CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA (original) (raw)
. Author manuscript; available in PMC: 2009 Jun 12.
Published in final edited form as: Science. 2008 Dec 19;322(5909):1843–1845. doi: 10.1126/science.1165771
Abstract
Horizontal gene transfer (HGT) in bacteria and archaea occurs through phage transduction, transformation, or conjugation, and the latter is particularly important for the spread of antibiotic resistance. Clustered, regularly interspaced short palindromic repeats (CRISPR) loci confer sequence-directed immunity against phages. A clinical isolate of Staphylococcus epidermidis harbors a CRISPR spacer that matches the nickase gene present in nearly all staphylococcal conjugative plasmids. Here we show that CRISPR interference prevents conjugation and plasmid transformation in S. epidermidis. Insertion of a self-splicing intron into nickase blocks interference despite the reconstitution of the target sequence in the spliced mRNA, indicating that the interference machinery targets DNA directly. We conclude that CRISPR loci counteract multiple routes of HGT and can limit the spread of antibiotic resistance in pathogenic bacteria.
CRISPR loci are present in ~40% of eubacterial genomes and nearly all archaeal genomes sequenced to date, and consist of short (~24–48 nucleotide) repeats separated by similarly sized, unique spacers (1, 2). They are generally flanked by a set of CRISPR-associated (cas) protein-coding genes (3–5). The CRISPR spacers and repeats are transcribed and processed into small CRISPR RNAs (crRNAs) (4, 6–8) that specify acquired immunity against bacteriophage infection by a mechanism that relies on the strict identity between CRISPR spacers and phage targets (3, 4).
The rise of hospital- and community-acquired methicillin- and vancomycin-resistant Staphylococcus aureus (MRSA and VRSA, respectively) is directly linked to the horizontal transfer of antibiotic resistance genes by plasmid conjugation (9, 10). S. aureus and S. epidermidis strains are the most common causes of nosocomial infections (11–13), and conjugative plasmids can spread from one species to the other. While the S. epidermidis strain ATCC 12228 (14) lacks CRISPR sequences, the clinically isolated strain RP62a (15) contains a CRISPR locus (Fig. 1A and Fig. S1A) that includes a spacer (spc1) that is homologous to a region of the nickase (nes) gene found in all sequenced staphylococcal conjugative plasmids (Fig. S1B), including those from MRSA and VRSA strains (9, 16, 17).
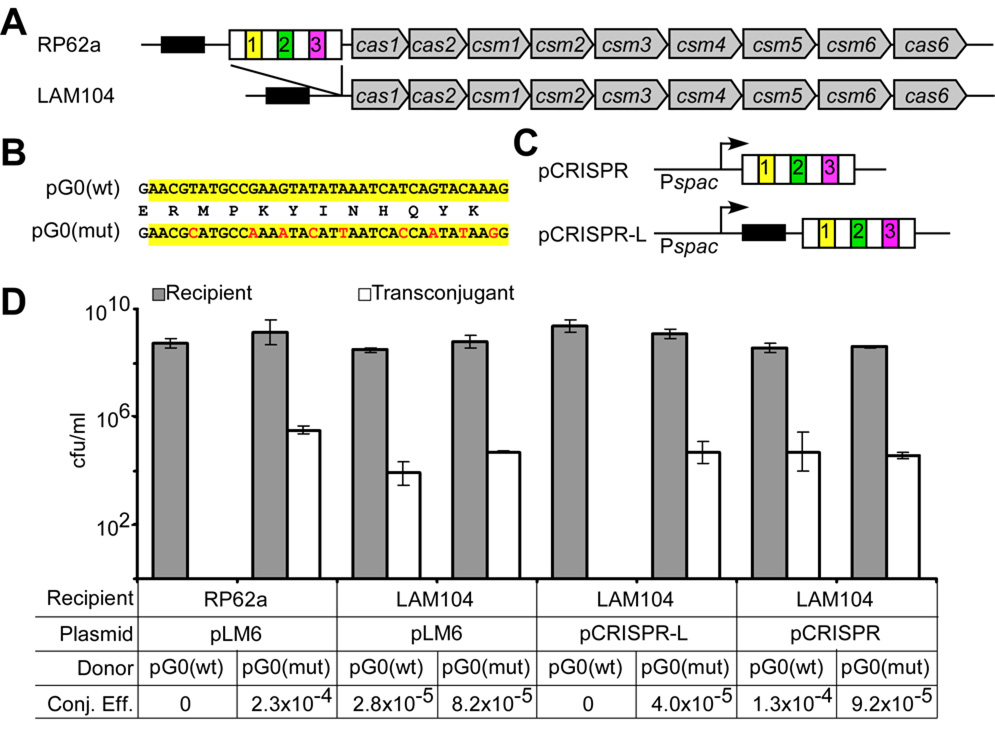
Figure 1.
A CRISPR locus provides immunity against plasmid conjugation in S. epidermidis. (A) Organization of the RP62a CRISPR locus. Repeats and spacers (colored boxes) are followed by CRISPR associated genes (cas1, cas2, cas6) and cas subtype M. tuberculosis genes (csm1 to csm6) (5). An AT-rich “leader” sequence precedes the repeat-spacer region (black box). LAM104 is an isogenic Δ_crispr_ strain lacking only the repeat and spacer sequences. (B) The staphylococcal conjugative plasmid pG0400 spc1 target sequence [pG0(wt), highlighted in yellow] is shown on the top. This sequence was altered to introduce synonymous mutations to generate pG0(mut), with changes shown in red. (C) To restore interference in strain LAM104, two plasmids were introduced: pCRISPR and pCRISPR-L. (D) Conjugation was carried out by filter mating in triplicate; the cfu/ml values (mean +/− SD) obtained for recipients and transconjugants are shown. Recipient strains, complementing plasmids, and donor conjugative plasmids are indicated. Conjugation efficiency (Conj. Eff.) was calculated as the recipients/transconjugants ratio.
To test whether spc1 prevents plasmid conjugation into S. epidermidis RP62a, we disrupted the sequence match by introducing nine silent mutations into the nes target in the conjugative plasmid pG0400 (18), generating pG0(mut) (Fig. 1B) (19). Both wild-type and mutant pG0400 were tested for transfer from S. aureus strain RN4220 (20) into either of the two S. epidermidis strains (Fig. 1D and Fig. S1C). While the conjugation frequency of both plasmids was similar for the CRISPR-negative ATCC 12228 strain, only pG0(mut) was transferred into the CRISPR-positive RP62a strain, and with a frequency similar to that of wild-type pG0400 in the control ATCC 12228 strain. These results indicate that CRISPR interference can prevent plasmid conjugation in a manner that is specified by sequence identity between a spacer and a plasmid target sequence.
To test this conclusion more rigorously, and to determine whether the CRISPR sequences themselves are responsible for the observed interference, we deleted the four repeats and three spacers present in the RP62a locus to generate the isogenic Δ_crispr_ strain LAM104 (Fig. 1A), and tested its ability to act as recipient for the conjugative transfer of pG0400. Again, wild-type RP62a was refractory to pG0400 transfer, whereas the conjugation efficiency for the LAM104 strain was similar to that obtained for S. epidermidis ATCC 12228 (Fig. 1D and Fig. S1C). pG0(mut) transfer was similar in both strains. To restore interference in the Δ_crispr_ mutant, LAM104 was transformed with plasmid pCRISPR or pCRISPR-L (Fig. 1C). Both of these plasmids contain the RP62a CRISPR repeats and spacers downstream of an IPTG-inducible promoter, but they differ in the amount of upstream flanking sequence included. The appearance of the spc1 crRNA in pCRISPR-L-containing LAM104 cells (Fig. S2) was IPTG-independent, indicating that the insert includes a natural CRISPR promoter. pCRISPR-L restored interference in the Δ_crispr_ strain but pCRISPR did not, even in the presence of IPTG (Fig. S1D). This suggests a role for the leader sequence (Fig. 1A) or other upstream sequences in cis during CRISPR interference, perhaps for processing of the crRNA precursor by Cascade proteins (4). Introduction of pCRISPR-L into strain ATCC 12228 did not alter the conjugation efficiency of pG0400 (Fig. S1D), as expected due to the lack of cas genes in this strain.
The nature of the crRNA targeting event (RNA-RNA or RNA-DNA) is not known. The requirement for nickase activity only in the donor cell (16) implies that interference with nes mRNA and protein expression should block the ability of S. epidermidis RP62a to function as a donor. Instead, our data (Fig. 1 and Fig. S1) indicate that the CRISPR locus limits the ability of S. epidermidis RP62a to act as a plasmid recipient, and therefore suggest that spc1_–directed interference does not target the nes transcript. Consistent with this, the orientation of spc1 leads to the expression of a crRNA that is identical with, rather than complementary to, the nes mRNA target sequence, and we have found no evidence for the expression of RNA from the opposite strand (Fig. S2) (4). Alternatively, the spc1 crRNA may target the incoming DNA. To test this, we interrupted the target sequence of the pG0400 nes gene with the orf142-I2 self-splicing group I intron from the staphylococcal Twort phage (21, 22). The mutant conjugative plasmid, pG0(I2), lacks an intact spc1 target DNA sequence (Fig. 2A), but the spc1 target sequence is regenerated in the nes mRNA after transcription and rapid splicing. When tested in conjugation assays, pG0(I2) was transferred to wild-type and Δ_crispr strains with equal efficiencies (Fig. 2B). This observation reflects an evasion of CRISPR activity when the intron is present in the plasmid, and therefore indicates that an intact target site is required in the nes DNA, but not mRNA, for interference to occur. To confirm that splicing is required for nes function, we tested the conjugation of pG0(dI2), a derivative of pG0(I2) containing a three-nucleotide deletion within the intron that inactivates self-splicing (21) (Fig. S3A). The pG0(dI2) plasmid was unable to transfer to the RP62a strain (Fig. S3B), indicating a lack of nickase activity in the presence of the unspliced intron.
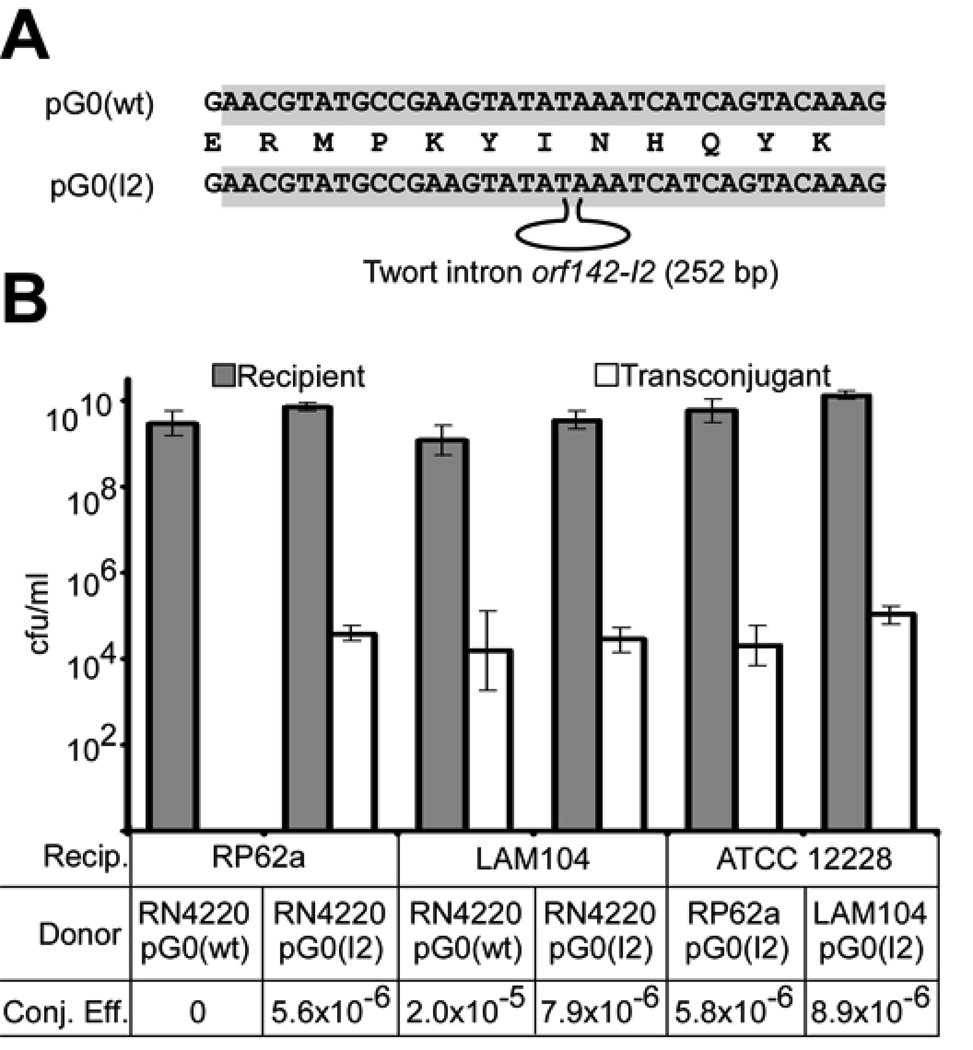
Figure 2.
CRISPR interference requires an intact target sequence in plasmid DNA but not mRNA. (A) Disruption of the pG0400 nes target sequence with the orf142-I2 self-splicing intron, generating the conjugative plasmid pG0(I2). (B) Conjugation efficiency was measured as in Fig. 1D, using RP62a and the Δ_crispr_ mutant LAM104 as recipients for pG0(wt) and pG0(I2). To test for interference with spliced nes mRNA, RP62a and LAM104 transconjugants were also used as donors of pG0(I2) to S. epidermidis ATCC 12228.
The requirement for nes transcription, splicing, and translation in the donor cell during conjugation (16), and our ability to obtain RP62a transconjugants with the intron-containing plasmid, allowed us to test the capacity of the CRISPR system to target intact, spliced nes mRNA by using RP62a as a pG0(I2) donor. pG0(I2) conjugative transfer was just as efficient from RP62a as from the isogenic Δ_crispr_ strain LAM104 (Fig. 2), indicating that spliced, functional nes mRNA (which must be present for conjugation to occur) is not targeted during CRISPR interference. RT-PCR assays confirmed the splicing of the nes pre-mRNA in RP62a cells carrying pG0(I2) (Fig. S3C). While our observations do not formally exclude an RNA targeting event that is somehow restricted to nascent, transient, unspliced transcripts, they provide strong evidence that DNA rather than mRNA is the likely crRNA target during CRISPR interference.
CRISPR activity against phage and conjugative plasmid DNA molecules suggests that CRISPR systems may also prevent plasmid DNA transformation. We therefore introduced pG0(wt) and pG0(mut) nes target and flanking sequences (200 bp) in either orientation into the staphylococcal plasmid pC194 (23), generating pNes(wt) and pNes(mut), respectively (d, direct insertion; i, inverted insertion; Fig. 3A). Flanking DNA was included in the inserts to ensure the presence of any sequences outside of the target that may contribute to CRISPR interference (24). Plasmids were transformed by electroporation into wild-type RP62a and isogenic Δ_crispr_ LAM104 strains. pC194 and both pNes(mut) plasmids were transformed into both strains, whereas the pNes(wt) plasmids were only transformed into the Δ_crispr_ mutant (Fig. 3B). We also performed pNes(wt)/pNes(mut) mixed transformations of RP62a or LAM104 strains to test interference in an internally controlled fashion. Again, only pNes(mut) plasmids were recovered from RP62a transformants, whereas pNes(wt) and pNes(mut) plasmids were found in LAM104 transformant colonies (Fig. S4). It remains to be established whether natural transformation, which involves the uptake of a single DNA strand (25), is subject to CRISPR interference. Nonetheless, our experiments suggest that CRISPR systems can counteract multiple routes of plasmid transfer.
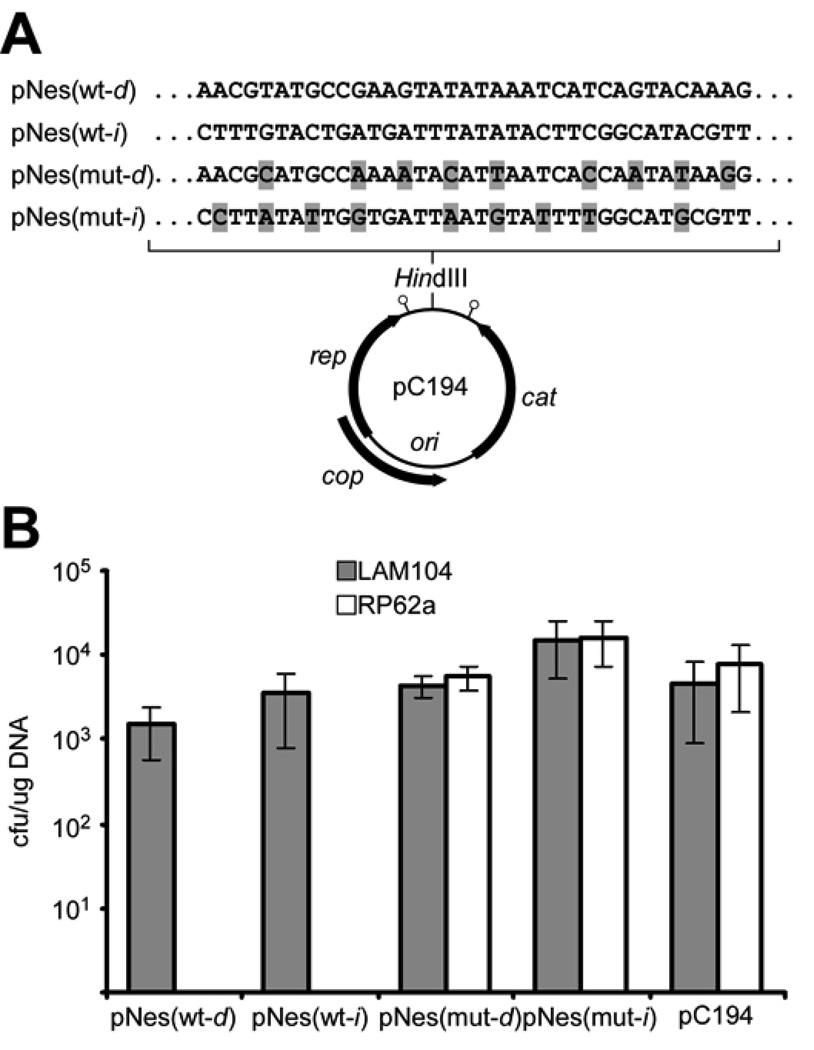
Figure 3.
Plasmid transformation is subject to CRISPR interference. (A) Introduction of the wild-type and mutant nes target sequences (mutations highlighted in grey) into the plasmid pC194 (d, direct; i, inverted). The origin of replication (ori) as well as protein-coding genes are indicated. Stem-loops denote the rep and cat transcriptional terminators. (B) RP62a and the Δ_crispr_ mutant LAM104 were transformed in triplicate with the plasmids described in (A). Transformation efficiency was calculated as cfu/µg DNA (mean +/− SD).
These transformation data provide additional evidence that crRNAs target DNA molecules. First, interference occurred regardless of the insert orientation in pNes(wt); this, combined with the lack of compelling evidence for CRISPR-derived double-stranded RNA (Fig. S2 and refs. (4, 6, 7)), is consistent with spc1 targeting either DNA strand rather than a unidirectional transcript. Second, the target sites in the pNes(wt) and pNes(mut) plasmids are located between the transcriptional terminators of the rep and cat genes (Fig. 3A) (23, 26, 27). This minimizes the likelihood that this region of the plasmid is even transcribed, consistent with its dispensability for plasmid maintenance (23, 28).
Altogether, these data provide strong functional evidence that CRISPR interference acts at the DNA level, and therefore differs fundamentally from the RNAi phenomenon observed in eukaryotes and to which CRISPR activity was originally compared (29). A DNA targeting mechanism for CRISPR interference implies a means to prevent its action at the encoding CRISPR locus itself, as well as other potential chromosomal loci such as prophage sequences. Little information exists to suggest how crRNAs would avoid targeting “self” DNA, though the role of flanking sequences during CRISPR interference (24) could contribute to target specificity. From a practical standpoint, the ability to direct the specific, addressable destruction of DNA that contains any given 24–48 nucleotide target sequence could have considerable functional utility, especially if the system can function outside of its native bacterial or archaeal context. Furthermore, our results demonstrate that CRISPR function is not limited to phage defense, but instead encompasses a more general role in the prevention of HGT and the maintenance of genetic identity, as with restriction-modification systems. A primary difference between restriction-modification and CRISPR interference is that the latter can be programmed by a suitable effector crRNA. If CRISPR interference could be manipulated in a clinical setting, it would provide a means to impede the ever-worsening spread of antibiotic resistance genes and virulence factors in staphylococci and other bacterial pathogens.
Supplementary Material
01
References and Notes
- 1.Grissa I, Vergnaud G, Pourcel C. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek R, Kunin V, Hugenholtz P. Nat. Rev. Microbiol. 2008;6:181. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, et al. Science. 2007;315:1709. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Brouns SJ, et al. Science. 2008;321:960. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haft DH, Selengut J, Mongodin EF, Nelson KE. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang TH, et al. Proc. Natl. Acad. Sci. USA. 2002;99:7536. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang TH, et al. Mol Microbiol. 2005;55:469. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 8.Hale C, Kleppe K, Terns RM, Terns MP. RNA. 2008 Oct 29; doi: 10.1261/rna.1246808. epub ahead of print (10.1261/rna.1246808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weigel LM, et al. Science. 2003;302:1569. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 10.Furuya EY, Lowy FD. Nat. Rev. Microbiol. 2006;4:36. doi: 10.1038/nrmicro1325. [DOI] [PubMed] [Google Scholar]
- 11.Lim SM, Webb SA. Anaesthesia. 2005;60:887. doi: 10.1111/j.1365-2044.2005.04220.x. [DOI] [PubMed] [Google Scholar]
- 12.Lowy FD. N. Engl. J. Med. 1998;339:520. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 13.von Eiff C, Peters G, Heilmann C. Lancet Infect. Dis. 2002;2:677. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YQ, et al. Mol. Microbiol. 2003;49:1577. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 15.Gill SR, et al. J. Bacteriol. 2005;187:2426. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Climo MW, Sharma VK, Archer GL. J. Bacteriol. 1996;178:4975. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diep BA, et al. Lancet. 2006;367:731. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 18.Morton TM, Johnston JL, Patterson J, Archer GL. Antimicrob. Agents. Chemother. 1995;39:1272. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material on Science Online.
- 20.Kreiswirth BN, et al. Nature. 1983;305:709. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 21.Golden BL, Kim H, Chase E. Nat. Struct. Mol. Biol. 2005;12:82. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 22.Landthaler M, Shub DA. Proc. Natl. Acad. Sci. USA. 1999;96:7005. doi: 10.1073/pnas.96.12.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horinouchi S, Weisblum B. J. Bacteriol. 1982;150:815. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deveau H, et al. J. Bacteriol. 2008;190:1390. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen I, Dubnau D. Nat. Rev. Microbiol. 2004;2:241. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 26.Byeon WH, Weisblum B. J. Bacteriol. 1984;158:543. doi: 10.1128/jb.158.2.543-550.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gros MF, te Riele H, Ehrlich SD. EMBO J. 1989;8:2711. doi: 10.1002/j.1460-2075.1989.tb08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gros MF, te Riele H, Ehrlich SD. EMBO J. 1987;6:3863. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.We are indebted to Olaf Schneewind (University of Chicago) for reagents and experimental assistance, Barbara Golden (Purdue University) for orf142-2 intron constructs and advice, and members of our laboratory for discussions and comments on the manuscript. L.A.M. is a Fellow of The Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by a grant (GM072830) from the National Institutes of Health to E.J.S.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01