Targeted Genetic Disruption of Peroxisome Proliferator–Activated Receptor-δ and Colonic Tumorigenesis (original) (raw)
Abstract
Peroxisome proliferator–activated receptor-delta (PPAR-δ) is overexpressed in human colon cancer, but its contribution to colonic tumorigenesis is controversial. We generated a mouse model in which PPAR-δ was genetically disrupted in colonic epithelial cells by targeted deletion of exon 4. Elimination of colon-specific PPAR-δ expression was confirmed by real-time reverse transcription–polymerase chain reaction (real-time RT-PCR), immunoblotting, and activity assays. Mice with and without targeted PPAR-δ genetic disruption (10–11 mice per group) were tested for incidence of azoxymethane-induced colon tumors. The effects of targeted PPAR-δ deletion on vascular endothelial growth factor expression were determined by real-time RT-PCR. Targeted PPAR-δ genetic disruption inhibited colonic carcinogenesis: Mice with PPAR-δ(−/−) colons developed 98.5% fewer tumors than wild-type mice (PPAR-δ(−/−) vs wild-type, mean = 0.1 tumors per mouse vs 6.6 tumors per mouse, difference = 6.5 tumors per mouse, 95% confidence interval = 4.9 to 8.0 tumors per mouse, P < .001, two-sided test). Increased expression of vascular endothelial growth factor in colon tumors vs normal colon was suppressed by loss of PPAR-δ expression. These findings indicate that PPAR-δ has a crucial role in promoting colonic tumorigenesis.
Peroxisome proliferator–activated receptor-delta (PPAR-δ), a member of the ligand-activated PPAR nuclear receptor family, is overexpressed in human colon cancers (1,2) and in experimentally induced colon tumors in rodents (2). However, the role of PPAR-δ in colon tumorigenesis remains controversial. Studies in human colon cancer cell models have indicated that PPAR-δ promotes the development of colon tumors. For example, xenografts of human HCT-116 colon cancer cells with a genetic deletion of PPAR-δ exons 4–6 form tumors more slowly than wild-type (WT) HCT-116 cells in immunodeficient mice (3). Inhibition of PPAR-δ by nonsteroidal anti-inflammatory drugs suppresses survival of HCT-116, SW480, RKO, and DLD-1 human colon cancer cells in vitro (1,4), and PPAR-δ promotes survival of HCT-116 and LoVo colorectal cancer cells by inhibiting PPAR-γ–mediated apoptosis (5). In contrast, studies of PPAR-δ in experimental mouse models of colonic tumorigenesis have yielded conflicting results. Activation of PPAR-δ using a synthetic ligand in _Apc_Min mice, a commonly used mouse model of intestinal tumorigenesis in which a mutation in the adenomatous polyposis coli (Apc) tumor suppressor gene markedly increases intestinal polyp formation, promotes intestinal tumorigenesis (6). By contrast, germline deletion of PPAR-δ exon 4 in _Apc_Min mice failed to reduce intestinal tumor incidence in one study (7), but germline deletion of PPAR-δ exon 8 in _Apc_Min mice increased intestinal tumor incidence in another (8). More recently, deletion of exons 4 and part of exon 5 of PPAR-δ was shown to inhibit intestinal tumorigenesis in _Apc_Min mice (9).
The conflicting results regarding the effect of PPAR-δ knockout on intestinal tumorigenesis in _Apc_Min mice have been attributed to differences in the PPAR-δ genetic disruption strategies (9). The strategy that targeted exon 4, which encodes an essential portion of the PPAR-δ DNA-binding domain, was predicted to disrupt PPAR-δ function as a nuclear transcriptional factor and inhibited tumorigenesis (9,10). The alternative strategy that targeted exon 8 (8,11), the last PPAR-δ exon, is postulated to produce a hypomorphic PPAR-δ protein that remains at least partly functional. This hypothesis is supported by the observation of high rates of embryonic mortality, subsequent to abnormal trophoblastic giant cell differentiation and abnormal placental development, when exon 4 but not exon 8 of PPAR-δ was deleted (7,10).
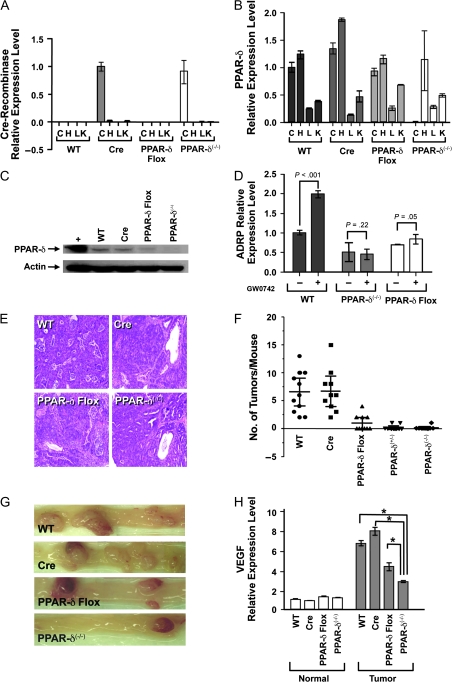
To further elucidate the role of PPAR-δ in intestinal tumorigenesis, we generated mice with a tissue-specific disruption of exon 4 of the gene for PPAR-δ in which deletion of PPAR-δ expression was targeted to the intestine (Supplementary Methods and Materials, available online). The advantage of this approach was that it would avoid the deleterious effects of unrestricted exon 4 deletion on mouse embryonic development and associated embryonic lethality (7,10). Briefly, mice that were heterozygous for the gene for Cre-recombinase directed expression by the intestine-specific villin promoter, and mice that were homozygous for a PPAR-δ allele with exon 4 flanked with loxP sites were mated to generate PPAR-δ(+/−) and PPAR-δ(−/−) mice in which tissue-specific expression of Cre-recombinase deleted PPAR-δ exon 4 in the mouse intestine. The villin–Cre-recombinase mice (n = 3) were obtained from the National Cancer Institute Mouse Models Repository (Frederick, MD), and the mice with the _loxP_-flanked exon 4 of PPAR-δ (n = 1) were a gift from Dr Ronald Evans (Salk Institute, La Jolla, CA). Both mouse strains were bred into a genetic background that was 95% or more FVB/N (FVB) because it is more efficient than other genetic backgrounds with respect to azoxymethane (AOM)-induced colonic tumorigenesis (12). Progeny mice (n = 4) were genotyped using polymerase chain reaction to amplify the PPAR-δ (WT and with _loxP_-flanked exon 4) and Cre-recombinase alleles from genomic DNA that was extracted from tail tissue (Supplementary Methods and Materials and Supplementary Figure 1, available online). To characterize PPAR-δ expression and function in the intestine, mice were killed by CO2 asphyxiation and the entire intestinal tract was removed from each mouse, colons were dissected longitudinally and washed in phosphate-buffered saline, further sectioned, and digested with dispase and collagenase at 37°C for 15 minutes to release colon crypts (Supplementary Methods and Materials, available online). Total RNA was isolated from colonic crypts, heart, liver, and kidney specimens from 6- to 8-week-old WT mice, mice with Cre but without the _loxP_-flanked (floxed) PPAR-δ exon 4 (Cre), mice with the floxed PPAR-δ exon 4 but not Cre (PPAR-δ Flox), or mice with Cre that were heterozygous or homozygous for PPAR-δ exon 4 deletion (PPAR-δ(+/−), PPAR-δ(−/−)) to measure relative levels of Cre-recombinase and PPAR-δ mRNA by real-time reverse transcription–polymerase chain reaction (RT-PCR). Targeting of Cre expression to the intestine (Figure 1, A) abolished the expression of PPAR-δ mRNA in the intestine but not other organs (Figure 1, B).
Figure 1.
Effects of targeted peroxisome proliferator–activated receptor-delta (PPAR-δ) genetic deletion on murine colonic epithelial cells and tumorigenesis. A) Cre-recombinase mRNA levels measured by quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR) in samples from isolated colonic crypts (C), heart (H), liver (L), and kidney (K) from wild-type (WT) FVB/N mice, mice with villin–Cre-recombinase but WT PPAR-δ alleles (Cre), mice with PPAR-δ exon 4 flanked by loxP sequence but no Cre-recombinase (PPAR-δ Flox), and mice with homozygous floxed PPAR-δ exon 4 deletion by Cre-recombinase (PPAR-δ(−/−)). Values are means with 95% confidence intervals of the means based on triplicate measurements from pooled samples from three mice (three repeated measurements showed similar results). The relative expression levels were calculated as the values relative to that of the calibrator sample (colon of Cre mice). Cre-recombinase expression levels in colonic epithelial cells were statistically significantly higher in Cre and PPAR-δ(−/−) mice than other mice or other organs (P < .001, two-way analysis of variance [ANOVA]). B) PPAR-δ mRNA expression levels, measured by quantitative real-time RT-PCR from the same RNA samples from the same groups of mice as in (A). Colonic epithelial cell PPAR-δ expression levels were statistically significantly lower in PPAR-δ(−/−) mice than in other organs or mouse groups (eg, WT, PPAR-δ Flox) (P < .001, two-way ANOVA). Values are means and 95% confidence intervals of triplicate measurements from pooled samples from three mice (three repeated measurements showed similar results). The relative expression levels were calculated as the values relative to that of the calibrator sample (colon of WT mice). C) PPAR-δ protein expression in dissected colonic epithelial cells from the indicated mice, as determined by immunoblots probed with a rabbit anti-mouse PPAR-δ antibody. Blots were stripped and probed with a rabbit polyclonal anti-actin antibody as a control for loading and transfer. + = positive control (mouse PPAR-δ expression vector-transfected cells). Three independent experiments were performed, and results were similar in each experiment. D) Effects of PPAR-δ genetic deletion on PPAR-δ activity. Primary colonic epithelial cells were isolated from the indicated mice, and the cells were treated with either PPAR-δ agonist (GW0742, +) or vehicle solution (control, –). Expression of the PPAR-δ target gene ADRP was measured by real-time RT-PCR. Values shown are the means and 95% confidence intervals of triplicate measurements from pooled samples of three mice (three repeated measurements showed similar results). P values for ADRP expression in GW0724-treated vs control cells, by two-way ANOVA, are as shown. E) Appearance of colon tumors in mice during genetic targeting of PPAR-δ. Mice of the indicated genotypes were treated with azoxymethane, killed 20 weeks later, and examined for colonic tumor formation. Pictures shown are from hematoxylin–eosin staining of colonic tumors at ×100 magnification. F) Scatter plot of number of tumors per mouse in mice treated as described in (E). For WT vs PPAR-δ Flox, PPAR-δ(+/−), and PPAR-δ(+/−), P < .001. For WT vs Cre, P = .897 (Poisson regression). G) Mouse colons after azoxymethane treatment as described in (E). Pictures taken with an SMZ800 stereoscopic zoom microscope (Nikon Instruments Inc, Melville, NY) at ×10 magnification. H) Effects of targeted PPAR-δ genetic disruption on vascular endothelial growth factor (VEGF) expression during azoxymethane-induced colonic tumorigenesis. Mice were treated with azoxymethane and killed 20 weeks later as described in (E). RNA was extracted from samples of normal and tumor colonic epithelial cells of isolated colonic crypts. VEGF mRNA levels were measured by quantitative real-time RT-PCR. The relative expression levels were calculated as the values relative to that of the calibrator sample (normal colon of WT mice). Values are means and 95% confidence intervals of the means based on triplicate measurements from pooled samples of 11 WT mice and 10 mice each in the other groups. Three repeated experiments showed similar results. *P < .001 (two-way ANOVA). All statistical tests were two-sided.
In additional experiments, protein was extracted from the isolated colonic crypts of mice of WT, Cre, PPAR-δ Flox, and PPAR-δ(−/−) genotypes, and immunoblots confirmed that PPAR-δ expression was absent in PPAR-δ(−/−) mice (Figure 1, C). Also, colonic crypt cells were plated in collagen-coated wells and were immediately treated with the PPAR-δ ligand GW0742 (1 μM for 24 hours) or vehicle (dimethyl sulfoxide) before RNA extraction and real-time RT-PCR to measure expression of the gene for adipose differentiation-related protein (ADRP), a known PPAR-δ target (10). The absence of PPAR-δ mRNA and protein expression (Figure 1, B and C) were associated with a statistically significant reduction in the baseline levels of ADRP expression (P < .001) and with a total loss of PPAR-δ agonist GW0742-stimulated ADRP expression in colonic epithelial cells (Figure 1, D). Interestingly, compared with WT or Cre mice, PPAR-δ Flox mice had similar PPAR-δ mRNA levels but markedly lower PPAR-δ protein expression and activity in the colon (Figure 1, C and D). These findings indicated that the loxP sequence insertion flanking PPAR-δ exon 4 likely reduced PPAR-δ protein expression and activity through posttranscriptional mechanisms.
To test the effects of PPAR-δ genetic disruption on colonic tumorigenesis, mice were injected intraperitoneally with the carcinogen AOM, a method that simulates human colorectal tumorigenesis better than genetic methods (eg, _Apc_Min mutation) or other chemical methods (eg, methylnitrosourea-induced tumorigenesis) (13). At 6–8 weeks of age, 10–11 mice of each genotype (WT, Cre, PPAR-δ Flox, PPAR-δ(+/−), and PPAR-δ(−/−)) were administered 250 μL of isotonic saline containing 10 mg AOM per kg body weight once a week for 6 weeks or were given saline alone as a control (see Supplementary Methods and Materials for details, available online). In accordance with protocol, all mice were killed by CO2 asphyxiation at 20 weeks after the last dose of AOM. Colorectal tumorigenesis was evaluated both by counting the number of colon tumors per mouse and by histological examination of representative tumors.
Targeted PPAR-δ genetic disruption profoundly inhibited AOM-induced colonic tumorigenesis. In the various experimental groups (WT, Cre, PPAR-δ Flox, PPAR-δ(+/−), and PPAR-δ(−/−)), AOM induced colonic adenocarcinomas that were histologically very similar to human colonic adenocarcinoma (Figure 1, E). WT mice developed a mean of 6.6 tumors per mouse (Table 1; Figure 1, F and G). However, mice that were heterozygous for the exon 4 deletion of PPAR-δ (PPAR-δ(+/−)) developed, on average, only 3.0% as many tumors per mouse as WT control mice (mean = 0.2 tumors per PPAR-δ(+/−) mouse, difference = 6.4, 95% confidence interval [CI] = 4.8 to 7.9 tumors per mouse, P < .001), and mice that were homozygous for the exon 4 deletion developed only 1.5% as many tumors as WT control mice (mean = 0.1 tumors per PPAR-δ(−/−) mouse, difference = 6.5, 95% CI = 4.9 to 8.0 tumors per mouse, P < .001) (Table 1; Figure 1, F and G). In addition, colonic tumor formation was statistically significantly inhibited for all tumor size categories in the PPAR-δ(+/−) and PPAR-δ(−/−) mice (Table 1). PPAR-δ Flox mice, in which PPAR-δ protein expression and function were reduced but not absent, developed 15% as many tumors as WT control mice (mean = 1.0 tumor per PPAR-δ Flox mouse, difference [vs WT] = 5.6 tumors per mouse, 95% CI = 3.9 to 7.2 tumors per mouse, P < .001), but 5-fold more tumors than PPAR-δ(+/−) mice (difference = 0.8 tumors, 95% CI = 0.04 to 1.56 tumors per mouse, P = .038) and 10-fold more tumors than PPAR-δ(−/−) mice (difference = 0.9 tumors, 95% CI = 0.15 to 1.65 tumors per mouse, P = .028) (Figure 1, F and G and Table 1). These data clearly indicated that PPAR-δ has a positive influence on susceptibility to colonic tumorigenesis. Furthermore, these data indicated that the extent of colonic tumorigenesis depended on the amount of remaining PPAR-δ function.
Table 1.
Effects of peroxisome proliferator–activated receptor-delta (PPAR-δ) genetic disruption targeted to murine intestine on azoxymethane-induced colonic tumorigenesis*
| Distribution of tumors by tumor diameter, mm | VEGF mRNA relative expression level‡ | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mouse group | No. of mice | Total no. of tumors | Mean no. of tumors per mouse (95% CI)† | <2.0§ | 2.0–4.0‖ | >4.0¶ | Normal colon | Colon tumor | Tumor/normal# |
| WT | 11 | 72 | 6.6 (5.3 to 8.5) | 23 | 37 | 12 | 1.00 | 6.60 | 6.59 |
| Cre | 10 | 67 | 6.7 (5.2 to 8.3) | 29 | 29 | 9 | 0.80 | 7.80 | 9.86 |
| PPAR-δ Flox | 10 | 10 | 1.0 (0.54 to 1.9) | 1 | 5 | 4 | 1.25 | 4.29 | 3.42 |
| PPAR-δ(+/−) | 10 | 2 | 0.2 (0.05 to 0.8) | 0 | 1 | 1 | 1.00 | 3.21 | 3.21 |
| PPAR-δ(−/−) | 10 | 1 | 0.1 (0.01 to 0.7) | 0 | 1 | 0 | 1.13 | 2.79 | 2.47 |
Various molecular events have been implicated in the promotion of carcinogenesis by PPAR-δ, including induction of COX-2 expression and prostaglandin E2 (PGE2) production (14,15), PGE2 receptor subtype EP4 expression through phosphatidylinositol-3 kinase signaling (16), activation of extracellular signal-regulated kinases 1 and 2 (ERK1 and ERK2) (17), increased expression of vascular endothelial growth factor (VEGF) (9,18,19), reduction of the proapoptotic cell death–inducing DFF45-like effector A and B levels and increase of antiapoptotic inhibitor of caspase-activated deoxyribonuclease short and long forms (ICAD-S and ICAD-L) (20), activation of the antiapoptotic Akt1 pathway (21) via 3-phosphoinositide–dependent kinase-1 (PDK1) and integrin-linked kinase activity (22), increased NF-κB activation and matrix metalloproteinase-9 secretion (22), suppression of PTEN expression (23), and transcriptional activation of 14-3-3ϵ protein (24). VEGF is increasingly being recognized to promote tumorigenesis via both angiogenic and nonangiogenic mechanisms (25), and the effects of PPAR-δ on VEGF expression during tumorigenesis are controversial because of conflicting findings in the literature. In vitro, the PPAR-δ ligand GW501516 was reported to stimulate VEGF expression in bladder, breast, and colon cancer cell lines (9,18,19), and PPAR-δ was necessary for GW501516 to stimulate VEGF expression (9). In vivo, GW501516 increased VEGF protein expression in intestinal adenomas of _Apc_Min mice (9). However, others have reported that the PPAR-δ agonists GW0742 and GW501516 failed to increase VEGF expression in cancer cell lines in vitro and that GW0742 failed to increase VEGF in colonic polyps of _Apc_Min mice (26).
We therefore also examined the effects of PPAR-δ genetic disruption on VEGF expression in the AOM colonic tumorigenesis model. For these experiments, VEGF mRNA levels were measured by real-time RT-PCR from total RNA extracted from tumor vs normal colon tissue from mice of each genotype, as in previous experiments. VEGF relative expression levels were then calculated relative to levels in nonmalignant colon tissue from WT mice.
Mice in which PPAR-δ was genetically disrupted exhibited a smaller increase in VEGF mRNA expression in tumors compared with WT or Cre control mice—the ratio of the VEGF mRNA levels in tumor vs nonmalignant colon cells in PPAR-δ(−/−) mice was 37.5% of that in WT mice and 25% of that in Cre mice (Figure 1, H; Table 1). Relative to the calibrator sample (nonmalignant colon of WT mice), tumors of WT mice had 6.6-fold more VEGF mRNA, tumors of Cre mice had 7.8-fold more VEGF mRNA, but tumors of PPAR-δ(−/−) mice had only 2.8-fold more VEGF mRNA (Table 1) (WT vs PPAR-δ(−/−) tumors: difference = 3.8-fold, 95% CI = 3.5 to 4.1-fold, P < .001; Cre vs PPAR-δ(−/−) tumors: difference = 5.0-fold, 95% CI = 4.6 to 5.5-fold, P < .001). The PPAR-δ Flox mice, which also had decreased PPAR-δ protein expression and function, also had tumors containing somewhat less VEGF mRNA than tumors of WT mice or Cre control mice (Table 1). Relative to nonmalignant colon of WT mice, tumors of PPAR-δ Flox mice had 4.29-more VEGF mRNA (WT vs PPAR-δ Flox tumors: difference = 2.3-fold, 95% CI = 1.8 to 2.8-fold, P < .001; Cre vs PPAR-δ Flox tumors: difference = 3.5-fold, 95% CI = 3.0 to 4.1-fold, P < .001). Levels of VEGF mRNA in PPAR-δ Flox tumors were higher than those in PPAR-δ(−/−) tumors (Table 1) (difference = 1.5-fold, 95% CI = 1.1 to 1.91-fold, P < .001), indicating that the extent of VEGF expression was inversely proportional to the degree of reduction in PPAR-δ function. These findings strongly suggest that PPAR-δ expression in colon cancer cells promotes VEGF expression and thus challenge the previously proposed hypothesis that expression of PPAR-δ in stromal cells, not tumor cells, promotes tumor angiogenesis (27).
Our findings demonstrate the effects of PPAR-δ on colonic tumorigenesis in the first mouse model with genetic disruption of PPAR-δ targeted to colonic epithelial cells. Our targeted approach successfully abolished PPAR-δ expression and activity in the intestine and had no effects on embryonic mouse development (data not shown), in contrast to what was observed in mouse models that used germline genetic disruption of PPAR-δ exon 4 (7,10). A previous study (9) that had used germline genetic disruption of PPAR-δ exons 4 and 5 showed a threefold decrease in intestinal adenoma formation in _Apc_Min mice that were homozygous for the PPAR-δ deletion compared with _Apc_Min mice with WT PPAR-δ. The current study using intestine-specific deletion of PPAR-δ and AOM-induced colonic carcinogenesis, which is a more representative model of human colorectal tumorigenesis than the one using _Apc_Min mice (_Apc_Min mice usually develop intestinal adenomas, but the AOM-induced mice usually develop colonic adenocarcinomas), showed more profound effects. Tumor incidence in this study was 65-fold lower in PPAR-δ(−/−) mice than in WT mice.
In another study (8), PPAR-δ genetic disruption increased AOM-induced colonic tumorigenesis, in contrast to our current findings. The different mouse genetic backgrounds used in the two studies, FVB in the current study vs C57BL/6 in the previous report, could have contributed to the contrasting results. However, we believe that the most plausible explanation for the conflicting results is that in our model, PPAR-δ exon 4 deletion effectively abolished PPAR-δ function in colonic epithelial cells, whereas in the previous study (10), exon 8 was targeted, which might have inadequately inhibited PPAR-δ function. Also, it has been postulated that PPAR-δ, like other gene products (27), could have a different role in stromal cells as opposed to epithelial cells during colonic tumorigenesis. Germline deletion of PPAR-δ was used in the previous study (8), whereas in our study, PPAR-δ was selectively deleted from colonic epithelial cells. The current findings are novel in finding that PPAR-δ can act specifically in cancer cells to promote tumorigenesis and tumor angiogenesis.
This study has some potential limitations. We used a single type of genetic deletion in PPAR-δ in a single mouse genetic background, and of course, carcinogenesis could be different in mice than in humans. In addition, we targeted deletion of PPAR-δ to colonic epithelial cells, as opposed to the colon in general, which could be considered a limitation if carcinogenesis were to require the contribution of nonepithelial cells independently of cancer epithelial cells. The relative contribution of stromal cell–specific PPAR-δ expression to tumorigenesis and tumor angiogenesis will require future studies that selectively genetically delete PPAR-δ from tumor stromal cells in the colon.
Our study suggests that therapeutics that inhibit PPAR-δ function might inhibit the growth of colon cancers. To date, with the exception of one recent report that characterized a selective PPAR-δ antagonist in vitro (28), the field of PPAR-δ therapeutic targeting has been limited to the development of PPAR-δ agonists for treatment of diseases other than cancer (28). Our current demonstration of the substantial involvement of PPAR-δ in colonic tumorigenesis should stimulate future efforts to test PPAR-δ inhibitors for the treatment and prevention of colon cancer and should serve to caution against clinical testing of PPAR-δ agonists without a complete preclinical evaluation of their possible capacity to promote tumorigenesis.
Funding
National Cancer Institute (grant no. R01-CA106577 and R01-CA104278 to I.S.), a Cancer Research Foundation of America grant (to X.Z.), and the Caroline Wiess Law Endowment for Cancer Prevention.
Supplementary Material
[Supplementary Data]
Footnotes
We are very thankful to Dr Ronald Evans for providing us with the PPAR-δ Flox mice and to the National Cancer Institute/Mouse Models of Human Cancers Consortium for providing us with the villin-Cre mice, which were originally donated by Dr Sylvie Robine (Institute Curie, France). The sponsors had no role in the study design, the analysis and interpretation of the data, the preparation of the manuscript, or the decision to submit the manuscript for publication.
References
- 1.He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99(3):335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RA, Tan J, Krause WF, et al. Prostacyclin-mediated activation of peroxisome proliferator-activated receptor delta in colorectal cancer. Proc Natl Acad Sci USA. 2000;97(24):13275–13280. doi: 10.1073/pnas.97.24.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park BH, Vogelstein B, Kinzler KW. Genetic disruption of PPARdelta decreases the tumorigenicity of human colon cancer cells. Proc Natl Acad Sci USA. 2001;98(5):2598–2603. doi: 10.1073/pnas.051630998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shureiqi I, Jiang W, Zuo X, et al. The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-delta to induce apoptosis in colorectal cancer cells. Proc Natl Acad Sci USA. 2003;100(17):9968–9973. doi: 10.1073/pnas.1631086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo X, Wu Y, Morris JS, et al. Oxidative metabolism of linoleic acid modulates PPAR-beta/delta suppression of PPAR-gamma activity. Oncogene. 2006;25(8):1225–1241. doi: 10.1038/sj.onc.1209160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta RA, Wang D, Katkuri S, Wang H, Dey SK, DuBois RN. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-delta accelerates intestinal adenoma growth. Nat Med. 2004;10(3):245–247. doi: 10.1038/nm993. [DOI] [PubMed] [Google Scholar]
- 7.Barak Y, Liao D, He W, et al. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci USA. 2002;99(1):303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-delta attenuates colon carcinogenesis. Nat Med. 2004;10(5):481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Wang H, Guo Y, et al. Crosstalk between peroxisome proliferator-activated receptor delta and VEGF stimulates cancer progression. Proc Natl Acad Sci USA. 2006;103(50):19069–19074. doi: 10.1073/pnas.0607948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadra K, Anghel SI, Joye E, et al. Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor {beta}/{delta} Mol Cell Biol. 2006;26(8):3266–3281. doi: 10.1128/MCB.26.8.3266-3281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters JM, Lee SS, Li W, et al. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20(14):5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nambiar PR, Girnun G, Lillo NA, Guda K, Whiteley HE, Rosenberg DW. Preliminary analysis of azoxymethane induced colon tumors in inbred mice commonly used as transgenic/knockout progenitors. Int J Oncol. 2003;22(1):145–150. [PubMed] [Google Scholar]
- 13.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2(8):1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Han C, Lim K, Wu T. Cross-talk between peroxisome proliferator-activated receptor {delta} and cytosolic phospholipase A2{alpha}/cyclooxygenase-2/prostaglandin E2 signaling pathways in human hepatocellular carcinoma cells. Cancer Res. 2006;66(24):11859–11868. doi: 10.1158/0008-5472.CAN-06-1445. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, Han C, Wu T. A novel positive feedback loop between peroxisome proliferator-activated receptor-{delta} and prostaglandin E2 signaling pathways for human cholangiocarcinoma cell growth. J Biol Chem. 2006;281(45):33982–33996. doi: 10.1074/jbc.M600135200. [DOI] [PubMed] [Google Scholar]
- 16.Han S, Ritzenthaler JD, Wingerd B, Roman J. Activation of peroxisome proliferator-activated receptor {beta}/{delta} (PPAR{beta}/{delta}) increases the expression of prostaglandin E2 receptor subtype EP4. The roles of phosphatidylinositol 3-kinase and CCAAT/enhancer-binding protein {beta} J Biol Chem. 2005;280(39):33240–33249. doi: 10.1074/jbc.M507617200. [DOI] [PubMed] [Google Scholar]
- 17.Daikoku T, Tranguch S, Chakrabarty A, et al. Extracellular signal-regulated kinase is a target of cyclooxygenase-1-peroxisome proliferator-activated receptor-{delta} signaling in epithelial ovarian cancer. Cancer Res. 2007;67(11):5285–5292. doi: 10.1158/0008-5472.CAN-07-0828. [DOI] [PubMed] [Google Scholar]
- 18.Stephen RL, Gustafsson MCU, Jarvis M, et al. Activation of peroxisome proliferator-activated receptor {delta} stimulates the proliferation of human breast and prostate cancer cell lines. Cancer Res. 2004;64(9):3162–3170. doi: 10.1158/0008-5472.can-03-2760. [DOI] [PubMed] [Google Scholar]
- 19.Fauconnet S, Lascombe I, Chabannes E, et al. Differential regulation of vascular endothelial growth factor expression by peroxisome proliferator-activated receptors in bladder cancer cells. J Biol Chem. 2002;277(26):23534–23543. doi: 10.1074/jbc.M200172200. [DOI] [PubMed] [Google Scholar]
- 20.Tan NS, Michalik L, Noy N, et al. Critical roles of PPARbeta/delta in keratinocyte response to inflammation. Genes Dev. 2001;15(24):3263–3277. doi: 10.1101/gad.207501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di-Poi N, Ng CY, Tan NS, et al. Epithelium-mesenchyme interactions control the activity of peroxisome proliferator-activated receptor {beta}/{delta} during hair follicle development. Mol Cell Biol. 2005;25(5):1696–1712. doi: 10.1128/MCB.25.5.1696-1712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di-Poï N, Tan NS, Michalik L, Wahli W, Desvergne B. Antiapoptotic role of PPAR[beta] in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol Cell. 2002;10(4):721–733. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- 23.Han S, Ritzenthaler JD, Zheng Y, Roman J. PPAR{beta}/{delta} agonist stimulates human lung carcinoma cell growth through inhibition of PTEN expression: the involvement of PI3K and NF-{kappa}B signals. Am J Physiol Lung Cell Mol Physiol. 2008;294(6):L1238–L1249. doi: 10.1152/ajplung.00017.2008. [DOI] [PubMed] [Google Scholar]
- 24.Liou J-Y, Ghelani D, Yeh S, Wu KK. Nonsteroidal anti-inflammatory drugs induce colorectal cancer cell apoptosis by suppressing 14-3-3{varepsilon} Cancer Res. 2007;67(7):3185–3191. doi: 10.1158/0008-5472.CAN-06-3431. [DOI] [PubMed] [Google Scholar]
- 25.Epstein RJ. VEGF signaling inhibitors: more pro-apoptotic than anti-angiogenic. Cancer Metastasis Rev. 2007;26(3–4):443–452. doi: 10.1007/s10555-007-9071-1. [DOI] [PubMed] [Google Scholar]
- 26.Hollingshead HE, Killins RL, Borland MG, et al. Peroxisome proliferator-activated receptor-/{delta} (PPAR/{delta}) ligands do not potentiate growth of human cancer cell lines. Carcinogenesis. 2007;28(12):2641–2649. doi: 10.1093/carcin/bgm183. [DOI] [PubMed] [Google Scholar]
- 27.Muller-Brusselbach S, Komhoff M, Rieck M, et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. EMBO J. 2007;26(15):3686–3698. doi: 10.1038/sj.emboj.7601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shearer BG, Steger DJ, Way JM, et al. Identification and characterization of a selective peroxisome proliferator-activated receptor {beta}/{delta} (NR1C2) antagonist. Mol Endocrinol. 2008;22(2):523–529. doi: 10.1210/me.2007-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]