in chemotactic for fibroblasts. J. Cell Biol. 92584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu, Y., D. Thumkeo, J. Keel, T. Ishizaki, H. Oshima, M. Oshima, Y. Noda, F. Matsumura, M. M. Taketo, and S. Narumiya. 2005. ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles. J. Cell Biol. 168941-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan, S., F. Wang, S. Glavas, A. Ott, F. Hofmann, K. Aktories, D. Kalman, and H. R. Bourne. 2003. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J. Cell Biol. 160375-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stramer, B., W. Wood, M. J. Galko, M. J. Redd, A. Jacinto, S. M. Parkhurst, and P. Martin. 2005. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzircotis, G., R. F. Thorne, and C. M. Isacke. 2005. Chemotaxis toward hyaluronan is dependent on CD44 expression and modulated by cell type variation in CD44-hyaluronan binding. J. Cell Sci. 1185119-5128. [DOI] [PubMed] [Google Scholar]
- 26.VanCompernolle, S. E., K. L. Clark, K. A. Rummel, and S. C. Todd. 2003. Expression and function of formyl peptide receptors on human fibroblast cells. J. Immunol. 1712050-2056. [DOI] [PubMed] [Google Scholar]
- 27.van Haastert, P. J., I. Keizer-Gunnink, and A. Kortholt. 2007. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J. Cell Biol. 177809-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidali, L., F. Chen, G. Cicchetti, Y. Ohta, and D. J. Kwiatkowski. 2006. Rac1-null mouse embryonic fibroblasts are motile and respond to platelet-derived growth factor. Mol. Biol. Cell 172377-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb, S. E., J. W. Pollard, and G. E. Jones. 1996. Direct observation and quantification of macrophage chemoattraction to the growth factor CSF-1. J. Cell Sci. 109(Pt. 4)793-803. [DOI] [PubMed] [Google Scholar]
- 30.Wells, C. M., M. Walmsley, S. Ooi, V. Tybulewicz, and A. J. Ridley. 2004. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J. Cell Sci. 1171259-1268. [DOI] [PubMed] [Google Scholar]
- 31.Yang, L., L. Wang, and Y. Zheng. 2006. Gene targeting of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in filopodia induction, directed migration, and proliferation in primary mouse embryonic fibroblasts. Mol. Biol. Cell 174675-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yonemura, S., T. Matsui, and S. Tsukita. 2002. Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J. Cell Sci. 1152569-2580. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, H., L. Vutskits, V. Calaora, P. Durbec, and J. Z. Kiss. 2004. A role for the polysialic acid-neural cell adhesion molecule in PDGF-induced chemotaxis of oligodendrocyte precursor cells. J. Cell Sci. 11793-103. [DOI] [PubMed] [Google Scholar]
- 34.Zicha, D., and G. A. Dunn. 1995. Are growth factors chemotactic agents? Exp. Cell Res. 221526-529. [DOI] [PubMed] [Google Scholar]
- 35.Zicha, D., G. A. Dunn, and A. F. Brown. 1991. A new direct-viewing chemotaxis chamber. J. Cell Sci. 99(Pt. 4)769-775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]