Polynesian origins: Insights from the Y chromosome (original) (raw)
Abstract
The question surrounding the colonization of Polynesia has remained controversial. Two hypotheses, one postulating Taiwan as the putative homeland and the other asserting a Melanesian origin of the Polynesian people, have received considerable attention. In this work, we present haplotype data based on the distribution of 19 biallelic polymorphisms on the Y chromosome in a sample of 551 male individuals from 36 populations living in Southeast Asia, Taiwan, Micronesia, Melanesia, and Polynesia. Surprisingly, nearly none of the Taiwanese Y haplotypes were found in Micronesia and Polynesia. Likewise, a Melanesian-specific haplotype was not found among the Polynesians. However, all of the Polynesian, Micronesian, and Taiwanese haplotypes are present in the extant Southeast Asian populations. Evidently, the Y-chromosome data do not lend support to either of the prevailing hypotheses. Rather, we postulate that Southeast Asia provided a genetic source for two independent migrations, one toward Taiwan and the other toward Polynesia through island Southeast Asia.
The major prehistoric events leading to the settlement of Polynesia have been examined from various perspectives, and two different models of population movements are proposed. The first of these, dubbed the “express train” model (1), based primarily on archeological and linguistic evidence (2), claims that about 4,000 to 5,000 years B.P. a rapid eastward migration of humans began in Southern China spreading Austronesian language and the associated Lapita culture through the Pacific islands and culminating in the colonization of Polynesia. In this model, Taiwan, which is adjacent to the Asian mainland, was colonized first. This hypothesis is supported by recent mtDNA data (3–6), which tie the Taiwanese aborigines with the Polynesians. The second hypothesis proposed by Terrell (7) asserts a neighboring homeland of the Polynesians in Melanesia, in which the Polynesians evolved in a complex nexus of interactions among the already settled Pacific islanders.
Although most genetic evidence favors the former hypothesis, the debate continues and other plausible scenarios are being examined as well. Notably, Richards et al. (8) recently suggested that evidence from mtDNA data is, in fact, more consistent with an origin in eastern Indonesia.
In recent years, the power of Y-chromosome markers in resolving evolutionary histories of human populations has been greatly recognized (9, 10). This is so because markers on the nonrecombining portion of the Y chromosome allow construction of intact haplotypes and thus, male-mediated migration can be readily recognized. Identification of a large number of biallelic markers on the Y chromosome (12, 13)¶¶ has augmented such studies. In this study, we have reviewed the origin of the Polynesian people from this angle through an analysis of 19 biallelic markers on 551 males derived from 36 populations living in greater Southeast Asia, Micronesia, Melanesia, and Polynesia.
Materials and Methods
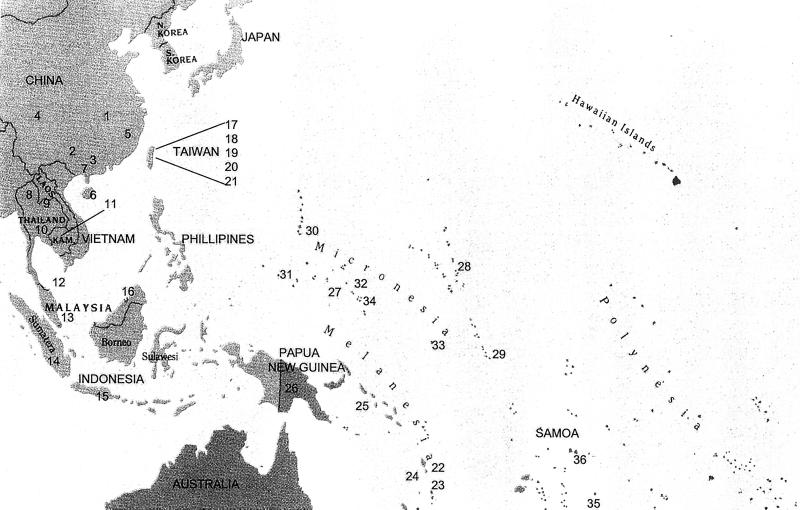
Names, sample sizes, and geographic locations of the studied populations are given in Table 1 and Fig. 1. Some of the published data (14) on Southeast Asian populations have been incorporated in this study for purposes of comparison. The details of the 19 biallelic markers, PCR amplification protocols, haplotype construction, and nomenclature are given in Su et al. (14). Primer sequences and protocols for PCR amplification and analysis of these markers are available on request. Haplotype diversity was calculated following the method of Nei (15), and genetic distances were computed following Nei (15) and Reynolds et al. (16).
Table 1.
Y-chromosome haplotype frequency distribution in Asian and Oceanic populations
| Populations | N | Haplotypes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H14 | H16 | H17 |
| Southeast Asia | ||||||||||||||
| Tujia (1)* | 10 | 10 | 20 | 30 | 10 | 20 | 10 | |||||||
| Yao (2)* | 20 | 35 | 15 | 10 | 15 | 15 | 20 | |||||||
| Dong (3)* | 10 | 20 | 10 | 20 | 20 | 10 | 20 | |||||||
| Yi (4)* | 14 | 14.3 | 42.9 | 21.4 | 7.1 | 14.3 | ||||||||
| She (5)* | 11 | 18.2 | 9.1 | 18.2 | 27.3 | 18.2 | 9.1 | |||||||
| Li (6)* | 11 | 9.1 | 27.3 | 54.5 | 9.1 | |||||||||
| Zhuang (7)* | 28 | 3.6 | 3.6 | 7.1 | 3.6 | 3.6 | 25 | 17.9 | 25 | 10.7 | ||||
| North Thai (8) | 20 | 20 | 5 | 30 | 20 | 20 | 5 | |||||||
| Northeast Thai (9)* | 20 | 5 | 5 | 5 | 5 | 5 | 5 | 45 | 20 | 5 | ||||
| So (10) | 5 | 20 | 20 | 40 | 20 | |||||||||
| Cambodian (11)* | 26 | 3.8 | 3.8 | 11.5 | 11.5 | 3.8 | 15.4 | 3.8 | 3.8 | 23.1 | 11.5 | 3.8 | 3.8 | |
| Orang Asli (12) | 17 | 23.5 | 5.9 | 5.9 | 64.7 | |||||||||
| Malay (13) | 27 | 3.7 | 18.5 | 33.3 | 22.2 | 3.7 | 14.8 | 3.7 | ||||||
| Batak (14)* | 18 | 5.6 | 5.6 | 11.1 | 11.1 | 16.7 | 22.2 | 27.8 | ||||||
| Javanese (15)* | 11 | 9.1 | 9.1 | 27.3 | 9.1 | 18.2 | 9.1 | 18.2 | ||||||
| Kota Kinabalu (16) | 19 | 10.5 | 5.3 | 10.5 | 31.6 | 10.5 | 26.3 | 5.3 | ||||||
| Taiwan | ||||||||||||||
| Bunun (17) | 9 | 11.1 | 66.7 | 22.2 | ||||||||||
| Atayal (18)* | 24 | 29.2 | 4.2 | 4.2 | 54.2 | 8.3 | ||||||||
| Yami (19)* | 8 | 25 | 75 | |||||||||||
| Paiwan (20)* | 11 | 18.2 | 54.5 | 27.3 | ||||||||||
| Ami (21)* | 6 | 100 | ||||||||||||
| Melanesia | ||||||||||||||
| Bankes & Torres (22) | 6 | 33.3 | 33.3 | 16.7 | 16.7 | |||||||||
| Maewo (23) | 10 | 60 | 20 | 20 | ||||||||||
| Santo (24) | 4 | 100 | ||||||||||||
| Nasioi Melanesian (25) | 3 | 100 | ||||||||||||
| New Guinea (26) | 90 | 15.5 | 2.2 | 43.3 | 38.9 | |||||||||
| Micronesia | ||||||||||||||
| Truk (27) | 17 | 5.9 | 64.7 | 5.9 | 5.9 | 5.9 | 11.8 | |||||||
| Majuro (28) | 9 | 11.1 | 66.7 | 22.2 | ||||||||||
| Kiribati (29) | 11 | 63.6 | 9.1 | 27.3 | ||||||||||
| Guam (30) | 6 | 16.7 | 16.7 | 33.3 | 33.3 | |||||||||
| Palau (31) | 13 | 7.7 | 7.7 | 61.5 | 23.1 | |||||||||
| Phonpei (32) | 10 | 30 | 70 | |||||||||||
| Nauru (33) | 7 | 28.6 | 71.4 | |||||||||||
| Polynesia | ||||||||||||||
| Kapingamarangi (34) | 10 | 30 | 70 | |||||||||||
| Tonga (35) | 1 | 100 | ||||||||||||
| Samoan (36) | 29 | 48.3 | 6.9 | 41.4 | 3.5 |
Figure 1.
Map showing the locations of the studied populations. The numbers correspond to the population names and respective numbers in parentheses in Table 1. Note that the Polynesian population Kapingamarangi (labeled 34) is geographically located in Micronesia.
Results and Discussion
Using 19 biallelic markers, we identified 15 haplotypes in the total sample of 551 Y chromosomes. The haplotype frequencies in various populations are shown in Table 1. Earlier we had presented a parsimonious phylogenetic tree of the haplotypes based on the 19 markers (14), in which H1 was considered as the ancestral haplotype because of its appearance in chimpanzees. Among the other haplotypes, H2 is also relatively ancient with its occurrence in both African and non-African populations; and, H5 appeared as the common ancestor of all other non-African haplotypes, which are regionally distributed. The Southeast Asians, with a total of 14 haplotypes and haplotypic diversity of 0.88, are by far the most diverse among all of the studied populations. The only missing haplotype is H17, which is apparently Melanesian-specific. The pooled 58 aboriginal Taiwanese males share seven of these haplotypes (H6–H12), with a diversity of 0.70. Two of these haplotypes (H6 and H7) were observed only in the Atayal population. In the pooled sample of 113 Micronesian and Polynesian individuals, we identified 10 haplotypes (haplotype diversity of 0.72), of which nine (H1, H2, H4, H5, H6, H8, H10, H12, and H14) are shared with the Southeast Asians. The tenth haplotype is H17, which was found only in two Trukese individuals.
A comparison of the haplotype distributions among the Micronesians and Polynesians with those in the Taiwanese populations is noteworthy. With the exception of H6, these two groups of populations harbor two independent sets of haplotypes. H1, H2, H4, and H5 are present exclusively in Micronesia and Polynesia. Likewise, Taiwanese haplotypes H7, H8, H9, H10, H11, and H12 are absent in Polynesia and comparatively rare in Micronesia. In fact, Micronesians and Polynesians together share only four haplotypes with the Taiwanese, three of which were found in Micronesia but not in Polynesia, which may reflect a more recent gene flow from Southeast Asia into Micronesia. Conspicuously, both H1 (the ancestral haplotype) and H5 (common ancestor of non-African haplotypes) are absent in the aboriginal Taiwanese populations, whereas they are present in appreciable frequencies in most Micronesian and Polynesian populations. It is evident that the Taiwanese aboriginal populations on one hand, and the Micronesians and Polynesians on the other, carry two different subsets of haplotypes found in the extant Southeast Asian populations. Genetic distance calculations (Table 2) show that the divergence between Taiwanese and Micronesian/Polynesian populations is twice as great as the divergence of either population groups from Southeast Asians.
Table 2.
Genetic distances
| Populations | _D_m* | _D_ST† | _F_ST† |
|---|---|---|---|
| SE Asia - Taiwan | 0.108 | 0.268 | 0.098 |
| SE Asia - Micronesia/Polynesia | 0.103 | 0.279 | 0.097 |
| Taiwan - Micronesia/Polynesia | 0.250 | 0.876 | 0.203 |
The express train model views that the settlement of Taiwan had occurred about 5,000 to 6,000 years ago through a proto-Austronesian migration from coastal Southern China. Although Austronesian languages are no longer spoken in southern parts of China, our data do not preclude the possibility of the coastal southern Chinese being the founders of the present-day Taiwanese aborigines for the reason, as noted above, that Taiwanese Y-chromosome haplotypes represent a subset that is observed in greater southeast Asia including southern China. We should note that several of the coastal Chinese populations (namely, Tujia, Yao, Dong, She, Li, and Zhuang) cited in this study, who, with the expansion of Han Chinese during the past 2,000 years, had moved to the southwest part of China (namely, Yunnan, Guizhou, and Sichuan), share a similar Y-haplotype profile with the other southeast Asian populations. Evidently the distribution of Y-chromosome haplotypes strongly suggests a genetic continuum throughout greater Southeast Asia including southern China, mainland, and insular Southeast Asia. This continuum is apparent across populations irrespective of their linguistic affiliations, namely, Sino-Tibetan, Hmong Mien, Austroasiatic, and Austronesian speakers. Notwithstanding these observations, the Y-chromosome data do not favor a Polynesian homeland in Taiwan. However, our findings do not refute the fast train model in its entirety, which contends the spread of Austronesian language and Lapita culture from Southeast Asia. Furthermore, the extent of haplotype diversity is the highest in Southeast Asia compared with any other populations further complementing the notion of a Southeast Asian homeland. The most plausible explanation of our data is that both the Taiwanese and the Polynesian populations derive their ancestry in Southeast Asia. Nonetheless, colonization of Polynesia had likely occurred via a route out of Southeast Asia independent of the expansion toward Taiwan. At this time, however, it is not possible to conclusively localize the center of origin of the Polynesian ancestors. Alternatively our data could be viewed as a spread of Y-chromosome haplotypes out of China to Taiwan and then to Micronesia/Polynesia; and in this model, the haplotypes became distinguished on account of random extinction of Y-chromosome lineages in different geographic regions. However, this model seems unlikely in view of the fact that multiple populations from the same geographical region in our study tend to show a similar distribution of haplotypes. It is worth noting that two of the predominant haplotypes, H1 and H5, are completely absent in all five Taiwanese populations.
The extent of any contribution from Melanesian populations in the peopling of Polynesia and Micronesia has been controversial (7, 17). Our finding of H17 being almost exclusively restricted to Melanesian populations, with an appearance elsewhere in only one Micronesian group (12% in Trukese), is highly noteworthy. The absence of H17 from Polynesian populations suggests that the contribution of Melanesian Y-chromosomal haplotypes to the Polynesian expansion is very low or negligible, in contrast to the higher proportion of Melanesian alleles seen at nuclear and mtDNA loci (3, 18). The reason for this discrepancy is not clear, but the forces of drift would have different effects on different loci, especially during the population bottlenecks involved in the settlement of Polynesia (19). Further, sex-dependent migration could have played an important role in the process of Pacific colonization (17) as also has been implicated in geographic dispersal of human populations elsewhere (20, 21).
In the context of recent Polynesian history, the Y-chromosome study by Hurles et al. (11) should be noted. They reported extensive European admixture in Polynesia. Our data, however, do not show any significant European contribution. The European-specific haplotype H14 (14) was observed only in two individuals, one Micronesian and one Polynesian. The difference in these two studies could be attributed to differences in the studied populations. The Polynesian samples in Hurles et al.'s study were collected from Rarotonga in the Cook Islands. It also should be noted that European admixture is a relatively more recent historical event and possibly has not reached all of the populations of the region, which do not mask the genetic trail of the original prehistoric migration of the Polynesians.
mtDNA data suggested a spread of humans from Taiwan to Polynesia by way of a corridor through the Philippines and Indonesia. A pattern involving nucleotide substitutions in the control region of the mtDNA genome, dubbed the Polynesian motif (5, 6), was found in high frequencies in this corridor with the highest in Polynesia. Related types of this motif also were found in appreciable frequencies in this area of distribution and the Taiwanese populations showed the highest diversity. Based on these observations, the origin of the Polynesian motif was traced to Taiwan (6), which seemed to provide strong genetic support to the express train hypothesis. A recent reanalysis (8) of the published mtDNA data (3, 5), however, questioned the validity of this proposition. Richards et al. (8) argued that, based on assessment of divergence times for the motif and age estimates of the relevant populations, mtDNA data do not support a Taiwanese origin of the Polynesians. Rather, the evidence is more consistent with an island Southeast Asian ancestry, the homeland being in eastern Indonesia. Although our findings are more in line with this general position, the Y-chromosome data do not unequivocally point to “a center of origin” of the Polynesian people, and island Southeast Asia emerges more likely as a midway station en route to Polynesia. Further in the context of the Polynesian motif, we found that one of the derivatives, a CAT substitution, is distributed all over Southeast Asia including southern China (data not shown). With the Polynesian motif not being discretely associated with the Taiwanese and island Southeast Asians alone on one hand, and the distribution pattern of the Y-chromosome haplotypes presented here on the other, the greater Southeast Asian enclave assumes the ancestral position in the cascade of Pacific colonization.
Acknowledgments
We thank Dr. S. H. Chen for providing some of the DNA samples. This research and collection of samples was supported by the Chinese National Natural Science Foundation and the Li Foundation (to L.J.), grants from the National Science Foundation (to R.D.), the National Institutes of Health (to R.D., L.J., R.C., S.T.M., and P.U.), and the Welcome Trust (J.M.).
Footnotes
¶¶
Oefner, P. J. & Underhill, P. A. (1995) Am. J. Hum. Genet. 57, A266 (abstr.).
References
- 1.Diamond J M. Nature (London) 1988;336:307–308. [Google Scholar]
- 2.Bellwood P. Man's Conquest of the Pacific: The Prehistory of Southeast Asia and Oceania. New York: Oxford Univ. Press; 1978. [Google Scholar]
- 3.Sykes B C, Leiboff A, Low-Beer J, Tetzner S, Richards M. Am J Hum Genet. 1995;57:1463–1475. [PMC free article] [PubMed] [Google Scholar]
- 4.Melton T, Peterson R, Redd A J, Saha N, Sofro A S M, Martinson J, Stoneking M. Am J Hum Genet. 1995;57:403–414. [PMC free article] [PubMed] [Google Scholar]
- 5.Redd A J, Takezaki N, Sherry S T, McGarvey S T, Sofro A S M, Stoneking M. Mol Biol Evol. 1995;12:604–615. doi: 10.1093/oxfordjournals.molbev.a040240. [DOI] [PubMed] [Google Scholar]
- 6.Melton T, Clifford S, Martinson J, Batzer M, Stoneking M. Am J Hum Genet. 1998;63:1807–1823. doi: 10.1086/302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terrell J. Antiquity. 1988;62:642–657. [Google Scholar]
- 8.Richards M, Oppenheimer S, Sykes B. Am J Hum Genet. 1998;63:1234–1236. doi: 10.1086/302043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jobling M A, Tyler-Smith C. Trends Genet. 1995;11:449–455. doi: 10.1016/s0168-9525(00)89144-1. [DOI] [PubMed] [Google Scholar]
- 10.Hammer M F. Nature (London) 1995;378:376–378. doi: 10.1038/378376a0. [DOI] [PubMed] [Google Scholar]
- 11.Hurles M E, Irven C, Nicholson J, Taylor P G, Santos F R, Loughlin J, Jobling M A, Sykes B C. Am J Hum Genet. 1998;63:1793–1806. doi: 10.1086/302147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Underhill P A, Jin L, Zemans R, Oefner P A, Cavalli-Sforza L L. Proc Natl Acad Sci USA. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Underhill P A, Jin L, Linn A A, Mehdi S Q, Jenkins T, Vollarth D, Davis R W, Cavalli-Sforza L L, Oefner P J. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, et al. Am J Hum Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds J, Weir B S, Cockerham C C. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lum J K, Cann R L, Martinson J J, Jorde L B. Am J Hum Genet. 1998;63:613–624. doi: 10.1086/301949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts-Thomson J M, Martinson J J, Norwich J T, Harding R M, Clegg J B, Boettcher B. Am J Hum Genet. 1996;58:1017–1024. [PMC free article] [PubMed] [Google Scholar]
- 19.Flint J, Boyce A J, Martinson J J, Clegg J B. Hum Genet. 1989;83:257–263. doi: 10.1007/BF00285167. [DOI] [PubMed] [Google Scholar]
- 20.Seielstad M T, Minch E, Cavalli-Sforza L L. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Lezaun A, Calafell F, Comas D, Mateu E, Bosch E, Martínez-Arias R, Clarimón J, Fiori G, Luiselli D, Facchini F, et al. Am J Hum Genet. 1999;65:208–219. doi: 10.1086/302451. [DOI] [PMC free article] [PubMed] [Google Scholar]