Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice (original) (raw)
Abstract
Influenza-related complications continue to be a major cause of mortality worldwide. Due to unclear mechanisms, a substantial number of influenza-related deaths result from bacterial superinfections, particularly secondary pneumococcal pneumonia. Here, we report what we believe to be a novel mechanism by which influenza-induced type I IFNs sensitize hosts to secondary bacterial infections. Influenza-infected mice deficient for type I IFN-α/β receptor signaling (Ifnar–/– mice) had improved survival and clearance of secondary Streptococcus pneumoniae infection from the lungs and blood, as compared with similarly infected wild-type animals. The less effective response in wild-type mice seemed to be attributable to impaired production of neutrophil chemoattractants KC (also known as Cxcl1) and Mip2 (also known as Cxcl2) following secondary challenge with S. pneumoniae. This resulted in inadequate neutrophil responses during the early phase of host defense against secondary bacterial infection. Indeed, influenza-infected wild-type mice cleared secondary pneumococcal pneumonia after pulmonary administration of exogenous KC and Mip2, whereas neutralization of Cxcr2, the common receptor for KC and Mip2, reversed the protective phenotype observed in Ifnar–/– mice. These data may underscore the importance of the type I IFN inhibitory pathway on CXC chemokine production. Collectively, these findings highlight what we believe to be a novel mechanism by which the antiviral response to influenza sensitizes hosts to secondary bacterial pneumonia.
Introduction
Influenza pneumonia is the leading cause of death from an infectious cause and the 8th overall cause of death annually in the United States (1). While influenza infection can be lethal in and of itself, a substantial number of postinfluenza deaths are due to secondary bacterial pneumonias, most commonly caused by Streptococcus pneumoniae, Staphylococcus aureus, and Klebsiella pneumoniae (2–4). However, the mechanisms by which influenza sensitizes patients to secondary bacterial infections are poorly understood. Given the imminent threat of an influenza pandemic and the increasing rates of antibiotic resistance, the identification of immune targets to prevent postinfluenza bacterial pneumonias has significant clinical ramifications.
Intact innate immune responses, including those mediated by resident alveolar macrophages and recruited neutrophils, are essential to the clearance of bacterial pathogens from the lung (5–7). Earlier studies have reported impairment in macrophage and neutrophil responses following influenza infection (8–18), but the molecular pathways underlying these defects have not been fully elucidated. Although various factors, including upregulation of platelet-activating factor receptor and the antiinflammatory cytokine IL-10 during influenza infection, have been implicated in promoting postinfluenza secondary pneumococcal pneumonia, attempts at modifying these factors have had limited effects on bacterial clearance (19–21).
Type I IFNs, which are central to antiviral defenses, are a large family of antiviral cytokines that include multiple IFN-α proteins and a single IFN-β protein. Type I IFNs signal through a common receptor, IFN-α/β receptor (IFNAR), resulting in the expression of proinflammatory genes that not only inhibit viral replication, but also augment various aspects of adaptive immunity (22–25). While the importance of type I IFNs to antiviral defenses is well established, their role in bacterial defenses is more ambiguous. We therefore established a model of sequential influenza and pneumococcus lung infection in our laboratory using genetically modified animals with defective IFNAR signaling (_Ifnar_–/– mice) to investigate the effects of type I IFNs induced during influenza infection on pulmonary host defense against bacteria. Interestingly, we found that induction of type I IFNs in the lung during influenza pneumonia had marked deleterious effects on the clearance and survival following secondary pneumococcal challenge. This appeared to be attributable at least in part to type I IFN-mediated attenuation of KC (also known as Cxcl1) and Mip2 (also known as Cxcl2) production, leading to impaired neutrophil responses. Thus, we found that antiviral host responses counteract the host’s ability to clear subsequent bacterial challenge through specific regulation of particular inflammatory genes.
Results
Influenza enhances sensitivity of mice to secondary infections with S. pneumoniae.
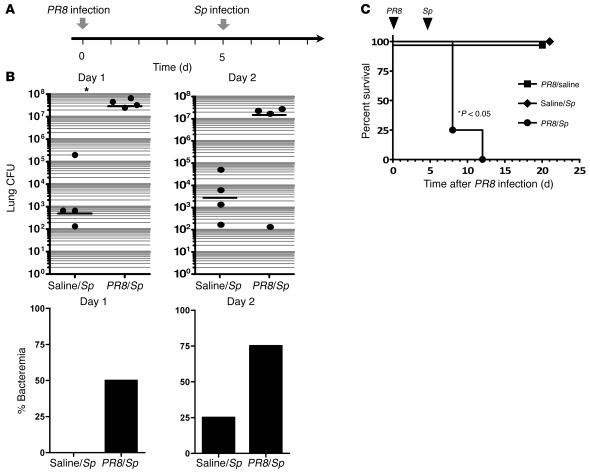
To characterize our murine model of influenza pneumonia, C57BL/6 mice were infected intratracheally (i.t.) with the PR8 strain of influenza virus at various doses and their survival examined. We found that i.t. administration of 200 infectious units of the PR8 strain of influenza virus reproducibly resulted in sublethal pneumonia. Prior work in the field has indicated that secondary infection with S. pneumoniae is most lethal between 5 and 7 days following the initial influenza infection (15, 20). Furthermore, most secondary bacterial infections occur within the first 2 weeks of the primary influenza infection (26). Therefore, we set up a combinatorial infection model in which, 5 days after the initial influenza infection, animals were administered S. pneumoniae i.t. (Figure 1A). Our preliminary studies demonstrated that in naive animals, a dose of 2,000 CFU was sublethal (<20% mortality) but still sufficient to lead to a mild inflammatory influx that was representative of that observed in milder cases of pneumococcal pneumonia in patients (data not shown). In animals with prior influenza infection, however, following a bacterial challenge of 2,000 CFU of S. pneumoniae, a 4-log increase in pulmonary bacterial burden was detected as early as 24 hours after secondary infection with S. pneumoniae, with striking differences persisting for up to 48 hours (Figure 1B). Similarly, markedly higher rates of bacteremia were noted in double-infected animals (rates of bacteremia in _PR8_- versus saline-pretreated animals were 50% vs. 0% at 24 hours and 75% vs. 25% at 48 hours) (Figure 1B). Furthermore, significant mortality was observed in _PR8/S. pneumoniae_–infected animals and not in saline/_S. pneumoniae_– or PR8/saline-treated animals (Figure 1C). Enhanced mortality was observed in animals with prior influenza infections when subsequently challenged with pneumococcal inocula as low as 100 CFU (data not shown).
Figure 1. Primary influenza pneumonia enhances sensitivity to secondary pneumococcal infection.
(A) Experimental setup for combinatorial infection model of wild-type C57BL/6 animals. Mice were administered i.t. influenza (PR8 strain, 200 PFU) or saline, followed 5 days later by i.t. S. pneumoniae (Sp) or saline. (B) Pulmonary bacterial burden and incidence of bacteremia were measured in saline/_S. pneumoniae_– and PR8/_S. pneumoniae_–infected animals on days 1 and 2 after secondary bacterial challenge (2,000 CFU). (C) Survival was examined for 3 weeks after the initial i.t. challenge for the following 3 groups: PR8/saline, saline/S. pneumoniae, and PR8/S. pneumoniae. n = 4–8 animals per group. Data are representative of 2 independent experiments.
Influenza-infected Ifnar–/– mice are resistant to secondary bacterial pneumonia.
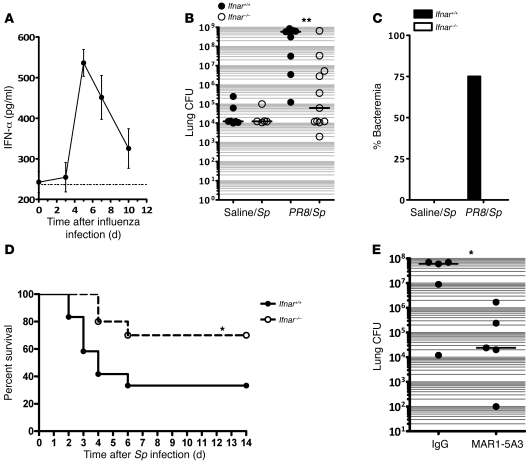
To better understand the mechanisms by which influenza sensitized mice to secondary pneumococcal pneumonia, we analyzed the kinetics of the immune response to viral infection in vivo. We first examined induction of type I IFN in the lung and found that levels of IFN-α peaked on day 5 after PR8 infection (Figure 2A), with elevated levels persisting to day 10, which correlated in prior studies with the timing of maximum susceptibility to secondary S. pneumoniae infection (i.e., 5–7 days after influenza infection) (15, 20). Although type I IFNs are considered important activators of innate and adaptive immune responses in response to infection, particularly during viral infection, we wished to determine whether the induction of type I IFNs in the lungs of influenza-infected animals paradoxically increased sensitivity to secondary bacterial pneumonia. Therefore, animals with a targeted deletion of the common type I IFN receptor (_Ifnar_–/–) and age- and sex-matched Ifnar+/+ animals were infected with PR8 and then challenged on day 5 with S. pneumoniae. We also infected Ifnar+/+ and Ifnar–/– animals with either PR8 or S. pneumoniae alone. Similar to prior reports, we found _Ifnar_–/– mice did not have significant differences in lung viral burdens at day 5 or day 7 or in other parameters of illness such as weight loss following influenza infection, when compared with their Ifnar+/+ counterparts after PR8 infection (27) (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI35412DS1), demonstrating redundancy in type I IFN–mediated responses in terms of viral clearance. Furthermore, no appreciable differences were observed in lung bacterial burdens of Ifnar+/+ and _Ifnar_–/– animals at 48 hours following S. pneumoniae infection alone (Figure 2B). Following sequential infection of PR8 and S. pneumoniae, however, _Ifnar_–/– mice appeared less ill (e.g., less lethargy and ruffled fur) compared with their wild-type counterparts. At 48 hours following i.t. S. pneumoniae infection, lungs from _PR8/S. pneumoniae_–infected _Ifnar_–/– mice contained nearly 4-log–fold fewer CFUs of S. pneumoniae as compared with Ifnar+/+ animals (Figure 2B; P < 0.01). At this time point, we were unable to detect positive blood cultures for S. pneumoniae in _PR8/S. pneumoniae_–infected _Ifnar_–/– mice, whereas 75% of Ifnar+/+ animals were bacteremic (Figure 2C; P < 0.05). These differences in pulmonary and systemic bacterial loads persisted to day 4 (Supplemental Figure 2) and were associated with increased mortality in Ifnar+/+ animals (Figure 2D). The animals that survived to day 7 started to demonstrate clearance of bacteria from lungs and blood, although there was still a slight trend toward prolonged bacteremia in the Ifnar+/+ group (Supplemental Figure 2). Thus, the presence of type I IFN signaling markedly increases susceptibility to secondary bacterial infections of the lung in influenza-infected hosts.
Figure 2. Influenza-infected _Ifnar–/_– mice are resistant to secondary pneumococcal pneumonia.
(A) Kinetics of IFN-α induction after influenza following i.t. PR8. Ifnar+/+ C57BL/6 mice were administered i.t. PR8 on day 0, and lungs were harvested at the designated time points for assessment of IFN-α levels by ELISA in lung homogenates. (B and C) Clearance of S. pneumoniae by influenza-infected Ifnar+/+ and Ifnar–/– animals. Age- and sex-matched animals of both genotypes were administered i.t. PR8 or saline, followed 5 days later with i.t. S. pneumoniae (2,000 CFU). Lungs (B) and blood (C) were harvested on day 2 following i.t. S. pneumoniae infection, for assessment of CFU. n = 8–11 animals per group (B) and n = 4 animals per group (C). **P < 0.01. Data are representative of 3 independent experiments. (D) Following i.t. PR8 and S. pneumoniae administration 5 days apart, survival was assessed for 14 days following S. pneumoniae infection. Survival rates were 70% for _Ifnar_–/– mice compared with 33% for Ifnar+/+ mice at 14 days. *P = 0.05, log-rank test. n = 10–12 mice per group. Data were combined from 2 separate experiments. (E) Lung CFU in animals following IFNAR neutralization of wild-type animals with MAR1-5A3. P < 0.05, 1-tailed Mann-Whitney U test. The horizontal lines represent the statistical medians.
In order to rule out developmental abnormalities in _Ifnar_–/– mice as a confounding factor, wild-type animals were treated with an IFNAR-neutralizing antibody (MAR1-5A3) (28) or mouse IgG and then infected as described above. Following i.t. PR8 infection, MAR1-5A3–treated mice had markedly enhanced pulmonary clearance of secondary pneumococcal challenge as compared with IgG-treated animals (Figure 2E) at 48 hours, which is consistent with what was observed in _Ifnar_–/– animals.
Because we have recently demonstrated that type I IFNs mediate the upregulation of IL-10 following LPS stimulation in macrophages (29), we wished to examine whether IL-10 was responsible for the detrimental effects of type I IFNs observed in our model. Our preliminary studies found that influenza infection led to upregulation of IL-10 production, peaking at day 7 following PR8 infection (Supplemental Figure 3A). Taken together with previously published reports showing that IL-10 can play a detrimental role for clearance of S. pneumoniae (19, 30), this led us to examine whether Il10–/– animals were able to clear bacteria in the post-influenza setting as well as Ifnar–/– animals did. We were unable to discern significant differences between Il10+/+ and Il10–/– animals in pneumococcal lung burden following i.t. administration of this infectious dose (2,000 CFU), whether they were naive (i.e., saline pretreated at 5 days prior to i.t. S. pneumoniae) or previously influenza-infected (i.e., administered i.t. PR8 5 days prior to i.t. S. pneumoniae; Supplemental Figure 3B). Therefore, these data suggest that type I IFN–mediated upregulation of IL-10 is unlikely to be responsible for the effects of type I IFNs on antibacterial immunity in our model.
Influenza-infected Ifnar–/– animals have higher levels of KC and Mip2 as compared with Ifnar+/+ animals at early time points following secondary pneumococcal infection.
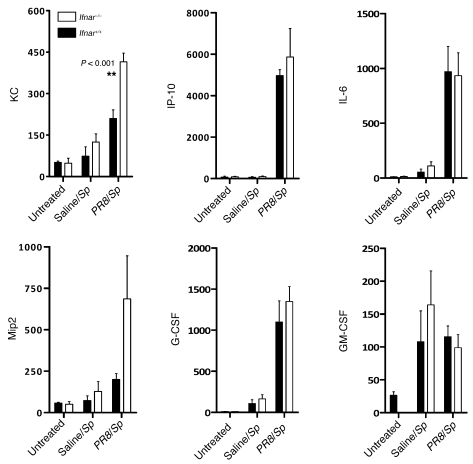
In light of our findings that type I IFNs sensitized mice to secondary bacterial pneumonia, we next examined whether we could find any differences in cytokine levels in the lungs of _PR8_-infected Ifnar+/+ and Ifnar–/– animals in response to challenge with S. pneumoniae. Since marked differences in lung bacterial burdens were noted 48 hours after pneumococcal infection, which might impact cytokine levels, we focused on differences in inflammatory cytokine responses at earlier time points, when significant differences between Ifnar+/+ and Ifnar–/– animals in bacterial burden were less likely to be present (i.e., at 16 hours). Lung homogenates obtained from saline-pretreated or influenza-infected Ifnar+/+ and Ifnar–/– animals were analyzed for cytokine and chemokine expression at 16 hours following secondary pneumococcal infection. Consistent with earlier reports, higher levels of inflammatory cytokines were generally observed in animals with both influenza and pneumococcal infections, as compared with animals with only pneumococcal infection (31), which is likely due to the markedly higher lung bacterial burden in PR8/_S. pneumoniae_–infected animals compared with saline/_S. pneumoniae_–infected animals. Following S. pneumoniae infection alone, we did not detect significant differences in inflammatory cytokine production between Ifnar+/+ and Ifnar–/– animals. In influenza-infected animals, however, we found that Ifnar–/– animals were able to mount a much more robust KC and Mip2 chemokine response to secondary S. pneumoniae challenge, as compared with their Ifnar+/+ counterparts. Interestingly, we could not find significant differences in the expression of other inflammatory cytokines including GM-CSF, G-CSF, IL-6, and IP10, suggesting that type I IFNs appeared to lead to selective attenuation of KC and Mip2 (Figure 3). This was of particular importance, as these chemokines have been previously shown to be essential to neutrophil recruitment and the clearance of bacterial pathogens from the lung, including pneumococcus (32). Therefore, in influenza-infected hosts, the presence of type I IFNs appear to attenuate KC and Mip2 responses during the early response to secondary bacterial infection in influenza-infected hosts.
Figure 3. Lung homogenates from _PR8/S. pneumoniae_–infected Ifnar–/– mice contain higher levels of KC and Mip2.
Production of KC and Mip2 in response to secondary pneumococcal infection, as determined by ELISA analysis of Ifnar+/+ and Ifnar–/– mice with prior influenza infection. Levels of other inflammatory cytokines were determined by multiplex cytokine protein analysis. Lung homogenates were obtained and analyzed 16 hours after secondary challenge with S. pneumoniae. n ≥ 4 mice per group.
Type I IFNs inhibit production of Kc and Mip2.
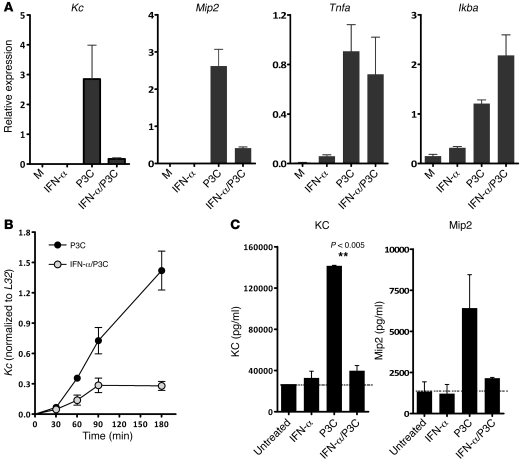
To our knowledge, regulation of KC and Mip2 expression by type I IFNs has not been previously described. Given the enhanced expression of these chemokines in _PR8/S. pneumoniae_–infected animals, we wished to determine whether type I IFNs could directly inhibit expression of KC and Mip2. Bone marrow–derived macrophages (BMMs) from wild-type animals were pretreated with IFN-α for 30 minutes and subsequently activated with Pam3Cys (P3C), a bacterial ligand for TLR2, which recognizes gram-positive bacteria. Interestingly, while IFN-α pretreatment had little effect on expression of most inflammatory molecules such as Tnfa, Il1b, and Ikba, we observed potent inhibition of P3C-induced Kc and Mip2 transcripts (Figure 4A). Reduced induction of the Kc transcript was noted at multiple time points (Figure 4B). Total KC and Mip2 protein production was also reduced as measured by ELISA analysis of cell supernatants 24 hours after stimulation with P3C (Figure 4C). Hence, type I IFNs do not appear to globally suppress the upregulation of inflammatory genes in response to bacterial ligands, but selectively impair the expression of Kc and Mip2 in response to TLR signaling in macrophages.
Figure 4. Type I IFNs inhibit production of Kc and Mip2 but not Tnfa or Ikba transcript in P3C-stimulated BMMs.
(A) Pretreatment of BMMs with IFN-α for 30 minutes, followed by stimulation with the TLR2 ligand P3C. Kc, Mip2, Tnfa, and Ikba expression was assessed quantitative PCR 4 hours after P3C stimulation. Data are representative of 4 independently performed experiments. M, media. (B) BMMs were exposed to recombinant IFN-α for 30 minutes, followed by P3C stimulation. Levels of KC expression were examined by quantitative PCR at various time points following P3C stimulation. All values were normalized to L32 expression levels. (C) Production of KC and Mip2 protein levels in cell culture supernatants were assessed by ELISA. BMMs were exposed to IFN-α, P3C, or IFN-α and P3C for 24 hours prior to collection of supernatants for analysis.
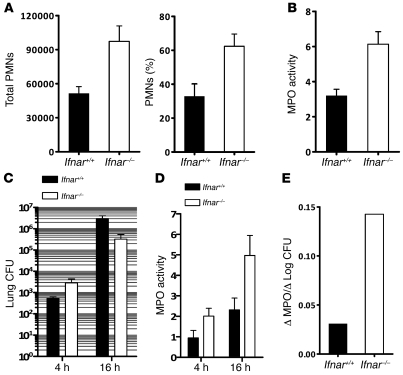
Influenza-infected Ifnar–/– mice had higher recruitment of PMNs into pulmonary tissues in response to S. pneumoniae.
We also sought to determine whether the impairment in KC and Mip2 expression was associated with functional differences in cellular inflammation between Ifnar+/+ and Ifnar–/– animals. To assess the cellular composition of airway cells, we performed bronchoalveolar lavage (BAL) in Ifnar+/+ and Ifnar–/– animals prior to infection and at 14 hours after i.t. administration of S. pneumoniae into lungs of _PR8_-infected animals. At baseline, untreated Ifnar+/+ and Ifnar–/– animals had similar total cell numbers. The cells were more than 95% macrophage in morphology (Supplemental Figure 4). Interestingly, BAL from PR8/_S. pneumoniae_–infected Ifnar–/– mice contained significantly higher numbers and percentages of PMNs compared with Ifnar+/+ mice (P < 0.05; Figure 5A and Supplemental Figure 4).
Figure 5. Ifnar–/– mice have higher numbers of airspace PMNs following i.t. PR8 and S. pneumoniae.
(A) Total number and percentage of infiltrating PMNs in doubly infected Ifnar+/+ and Ifnar–/– animals were analyzed in BAL 14 hours after secondary pneumococcal challenge. (B) MPO activity was assessed in lung homogenates of doubly infected _Ifnar_–/– and Ifnar+/+ mice 24 hours after secondary challenge with S. pneumoniae (P < 0.05). n = 3. Data are representative of at least 2 independently performed experiments. (C) Lung homogenate CFU of doubly infected Ifnar+/+ and _Ifnar_–/– animals at 4 and 16 hours following S. pneumoniae challenge. (D) MPO activity of lung homogenates from doubly infected Ifnar+/+ and _Ifnar_–/– animals were determined at 4 and 16 hours after secondary S. pneumoniae challenge. (E) Average change in MPO activity (16 hours – 4 hours) divided by the average change in Log CFU (16 hours – 4 hours).
In addition to BAL, myeloperoxidase (MPO) activity was also measured in lung homogenates of influenza-infected mice as another measure of pulmonary PMN activity following secondary pneumococcal infection. By 24 hours after infection in lung homogenates of doubly infected Ifnar–/– animals, MPO activity was significantly higher, compared with Ifnar+/+ animals (P < 0.01; Figure 5B). Since bacterial differences were more apparent at this time point, we examined MPO activity at earlier time points. Four hours after infection, before differences in bacterial burden became apparent (Figure 5C), _Ifnar_–/– mice had slightly higher levels of MPO activity (Figure 5D). By 16 hours following infection, _Ifnar_–/– mice displayed appreciably higher MPO levels as compared with their wild-type counterparts (Figure 5, C and D). In fact, for unit log-fold changes in CFU between 4 and 16 hours, Ifnar–/– mice upregulated MPO activity approximately 5 times better than similarly infected wild-type animals (Figure 5E).
H&E staining of lungs from doubly infected animals revealed that _Ifnar_–/– mice possessed a more robust inflammatory cellular infiltrate, compared with their Ifnar+/+ counterparts (Supplemental Figures 5 and 7). In contrast, a more modest inflammatory response was appreciated in mice that had received either PR8 or S. pneumoniae alone, with no differences observed between Ifnar+/+ and _Ifnar_–/– animals (data not shown). Staining of lung tissue sections for MPO revealed a higher number of MPO-positive cells in infected regions of Ifnar–/– mice at this time point (Supplemental Figure 7). Collectively, these studies demonstrate that _Ifnar_–/– animals with prior influenza infection are able to mount a more effective neutrophil response against rising bacterial titers following secondary pneumococcal challenge.
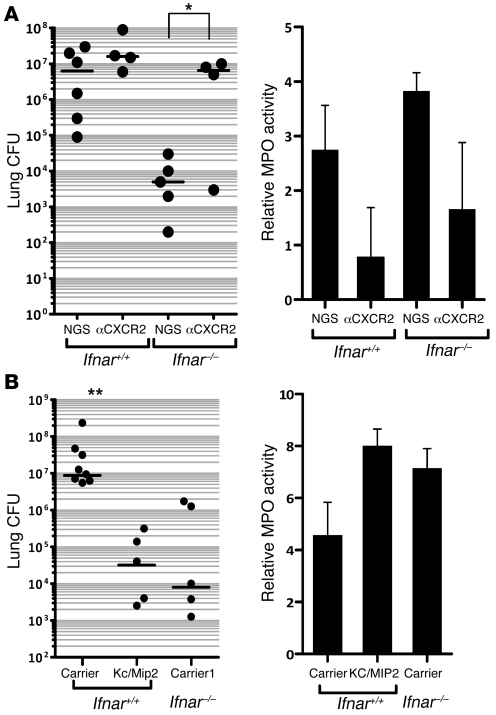
KC and Mip2 play a critical role for pulmonary clearance of S. pneumoniae in influenza-infected animals.
Having observed inhibitory effects of type I IFNs on production of KC and Mip2, along with decreased levels of these cytokines in doubly infected wild-type animals, it still remained unclear whether type I IFN–mediated suppression of KC and Mip2 was the major mechanism underlying the enhanced susceptibility of wild-type mice to secondary pneumococcal pneumonia. We predicted that if KC and Mip2 were significant mediators of enhanced clearance of secondary pneumococcal pneumonia observed in Ifnar–/– animals, neutralization of their receptor would dramatically increase sensitivity of influenza-infected _Ifnar_–/– mice. Previous reports have shown that injury-mediated recruitment of PMNs into the lung requires Cxcr2, the main receptor for both cytokines (33). Therefore, we used Cxcr2-blocking antibodies to measure the importance of these cytokines for bacterial clearance during secondary pneumococcal infections. To evaluate the effectiveness of the depletion, pulmonary MPO activity was measured and found to be decreased in animals treated with α-Cxcr2 antibodies (Figure 6A). Cxcr2 depletion resulted in a significant increase (over 3,000-fold; P < 0.05) in bacterial burden in the lungs of _Ifnar_–/– mice 24 hours following secondary S. pneumoniae challenge (Figure 6A). The bacterial counts in Cxcr2-depleted _Ifnar_–/– mice were strikingly close to those of _PR8/S. pneumoniae_–infected Ifnar+/+ mice treated with normal goat serum (NGS). Interestingly, inhibition of Cxcr2 did not significantly change pulmonary bacterial burden in Ifnar+/+ animals (~10-fold, P = 0.23; Figure 6A) suggesting that KC and Mip2 are expressed at functionally inadequate levels in wild-type animals.
Figure 6. KC and Mip2 play a critical role in the clearance of secondary bacterial infection.
(A) Ifnar+/+ and _Ifnar_–/– animals were administered i.t. PR8, followed 5 days later by i.t. S. pneumoniae. Anti-Cxcr2 antibody or normal goat serum were administered i.p. 24 hours prior to i.t. S. pneumoniae challenge. Left: Lung homogenate CFU was assessed 24 hours after secondary S. pneumoniae (*P < 0.05, n = 4–6 animals per group). Right: MPO activity of lung homogenates was assessed 24 hours after S. pneumoniae infection. NGS, normal goat serum. (B) Animals were administered i.t. PR8, followed 5 days later by i.t. S. pneumoniae. Recombinant murine KC and Mip2 were given i.t. at the time of pneumococcal administration. At 24 hours following secondary S. pneumoniae challenge, lungs were collected for homogenization and determination of CFU (left; **P < 0.01; n = 5–8 mice per group) and MPO activity (right). Data are representative of 2 independent experiments. Carrier, saline carrier. Horizontal bars in the left panels represent statistical medians.
Conversely, we wished to see whether exogenous KC and Mip2 would be sufficient to rescue influenza-infected Ifnar+/+ animals challenged secondarily with pneumococcus. Therefore, in separate experiments, Ifnar+/+ mice infected with influenza virus were concurrently treated i.t. with 2,000 CFU of S. pneumoniae and a single dose of KC plus Mip2 or carrier solution. For comparison, _PR8_-infected _Ifnar_–/– animals were treated with the carrier solution at the time of secondary bacterial infection. By 24 hours after infection, treatment with KC and Mip2 had upregulated expression of MPO in lung homogenates and had potently reduced bacterial burdens in the lungs of infected mice by 2 logs as compared with Ifnar+/+ mice treated with saline carrier (Figure 6B; P < 0.005). Interestingly, the observed bacterial load in KC plus Mip2–treated wild-type animals was very similar to that of carrier-treated _PR8/S. pneumoniae_–infected _Ifnar_–/– mice. Thus, treatment of influenza-infected Ifnar+/+ mice with a single dose of KC and Mip2 at an early time point was sufficient to restore resistance to secondary pneumococcal pneumonia to the levels observed in _Ifnar_–/– animals. Together, these findings highlight the critical importance of KC and Mip2 for clearance of secondary pneumococcal pneumonia and suggest that type I IFN–mediated inhibition of these cytokines is a major mechanism for enhanced susceptibility to secondary pneumococcal pneumonia.
Discussion
While infections with influenza are very common, mortalities purely attributable to the primary viral infection are relatively uncommon. Many influenza-related deaths are attributable to secondary bacterial infection with S. pneumoniae and other bacteria, which present a unique challenge due to the emergence of antibiotic resistance and increased severity of infection. Influenza infections have been shown to impair macrophage and neutrophil function in the lung, both of which are critical cellular elements of innate pulmonary host defense against bacterial pathogens. Here we report a mechanism by which a central component of the antiviral response, type I IFNs, impairs innate immune responses against secondary bacterial challenge by impairing neutrophil responses. Importantly, we found that _Ifnar_–/– mice were more resistant to secondary bacterial pneumonia in the post-influenza setting, with strikingly lower lung and blood bacterial burdens and increased survival compared with their doubly infected Ifnar+/+ counterparts. In addition, neutralization of the IFNAR is sufficient to protect influenza-infected hosts from secondary pneumococcal pneumonia. These findings demonstrate that type I IFNs induced during influenza infection in the lung play a significant role in sensitizing hosts to bacteria infections in the post-influenza period.
The inhibitory effects of type I IFNs appear to be mediated at least in part through selective impairment of KC and Mip2 production, subsequently leading to attenuated neutrophil responses following secondary bacterial challenge in vivo. Having an adequate neutrophil response is critical to an effective pulmonary host defense under circumstances in which the bacterial burden exceeds the antimicrobial capacity of alveolar macrophages. Recently, Sun and Metzger identified type II IFNs as a mediator of macrophage dysfunction post influenza infection, including downregulation of the scavenger receptor MARCO, and impairment of macrophage-mediated bacterial phagocytosis and clearance (34). Although we found that type I IFNs led to downregulation of MARCO expression on alveolar macrophages, we were unable to convincingly find that this resulted in altered macrophage phagocytosis either in vivo or ex vivo (our unpublished observations). While it remains to be determined whether type I IFNs promote susceptibility of influenza-infected hosts to capsular subtypes of S. pneumoniae that are more dependent upon macrophages for clearance, higher titers of bacterial infection generally require neutrophil recruitment for effective clearance (35). Cxcr2 and its ligands (KC and Mip2) appear to be important mediators of neutrophil recruitment in host defense against bacterial pathogens, including S. pneumoniae (36–39). Since alveolar macrophage function is attenuated during influenza infection, having sufficient neutrophils recruited to the lung to counteract a rising bacterial burden is essential. Hence, type I IFN–regulated impairment in neutrophil responses likely contributes significantly to the development of post-influenza bacterial lung infections, which tend to be more fulminant than de novo bacterial pneumonias.
Here we report what we believe is a novel negative regulatory role of type I IFNs on CXC chemokine production in response to TLR2 (gram-positive) bacterial ligand challenge in macrophages. In vivo, Ifnar+/+ animals failed to mount an adequate neutrophil response commensurate to their growing bacterial burden (Figure 5, C–E), whereas _Ifnar_–/– animals were capable of generating a more robust neutrophil response. Indeed, neutralization of Cxcr2 receptor in influenza-infected _Ifnar_–/– animals limited the ability of these animals to contain secondary bacterial infection, whereas administration of KC and Mip2 to Ifnar+/+ animals conferred a protective advantage against post-influenza pneumococcal infection, demonstrating that Cxcr2 activity indeed acts downstream of type I IFNs. This might provide a molecular basis for the interesting findings recently reported by Didierlaurent et al., who described influenza-induced desensitization of alveolar macrophages, including decreased Mip2 and KC production, in response to flagellin (TLR5 ligand) challenge following influenza infection (40). Collectively, the results of these studies suggest that the antiviral response to influenza appears to impair 2 critical antibacterial cellular lines of defense — macrophage impairment as mediated by type II IFNs and inadequate neutrophil responses attributable to type I IFN–mediated suppression of Cxcr2 chemokine expression.
In addition to post-influenza macrophage and neutrophil dysfunction, prior studies have identified several other factors thought to enhance susceptibility of influenza-infected mice to secondary bacterial infections. These include damage to the respiratory epithelium, upregulation of factors (e.g., platelet aggregating factor) that promote adhesion of S. pneumoniae to the respiratory mucosa, and production of IL-10 (15, 20, 21, 41). However, attempts at manipulating some of these changes have only had modest effects on the host’s ability to control the secondary pneumococcal infection (19, 20). In the current study, we found that _Il10_–/– and wild-type mice had comparable clearance of secondary pneumococcal lung infection following influenza infection (Supplemental Figure 3). This does not preclude the possibility that at time-points after influenza challenge, IL-10 may contribute to bacterial susceptibility of influenza-infected hosts, although recently published data suggest this may not be the case (34). It is likely that at different times following influenza infection, multiple mechanisms contribute to the sensitization of influenza-infected hosts to secondary bacterial infections, depending on the severity of the initial influenza infection and the nature of the host antiviral response. Further studies are needed to elucidate how the antiviral immune response alters antibacterial defenses over time.
Our studies also reveal the complex effects of type I IFNs on antimicrobial defenses. While type I IFNs play an important role for clearance of viral pathogens, they appear to have contradictory effects in the context of bacterial infections, promoting clearance of some intracellular pathogens (e.g., Salmonella, Legionella) while inhibiting clearance of others (e.g., Listeria, Mycobacteria) (42–44). Consistent with published studies, our data reveal that _Ifnar_–/– mice cleared the initial influenza infection from the lung in a manner comparable to Ifnar+/+ animals (Supplemental Figure 1), demonstrating the redundant nature of type I IFNs in antiviral defense against some viruses. In addition, the absence of IFNAR signaling did not significantly alter clearance of pneumococcus alone in the lung at sublethal inoculating doses (Figure 2B). Following influenza infection, however, type I IFNs appeared to have immunomodulatory effects that were detrimental to antibacterial defense in the lung. Type I IFNs have been shown to induce apoptosis in various cells, including splenocytes, which may contribute to impaired clearance of intracellular bacteria, such as Listeria (42). Our preliminary studies, however, indicate that no significant differences in apoptotic cells could be observed in lung histology sections of Ifnar–/– animals as compared with Ifnar+/+ animals following PR8 infection (Supplemental Figure 6). While apoptosis might play a role at higher viral inocula (studies are ongoing), it does not appear to contribute significantly to the detrimental effects of type I IFNs in our present model.
Although multiple mechanisms may contribute in aggregate to influenza-induced impairment in antibacterial innate immune responses, our findings reveal a what we believe is a novel mechanism by which sublethal influenza infections sensitize hosts to secondary bacterial infection, via type I IFN-mediated attenuation of neutrophil responses. Why this regulatory pathway exists is unclear. We postulate that despite the critical role of neutrophils in host defense against bacterial infections, their presence can also be detrimental, especially in the lung. If left unchecked, activated neutrophils can lead to considerable lung injury (45). Therefore, during viral infection, type I IFNs may be a potential mechanism by which the body can regulate neutrophilic responses. This suggests that following primary viral infection, the immune system attempts to control specific aspects of inflammation, rather than globally suppressing inflammation, as one would observe with endotoxin tolerance or during sepsis. Our findings support the following immune mechanistic model: during influenza infection, type I IFNs are induced as the host attempts to limit viral replication. While type I IFNs protect the host by upregulating antiviral genes, they seem to specifically suppress further production of KC and Mip2, which may be of little protective value for clearance of the primary viral infection (46) and may mediate lung injury. These changes, however, result in impairment of the host’s ability to mount an appropriate neutrophil response to incoming bacterial pathogens, culminating in secondary bacterial pneumonia.
Collectively, these findings provide a mechanism by which the immune response to influenza predisposes hosts to secondary bacterial pneumonia, thereby providing a potential therapeutic target for reversing viral-induced immunosuppression. Further studies examining how type I IFNs regulate neutrophil responses against primary influenza and secondary bacterial infections are needed. Recently, Th17 cytokines have been demonstrated to be important mediators of neutrophilic responses in the lung during bacterial pneumonia (47). We and others have recently reported that type I IFNs and STAT1, which is activated by IFNAR, inhibit Th17 cell development (48, 49). We are presently in the process of examining whether type I IFN–mediated impairment of Th17-type responses may be beneficial in limiting lung injury induced by the primary influenza infection, while impairing the host’s ability to counteract a secondary bacterial infection.
The mammalian innate immune system is constantly confronted with a diverse and complex array of pathogenic organisms. Although common mediators are induced following a viral or bacterial challenge, this study demonstrates that the antiviral and antibacterial response can frequently be at odds with one another. Nowhere is this more evident than in the setting of respiratory influenza infections, which are often complicated by secondary bacterial infections. Controlling type I IFN–mediated inhibition of antibacterial responses presents a unique direction for research in combating complicated influenza pneumonia and secondary bacterial infections.
Methods
Reagents.
Recombinant murine anti–TNF-α, –Mip2, –IL-12p70, -KC, –IL-1β, –IL-12, and –IFN-γ antibodies used in the ELISAs were purchased from R&D Systems. IFN-α antibody used throughout the studies was purchased from PBL Biomed. P3C was purchased from Sigma-Aldrich as previously described (50). The Myeloperoxidase Assay Kit was purchased from Cytostore and used according to the manufacturer’s protocol.
Mice.
A colony of _Ifnar_–/– mice bred on a C57BL/6 background was established in-house. Age- and sex-matched specific pathogen–free Ifnar+/+ mice were used in all experiments. All animals were housed in specific pathogen–free conditions within the UCLA animal facilities until the day of sacrifice. All animal experiments were performed in accordance with NIH policies regarding the humane care and use of laboratory animals and were approved by the UCLA Animal Research Committee.
Influenza virus and plaque assays.
The PR8 strain of influenza virus was grown on monolayers of MDCK cells in the presence of l-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK-treated) trypsin (Sigma-Aldrich). Three days later, overlay medium was cleared of cellular debris, aliquoted, and stored at –80°C. The 50% tissue culture infective dose (TCID50) was determined by plating MDCK cells in 96-well plates and performing 2-fold serial dilution of virus stock, which were then incubated with MDCK cells for 2 days in the presence of TPCK trypsin. Presence of virus was determined by performing hemagglutinin activity assays using chicken red blood cells. TCID50 was reported as the dilution in which 50% of the infected wells were positive for virus.
Plaque assay was performed by incubating MDCK monolayers in 6-well plates at 37°C for 1 hour with whole-lung homogenates, which were serially diluted in virus dilution buffer (PBS with 10% BSA, CaCl2, and MgCl2). Viral growth medium consisting of MEM (Invitrogen), BME vitamins (Sigma-Aldrich), NaHCO3, 10% BSA, 1% DEAE Dextran, and TPCK-treated trypsin (0.7 μg/ml) and 2% agarose overlay were then added to each well. Plates were incubated for 72 hours at 34°C and 5% CO2 prior to enumeration of plaques by crystal violet staining.
Bacterial preparation and i.t. inoculation.
Type 3 S. pneumoniae (ATCC 6303 clinical isolate with capsular serotype 3) was used in our studies. This serotype was chosen because it is virulent in mice and commonly causes human disease. Bacteria were grown in Todd-Hewitt broth with yeast extract (Difco Laboratories) at 37°C for 8 hours or until log phase. The concentration of bacteria in broth was determined by measuring the absorbance at 600 nm and then plotting the OD on a standard curve generated by known CFU values. The bacteria culture was then diluted to the desired concentration. Mice were anesthetized with an i.p. injection of ketamine and xylazine mixture. Next, the trachea was exposed, and 30 μl of inoculum was administered via a sterile 26-gauge needle. The skin incision was closed using sutures.
Whole-lung homogenization for CFU.
At the designated time points, mice were euthanized by i.p. pentobarbital. Before lung removal, the pulmonary vasculature was perfused by infusing 1 ml of PBS containing 5 mM EDTA into the right ventricle. Whole lungs were removed, taking care to dissect away lymph nodes. The lungs were homogenized in 1 ml of PBS supplemented with protease inhibitor cocktail (Roche Applied Science). Homogenates were serially diluted 1:5 in PBS and plated on blood agar to determine lung CFU.
Peripheral blood CFU determination.
Blood was collected in a heparinized syringe from the right ventricle at designated time points, serially diluted 1:2 with PBS, and plated on blood agar to determine blood CFU.
BAL.
BAL was performed for assessment of leukocyte accumulation and to obtain air space leukocytes for ex vivo studies. The trachea was exposed and intubated using a 1.7-mm outer diameter polyethylene catheter. BAL was performed by instilling 10 ml of PBS containing 5 mM EDTA in 1-ml aliquots, resulting in 8 ml of retrieved lavage. Lavaged cells from each group of animals were pooled and counted after lysis with hypotonic solution. Cytospins (Shandon Inc., Thermo Electron Corp.) were prepared for determination of BAL differentials using a modified Wright stain. Lavaged cells consisted of more than 95% alveolar macrophages for Ifnar+/+ and _Ifnar_–/– mice at baseline (data not shown).
HA activity assay.
HA activity assay was performed by incubating serial 2-fold dilutions of lung homogenates with 0.6% chicken red blood cells in a 1:1 vol/vol ratio. The activity was measured as the lowest dilution where red blood cell precipitation was inhibited.
Histology and immunofluorescence.
Formalin-fixed, paraffin-embedded 5-μm sections of lungs were used for immunohistochemistry. Following deparaffinization and rehydration, antigen retrieval was performed by boiling the sections in 10 mM sodium citrate buffer for 15 minutes. Sections were blocked for 60 minutes using horse serum (vector IMPRESS reagent anti-rabbit kit), incubated for 1 hour with rabbit anti-MPO (Abcam; catalog ab15484), and then incubated for 40 minutes with anti-rabbit secondary antibody (vector IMPRESS reagent anti-rabbit kit) at room temperature. Each step was followed by two 5-minute washes in PBS. Mounting medium for fluorescence containing DAPI (Vectashield, Vector Laboratories Inc.) was applied to the slides as the final step. TUNEL staining of 4-μm-thick lung sections was performed according to the manufacturer’s protocol (Texas Red kit; Roche).
Murine cytokine ELISA.
Mip2 and KC ELISA kits were purchased from R&D Biosciences. Cytokine levels were measured according to the manufacturer’s instructions. IFN-α ELISA was performed as previously described (50). Briefly, NUNC plates were coated with rat anti–IFN-α antibody (PBL Biomedical Laboratories). Plates were blocked with 1% BSA in PBS and 0.05% Tween, and samples were incubated overnight to allow for equilibrium binding by the capture antibody. Detection was performed using rabbit anti-mouse IFN-α antibody (PBL Biomedical Laboratories) for 1 hour, followed by incubation with HRP-conjugated goat anti-rabbit antibodies. Other cytokine analyses were performed using the Luminex 22-Plex Murine Cytokine Detection System (Millipore) according to the manufacturer’s protocol.
Isolation of mRNA and real-time RT-PCR.
Total cellular RNA from macrophages were extracted using TRIzol (Invitrogen), reversed transcribed into cDNA (iScript; BioRad), and then amplified using specific primers for the cytokines tested. All values were normalized against the expression of ribosomal protein L32. The primer sequences for the genes examined were as follows: mKC forward, CAAGAACATCCAGAGCTTGAAGGT; reverse, GTGGCTATGACTTCGGTTTGG; mMip2 forward, AGCTACATCCCACCCACACAG; reverse, AAAGCCATCCGACTGCATCT; mTNFα forward, GGTGCCTATGTCTCAGCCTCTT; reverse, CGATCACCCCGAAGTTCAGTA; Iκbα forward, CTGCAGGCCACCAACTACAA; reverse, CAGCACCCAAAGTCACCAAGT. Semi-quantitative PCR was performed using Applied Biosystems Power SYBR Green Supermix on a single-color real-time PCR machine. All experiments were performed in triplicate.
Bone marrow–derived macrophages.
Bone marrow was isolated from the tibias and femurs of mice and differentiated in the presence of M-CSF, in 10% FBS containing DMEM. On day 7, cells were stimulated as described in the text. Recombinant IFN-α4 (1,000 U/ml) was added to the medium 30 minutes before stimulation with either P3C (100 ng/ml) or CpG (100 nM). Total cellular RNA was harvested 4 hours later with Trizol reagent and isolated following extraction with chloroform and precipitation with isopropanol.
Antibody-mediated neutralizations and cytokine reconstitutions.
IFNAR1 neutralization was performed by administration of 1.5 mg of MAR1-5A3 (Leinco Technologies) antibody on day 0 (same day as influenza infection), followed by booster doses of 0.75 mg on days 2 and 4. Normal mouse IgG was used as a control. On day 5, mice were infected with S. pneumoniae, and lungs were harvested on day 6. For Cxcr2 neutralization experiments, influenza-infected mice were depleted with a single 300-μl i.p. injection of goat anti-Cxcr2 serum or non-specific goat serum as described previously (51–53). Mice were subsequently infected with S. pneumoniae the following day. Twenty-four hours after secondary bacterial infection, mice were sacrificed to measure pulmonary bacterial burden and perform MPO assay on lung homogenates. For Mip2 plus KC reconstitutions, influenza-infected animals were treated with a single dose of i.t. KC plus Mip2 (1 μg each) resuspended in 0.1% BSA in PBS. The KC and Mip2 contained less than 1.0 U endotoxin/1 μg cytokine, as determined by Limulus amoebocyte lysate assay (R&D Systems). Mock-treated animals received PBS and 0.1% BSA. Bacterial inoculums were administered concurrently. Pulmonary bacterial burdens and lung homogenate MPO activity were assessed 24 hours after infection.
Statistics.
Survival curves were compared using the log-rank test. For other data, statistical significance was determined using 2-tailed unpaired t test, 2-tailed Mann-Whitney U test (for CFU Data), or 1-way ANOVA corrected for multiple comparisons where appropriate. A P value of 0.05 or less was considered statistically significant. All calculations were performed using the Prism software program for Windows (GraphPad Software Inc.). All bars across CFU data are presented as medians of the biological replicates. Error bars in all graphs indicate SEM and represent biological replicates.
Supplementary Material
Supplemental data
Acknowledgments
This work was supported by NIH grants T32 AI007323-19 and GM08042 (to A. Shahangian), K08 HL081229 (to J.C. Deng), and 1R01 AI056154 and 1R01 AI069120 (to G. Cheng). We are grateful to Paul W. Dempsey for invaluable feedback and discussions and to Tess E. Lin for technical assistance with the studies.
Footnotes
Authorship note: Genhong Cheng and Jane C. Deng contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: BAL, bronchoalveolar lavage; BMM, bone marrow–derived macrophage; Ifnar, IFN-α/β receptor; MPO, myeloperoxidase.
Citation for this article: J. Clin. Invest. 119:1910–1920 (2009). doi:10.1172/JCI35412
References
- 1.Hamilton B.E., et al. Annual summary of vital statistics: 2005. Pediatrics. 2007;119:345–360. doi: 10.1542/peds.2006-3226. [DOI] [PubMed] [Google Scholar]
- 2.LeVine A.M., Koeningsknecht V., Stark J.M. Decreased pulmonary clearance of S. pneumoniae following influenza A infection in mice. J. Virol. Methods. 2001;94:173–186. doi: 10.1016/S0166-0934(01)00287-7. [DOI] [PubMed] [Google Scholar]
- 3.Sethi S. Bacterial pneumonia. Managing a deadly complication of influenza in older adults with comorbid disease. Geriatrics. 2002;57:56–61. [PubMed] [Google Scholar]
- 4.McCullers J.A. Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broug-Holub E., et al. Alveolar macrophages are required for protective pulmonary defenses in murine Klebsiella pneumonia: elimination of alveolar macrophages increases neutrophil recruitment but decreases bacterial clearance and survival. Infect. Immun. 1997;65:1139–1146. doi: 10.1128/iai.65.4.1139-1146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toews G.B., Hansen E.J., Strieter R.M. Pulmonary host defenses and oropharyngeal pathogens. Am. J. Med. 1990;88:20S–24S. doi: 10.1016/0002-9343(90)90256-d. [DOI] [PubMed] [Google Scholar]
- 7.Zhang P., Summer W.R., Bagby G.J., Nelson S. Innate immunity and pulmonary host defense. Immunol. Rev. 2000;173:39–51. doi: 10.1034/j.1600-065X.2000.917306.x. [DOI] [PubMed] [Google Scholar]
- 8.Abramson J.S., Giebink G.S., Mills E.L., Quie P.G. Polymorphonuclear leukocyte dysfunction during influenza virus infection in chinchillas. J. Infect. Dis. 1981;143:836–845. doi: 10.1093/infdis/143.6.836. [DOI] [PubMed] [Google Scholar]
- 9.Abramson J.S., et al. Inhibition of neutrophil lysosome-phagosome fusion associated with influenza virus infection in vitro. Role in depressed bactericidal activity. J. Clin. Invest. 1982;69:1393–1397. doi: 10.1172/JCI110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abramson J.S., Lyles D.S., Heller K.A., Bass D.A. Influenza A virus-induced polymorphonuclear leukocyte dysfunction. Infect. Immun. 1982;37:794–799. doi: 10.1128/iai.37.2.794-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abramson J.S., et al. Suppression of endocytosis in neutrophils by influenza A virus in vitro. J. Infect. Dis. 1986;154:456–463. doi: 10.1093/infdis/154.3.456. [DOI] [PubMed] [Google Scholar]
- 12.Astry C.L., Jakab G.J. Influenza virus-induced immune complexes suppress alveolar macrophage phagocytosis. J. Virol. 1984;50:287–292. doi: 10.1128/jvi.50.2.287-292.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakab G.J. Immune impairment of alveolar macrophage phagocytosis during influenza virus pneumonia. Am. Rev. Respir. Dis. 1982;126:778–782. doi: 10.1164/arrd.1982.126.5.778. [DOI] [PubMed] [Google Scholar]
- 14.Jakab G.J., Warr G.A., Knight M.E. Pulmonary and systemic defenses against challenge with Staphylococcus aureus in mice with pneumonia due to influenza A virus. J. Infect. Dis. 1979;140:105–108. doi: 10.1093/infdis/140.1.105. [DOI] [PubMed] [Google Scholar]
- 15.McNamee L.A., Harmsen A.G. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect. Immun. 2006;74:6707–6721. doi: 10.1128/IAI.00789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickerson C.L., Jakab G.J. Pulmonary antibacterial defenses during mild and severe influenza virus infection. Infect. Immun. 1990;58:2809–2814. doi: 10.1128/iai.58.9.2809-2814.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nugent K.M., Pesanti E.L. Effect of influenza infection on the phagocytic and bactericidal activities of pulmonary macrophages. Infect. Immun. 1979;26:651–657. doi: 10.1128/iai.26.2.651-657.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nugent K.M., Pesanti E.L. Staphylococcal clearance and pulmonary macrophage function during influenza infection. Infect. Immun. 1982;38:1256–1262. doi: 10.1128/iai.38.3.1256-1262.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Sluijs K.F., et al. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J. Immunol. 2004;172:7603–7609. doi: 10.4049/jimmunol.172.12.7603. [DOI] [PubMed] [Google Scholar]
- 20.McCullers J.A., Rehg J.E. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 2002;186:341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- 21.McCullers J.A., Bartmess K.C. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 2003;187:1000–1009. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- 22.Sprenger H., et al. Chemokines in the cerebrospinal fluid of patients with meningitis. Clin. Immunol. Immunopathol. 1996;80:155–161. doi: 10.1006/clin.1996.0109. [DOI] [PubMed] [Google Scholar]
- 23.Matikainen S., et al. Influenza A and sendai Viruses induce differential chemokine gene expression and transcription factor activation in human macrophages. Virology. 2000;276:138–147. doi: 10.1006/viro.2000.0542. [DOI] [PubMed] [Google Scholar]
- 24.Yoneyama H., et al. Plasmacytoid DCs help lymph node DCs to induce anti-HSV CTLs. J. Exp. Med. 2005;202:425–435. doi: 10.1084/jem.20041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tough D.F., Borrow P., Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- 26.Cox N.J., Fukuda K. Influenza. Infect. Dis. Clin. North Am. 1998;12:27–38. doi: 10.1016/S0891-5520(05)70406-2. [DOI] [PubMed] [Google Scholar]
- 27.Price G.E., Gaszewska-Mastarlarz A., Moskophidis D. The role of alpha/beta and gamma interferons in development of immunity to influenza A virus in mice. J. Virol. 2000;74:3996–4003. doi: 10.1128/JVI.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheehan K.C.F., et al. Blocking monoclonal antibodies specific for mouse IFN-a/β receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 29.Chang E.Y., Guo B., Doyle S.E., Cheng G. Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J. Immunol. 2007;178:6705–6709. doi: 10.4049/jimmunol.178.11.6705. [DOI] [PubMed] [Google Scholar]
- 30.van der Poll T., Marchant A., Keogh C.V., Goldman M., Lowry S.F. Interleukin-10 impairs host defense in murine pneumococcal pneumonia. J. Infect. Dis. 1996;174:994–1000. doi: 10.1093/infdis/174.5.994. [DOI] [PubMed] [Google Scholar]
- 31.Seki M., et al. Immunokinetics in severe pneumonia due to influenza virus and bacteria coinfection in mice. Eur. Respir. J. 2004;24:143–149. doi: 10.1183/09031936.04.00126103. [DOI] [PubMed] [Google Scholar]
- 32.Garvy B.A., Harmsen A.G. The importance of neutrophils in resistance to pneumococcal pneumonia in adult and neonatal mice. Inflammation. 1996;20:499–512. doi: 10.1007/BF01487042. [DOI] [PubMed] [Google Scholar]
- 33.Reutershan J., et al. Critical role of endothelial CXCR2 in LPS-induced neutrophil migration into the lung. J. Clin. Invest. 2006;116:695–702. doi: 10.1172/JCI27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun K., Metzger D.W. Inhibition of pulmonary antibacterial defense by interferon-[gamma] during recovery from influenza infection. Nat. Med. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 35.Dockrell D.H., et al. Alveolar macrophage apoptosis contributes to pneumococcal clearance in a resolving model of pulmonary infection. J. Immunol. 2003;171:5380–5388. doi: 10.4049/jimmunol.171.10.5380. [DOI] [PubMed] [Google Scholar]
- 36.Tateda K., et al. Chemokine-dependent neutrophil recruitment in a murine model of Legionella pneumonia: potential role of neutrophils as immunoregulatory cells. Infect. Immun. 2001;69:2017–2024. doi: 10.1128/IAI.69.4.2017-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai W.C., et al. CXC chemokine receptor CXCR2 is essential for protective innate host response in murine Pseudomonas aeruginosa pneumonia. Infect. Immun. 2000;68:4289–4296. doi: 10.1128/IAI.68.7.4289-4296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai W.C., et al. Lung-specific transgenic expression of KC enhances resistance to Klebsiella pneumoniae in mice. J. Immunol. 1998;161:2435–2440. [PubMed] [Google Scholar]
- 39.Sun K., Salmon S.L., Lotz S.A., Metzger D.W. Interleukin-12 promotes gamma interferon-dependent neutrophil recruitment in the lung and improves protection against respiratory Streptococcus pneumoniae infection. Infect. Immun. 2007;75:1196–1202. doi: 10.1128/IAI.01403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Didierlaurent A., et al. Sustained desensitization to bacterial Toll-like receptor ligands after resolution of respiratory influenza infection. J. Exp. Med. 2008;205:323–329. doi: 10.1084/jem.20070891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plotkowski M.C., Puchelle E., Beck G., Jacquot J., Hannoun C. Adherence of type I Streptococcus pneumoniae to tracheal epithelium of mice infected with influenza A/PR8 virus. Am. Rev. Respir. Dis. 1986;134:1040–1044. doi: 10.1164/arrd.1986.134.5.1040. [DOI] [PubMed] [Google Scholar]
- 42.O’Connell R.M., et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrero J.A., Calderon B., Unanue E.R. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auerbuch V., Brockstedt D.G., Meyer-Morse N., O’Riordan M., Portnoy D.A. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee W.L., Downey G.P. Neutrophil activation and acute lung injury. Curr. Opin. Crit. Care. 2001;7:1–7. doi: 10.1097/00075198-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 46.Wareing M.D., Shea A.L., Inglis C.A., Dias P.B., Sarawar S.R. CXCR2 is required for neutrophil recruitment to the lung during influenza virus infection, but is not essential for viral clearance. Viral Immunol. 2007;20:369–378. doi: 10.1089/vim.2006.0101. [DOI] [PubMed] [Google Scholar]
- 47.Ye P., et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amadi-Obi A., et al. TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat. Med. 2007;13:711–718. doi: 10.1038/nm1585. [DOI] [PubMed] [Google Scholar]
- 49.Guo B., Chang E.Y., Cheng G. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J. Clin. Invest. 2008;118:1680–1690. doi: 10.1172/JCI33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oganesyan G., et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 51.Belperio J.A., et al. Critical role for CXCR2 and CXCR2 ligands during the pathogenesis of ventilator-induced lung injury. J. Clin. Invest. 2002;110:1703–1716. doi: 10.1172/JCI15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keane M.P., Belperio J.A., Xue Y.Y., Burdick M.D., Strieter R.M. Depletion of CXCR2 inhibits tumor growth and angiogenesis in a murine model of lung cancer. J. Immunol. 2004;172:2853–2860. doi: 10.4049/jimmunol.172.5.2853. [DOI] [PubMed] [Google Scholar]
- 53.Addison C.L., et al. The CXC Chemokine Receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J. Immunol. 2000;165:5269–5277. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data