MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes (original) (raw)
Abstract
Polymerase I and transcript release factor (PTRF)/Cavin is a cytoplasmic protein whose expression is obligatory for caveola formation. Using biochemistry and fluorescence resonance energy transfer–based approaches, we now show that a family of related proteins, PTRF/Cavin-1, serum deprivation response (SDR)/Cavin-2, SDR-related gene product that binds to C kinase (SRBC)/Cavin-3, and muscle-restricted coiled-coil protein (MURC)/Cavin-4, forms a multiprotein complex that associates with caveolae. This complex can constitutively assemble in the cytosol and associate with caveolin at plasma membrane caveolae. Cavin-1, but not other cavins, can induce caveola formation in a heterologous system and is required for the recruitment of the cavin complex to caveolae. The tissue-restricted expression of cavins suggests that caveolae may perform tissue-specific functions regulated by the composition of the cavin complex. Cavin-4 is expressed predominantly in muscle, and its distribution is perturbed in human muscle disease associated with Caveolin-3 dysfunction, identifying Cavin-4 as a novel muscle disease candidate caveolar protein.
Introduction
Caveolae are a characteristic feature of most mammalian cell types (Parton and Simons, 2007). The major membrane protein components of caveolae, caveolins, are integral membrane proteins that bind cholesterol and form high molecular weight oligomers (Monier et al., 1996). Caveolin-1 (Cav1), the major nonmuscle isoform of the caveolin family, is essential for caveola formation (Fra et al., 1995; Drab et al., 2001). Loss of Cav1 in mice and human patients causes severe lipid dysregulation, including perturbation of lipid storage (Razani et al., 2002; Fernandez et al., 2006; Le Lay et al., 2006), disruption of lipolysis (Cohen et al., 2004), insulin resistance (Capozza et al., 2005), and a severe lipodystrophy (Garg and Agarwal, 2008; Kim et al., 2008). Caveolae have also been implicated in the regulation of cell proliferation, mechanosensation, and endocytosis of specific cargoes (Tagawa et al., 2005; Yao et al., 2005; Cheng et al., 2006; Parton and Simons, 2007). Caveolins have also been strongly linked to disease states including breast and prostate cancer (Lee et al., 1998; Tahir et al., 2008) and, in the case of the muscle-specific Caveolin-3 (Cav3), muscle diseases known as caveolinopathies (for a review see Woodman et al., 2004).
Until recently, studies of caveolae have invariably focused on caveolins in the absence of information on other bona fide caveolar machinery proteins. However, although caveolins are essential for biogenesis of caveolae, recent studies have highlighted additional levels of regulation of caveola formation and potentially function. These studies have shown a role for accessory proteins, such as polymerase I and transcript release factor (PTRF)/Cavin (Hill et al., 2008; Liu and Pilch, 2008; Liu et al., 2008), and kinases, including ARAF1 (Pelkmans and Zerial, 2005), which regulate caveola formation. This has started to change the perception of caveolae as a stable domain (Pelkmans et al., 2004).
PTRF/Cavin was originally described as a protein able to dissociate transcription complexes in vitro (Mason et al., 1997; Jansa et al., 1998, 2001) and was more recently shown to localize to caveolae (Vinten et al., 2001, 2005; Voldstedlund et al., 2001, 2003). PTRF/Cavin is enriched in caveolar fractions (Aboulaich et al., 2004), and monoclonal antibodies to PTRF/Cavin specifically labeled morphological caveolae in adipocytes (Vinten et al., 2001, 2005). PTRF/Cavin was subsequently shown to be essential for caveola formation in mammalian cells (Hill et al., 2008; Liu and Pilch, 2008; Liu et al., 2008), zebrafish (Hill et al., 2008), and mice (Liu et al., 2008). PTRF/Cavin shares sequence homology with a family of proteins classified as PTRF/SDR homology proteins in Swissprot (http://www.expasy.org). These include serum deprivation response (SDR) gene product (also called SDPR; Gustincich and Schneider, 1993) and SDR-related gene product that binds to C kinase (SRBC; Izumi et al., 1997). As well as showing significant sequence homology (Xu et al., 2001), all three proteins have been suggested to bind to phosphatidylserine (PS; Burgener et al., 1990; Izumi et al., 1997; Gustincich et al., 1999; Hill et al., 2008). In addition, several studies have linked SDR and SRBC to protein kinase C (Izumi et al., 1997; Mineo et al., 1998) and to caveolae, including a proteomic study of adipocyte caveolin-enriched fractions (Aboulaich et al., 2004), recruitment of SDR to detergent-resistant floating fractions by Cav1 (Hill et al., 2008), and immunolocalization of SDR to caveolae (Mineo et al., 1998). By analogy to PTRF/Cavin, these studies raise the possibility that these related proteins regulate some aspect of caveolar function.
We have now extensively characterized the members of the mammalian PTRF/SDR family of proteins, termed Cavins. Using several complementary approaches, we show that these proteins are coexpressed and coassociate in mammalian cells to form an oligomeric assembly that we term the Cavin complex. This complex is present in the cytosol, but in cells expressing caveolins, the cavin complex is associated with caveolae. Functional studies in a heterologous cell system show an essential role for PTRF/Cavin-1 but not the other identified family members in caveola formation and recruitment of the cavin complex to the plasma membrane (Hill et al., 2008; this study). The tissue-specific expression of cavins suggests that each member regulates tissue-specific caveolae functions. We also report that a fourth member of the family, previously proposed to be a purely cytosolic protein (Ogata et al., 2008; Tagawa et al., 2008), is a muscle-specific component of the cavin complex associated with the sarcolemmal caveolae of muscle cells. Moreover, we demonstrate that this protein, muscle-restricted coiled-coil protein (MURC)/Cavin-4, is a new candidate protein for caveolin-associated muscle disease.
Results
Identification of PTRF/Cavin-related proteins; MURC/Cavin-4 is a muscle-enriched PTRF family member
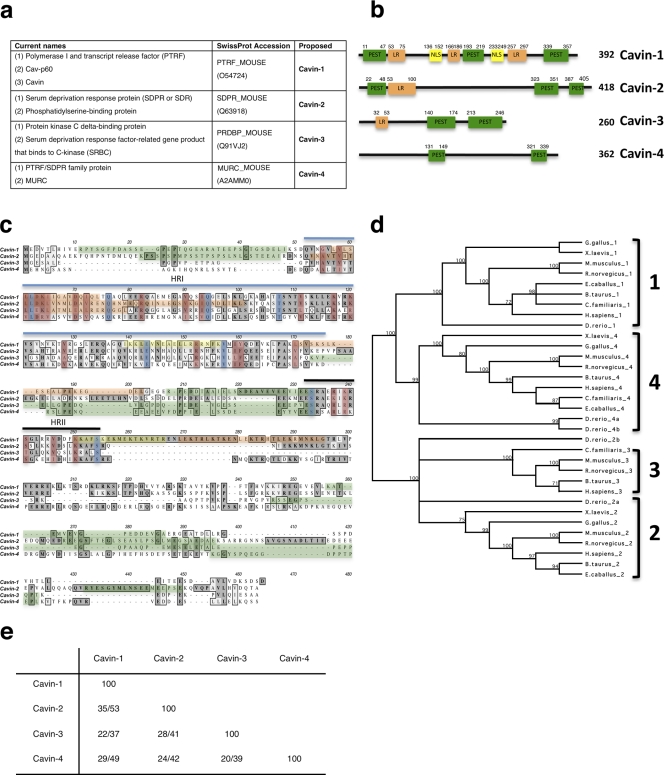
Through sequence homology searches, we identified a putative PTRF/SDR family member, which we term Cavin-4 (Fig. 1 a). Cavin-4 has been predicted by the Functional Annotation of the Mammalian cDNA Consortium, the Institute of Physical and Chemical Research Genome Exploration Research Group, and the Genome Science Group (Genome Network Project Core Group; cDNA clone 2310039E09; Carninci et al., 2005). Cavin-4 protein has subsequently been annotated by Swissprot (accession no. A2AMM0) and recently reported as MURC, a cardiac and skeletal muscle–specific protein (Ogata et al., 2008; Tagawa et al., 2008).
Figure 1.
The PTRF/Cavin family member proteins. (a) Summary of the names used in this paper. (b) Domain structure of cavin proteins in mouse. Putative PEST domains (green), leucine-rich regions (LR; orange), and nuclear localization sequences (NLS; yellow) are shown with amino acid numbers indicated. (c) Amino acid alignment of murine cavin proteins. Dark gray denotes identity, whereas light gray denotes similarity. Proposed cavin homology regions (HR) are marked with lines. Blue indicates identical amino acids, and red indicates conserved amino acids in evolution across all family members. Green, orange, and yellow are as indicated in b. (d) Midpoint-rooted tree showing several species containing cavin family members. Numbers represent the percentage of 1,000 bootstrap trials that supports the branch (numbers <70% are not depicted). (e) Percentage identity/similarity of murine cavin proteins.
The MURC/Cavin-4 gene localizes to human chromosome 9 (contig 102,380,247–102,388,554) and mouse chromosome 4 (contig 48,676,386–48,686,364), and in both species, the gene comprises two exons; this genomic organization seems to be conserved in all species identified. As shown in Fig. 1 (b and c), the mouse Cavin-4 sequence encodes a protein of 362 amino acids, which shares structural PEST (proline, glutamic acid, serine, and threonine-rich) domains with other cavins. Interestingly, all the family members are predicted to have regions of intrinsic disorder, which are coincident with the predicted PEST domains (disorder predictions made at http://bioinf.cs.ucl.ac.uk/disopred/index.html). In addition to the PEST domains, PTRF/Cavin-1, SDR/Cavin-2, and SRBC/Cavin-3 are also predicted to have leucine-rich regions (putative leucine zipper), and Cavin-1 contains polybasic regions (possible nuclear localization signals).
Orthologues of Cavin-4 were identified in mammals, fish, and frogs but not Caenorhabditis elegans. Murine Cavin-4 shows 29% homology and 49% similarity to Cavin-1, 24% homology and 42% similarity to Cavin-2, and 20% homology and 39% similarity to Cavin-3 (Fig. 1 e). Evolutionary analysis (Fig. 1 d) demonstrated distinct clades for the family members. The sequence homology between all the family members is higher in the N-terminal region (Fig. 1 c; Gustincich et al., 1999; Xu et al., 2001), and based on this, we propose that this family of proteins shares two homology regions (Fig. 1c) with 61–77% sequence similarity in region I and 78–87% in region II among the cavin family members.
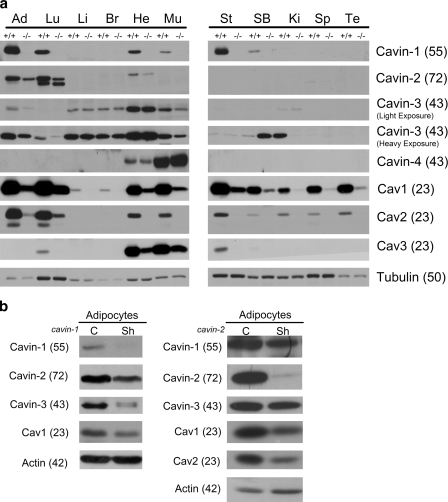
The tissue distribution of Cavin-1 closely parallels that of Cav1 (Hill et al., 2008; Liu et al., 2008). To examine the tissue distribution of the other family members, we performed immunoblotting using affinity-purified rabbit antibodies to each cavin. As shown in Fig. 2 a, the tissue distribution of Cavin-2 and -3 also correlates with Cav1 expression. For Cavin-4, in agreement with other data (Cancer Genome Anatomy Project virtual northern expression pattern for cluster UniGene accession no. Hs.99004; Tagawa et al., 2008), we detected a high expression level in skeletal and cardiac muscles (Fig. 2 a). We also examined the expression of Cavin-2 to -4 in tissues from _cavin-1_−/− mice (Liu et al., 2008). As shown in Fig. 2 a, the genetic loss of Cavin-1 results in a marked loss of Cav1-3 proteins in all tissues (80–90%; Liu et al., 2008). The levels of Cavin-2 and -3 proteins were also reduced in the tissues where Cavin-1 and other cavins are coexpressed, although this loss was not to the same extent as the reduction in caveolins. Cavin-3 and Cavin-4 proteins were generally the least affected by the absence of Cavin-1 (Fig. 2 a), and Cavin-3 is expressed in liver and brain where the other caveolae proteins are expressed at very low levels. Interestingly, Cavin-4 expression levels in cardiac and skeletal muscle were not affected by the absence of Cavin-1 (Fig. 2 a). Cavin-3, which is most highly expressed in cardiac muscle, was also not affected by the loss of Cavin-1 in this tissue (Fig. 2 a). Strengthening the link to caveolae, levels of Cavin-2 and -3 were also decreased in tissues from mice lacking Cav1, whereas Cavin-4 levels were decreased in cardiac muscle but not in skeletal muscle (unpublished data), where the isoform Cav3 is predominant over Cav1 (Way and Parton, 1995; Parton et al., 1997). Collectively, the results in Cavin-1−/− and Cav1−/− tissues suggest that these proteins are to some extent coregulated at the level of protein expression/stability. In addition, the results in Cavin-1−/− tissues suggest that the cavin proteins may have independent cellular functions.
Figure 2.
Cavin family members are down-regulated in cavin-1 knockout mice. (a) Tissue lysates from WT (+/+) and cavin-1 knockout mice (−/−) from the indicated tissues were prepared in RIPA buffer. Heavy and light exposures of Cavin-3 are shown to best demonstrate its distribution. Equal protein amounts were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. Ad, adipose tissue; Lu, lung; Li, liver; Br, brain; He, heart; Mu, skeletal muscle; St, stomach; SB, small bowel; Ki, kidney; Sp, spleen; Te, testis. (b) Cavin-1 to -3, Cav1, and Cav2 are down-regulated in shRNA (Sh) knockdown 3T3-L1 adipocytes. Whole cell lysates from Cavin-1–shRNA (Liu and Pilch, 2008) or Cavin-2–shRNA knockdown adipocytes were analyzed by Western blotting with the indicated antibodies. C, control. Molecular masses (in parentheses) are given in kilodaltons.
To further examine the interdependence of the cavin family members, we also determined the expression of cavins in 3T3-L1 adipocytes that have abundant caveolae (Fig. 2 b). Interestingly, in contrast to results from Cavin-1−/− tissues, our knockdown experiments in 3T3-L1 adipocytes showed that the loss of Cavin-1 caused a marked reduction in Cavin-3 protein. In contrast, knockdown of Cavin-2 in adipocytes resulted in a more modest reduction of Cavin-1 with relatively little effect on Cavin-3 (Fig. 2 b). We have not yet been able to achieve a significant knockdown of Cavin-3 in adipocytes, and at a 40–50% short hairpin RNA (shRNA)–mediated reduction in this protein, there is no apparent effect on the other caveolae proteins (unpublished data).
The cavin family proteins localize to caveolae and are dependent on Cav1 for membrane association
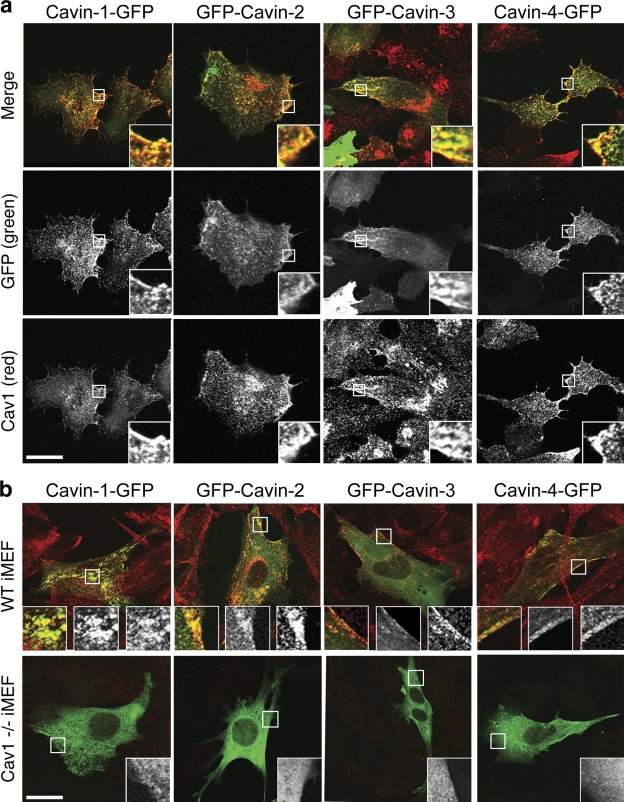
SDR/Cavin-2 and SRBC/Cavin-3 have been suggested to associate with caveolae (Mineo et al., 1996; Izumi et al., 1997; Gustincich et al., 1999). In addition, MURC/Cavin-4 has been proposed to interact with Cavin-2 (Ogata et al., 2008). In view of the sequence homology between the family members and the aforementioned findings, we postulated that all four family members associate with caveolae. Consistent with this hypothesis, all four cavins showed colocalization with Cav1 at the cell surface when expressed in BHK cells (Fig. 3 a). As observed previously with PTRF/Cavin-1 (Hill et al., 2008), Cavin-2, -3, and -4 did not associate with caveolin in the Golgi, which may indicate that these proteins recognize the plasma membrane environment generated by caveolin. Although plasma membrane Cav1 is organized in higher order oligomers, Golgi Cav1 is predominantly monomeric (Pol et al., 2005). Differences in oligomerization state may alter the lipid environment, leading to differential cavin recruitment. We next tested whether caveolin is necessary for recruitment of cavins to the plasma membrane using wild-type (WT) and Cav1−/− immortalized mouse embryonic fibroblasts (iMEFs). All four cavins showed plasma membrane association and colocalized with Cav1 in WT iMEFs, but in Cav1−/− iMEFs, the cavin proteins accumulated in the cytosol (Fig. 3 b).
Figure 3.
Cavins colocalize with caveolin and are dependent on caveolin for surface localization. (a) BHK cells transfected with GFP-tagged Cavin-1, -2, -3, or -4 (green) were PFA fixed and immunolabeled for endogenous Cav1 (red). Cavin family members and Cav1 extensively colocalized in a pattern consistent with caveolae distribution. Insets show enlargements of the selected areas. (b) WT and Cav1−/− iMEFs were transfected with GFP-tagged cavins (green) and labeled for endogenous Cav1 (red). In WT iMEFs, cavins colocalized with Cav1 on the plasma membrane. In Cav1−/− iMEFs, cavin proteins were mainly restricted to the cytosol, although in a small proportion of highly expressing cells, a surface pool of protein could be detected. Insets represent (from left to right) an enlargement of the selected area showing the merged image, GFP (green), and Cav1 (red). Bars, 10 µm.
PTRF/Cavin-1, SDR/Cavin-2, and SRBC/Cavin-3 form a caveolar complex in adipocytes
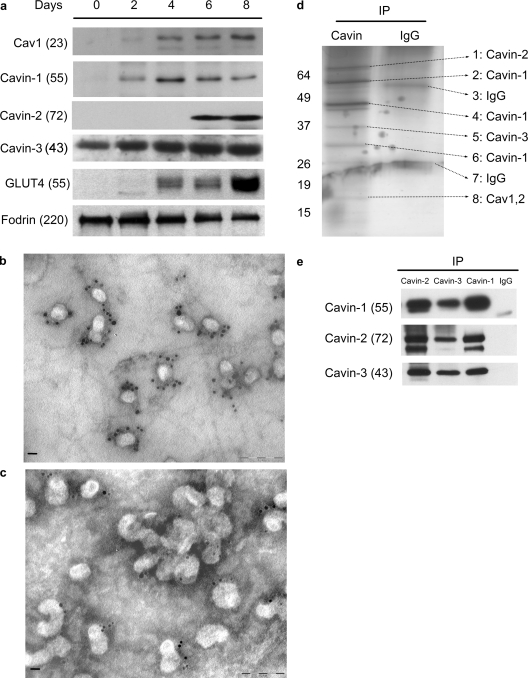
Adipocytes are a rich source of caveolae, and we have devised protocols for their rapid purification from this tissue (Souto et al., 2003), allowing us to identify by proteomic analysis PTRF/Cavin-1, SDR/Cavin-2, and SRBC/Cavin-3 (Fig. 4 d; Pilch et al., 2007). Cultured murine adipocytes such as 3T3-L1 cells acquire their fatty phenotype during differentiation (Green and Kehinde, 1976), which occurs at about the same time as a dramatic increase in Cav1 protein expression and caveola formation (Scherer et al., 1994; Kandror et al., 1995). Thus, we examined the expression of the Cavin-1 to -3 during the process of adipocyte differentiation. As shown in Fig. 4 a, Cavin-3 levels are relatively high even in the nondifferentiated cells (days 0 and 2) and do not significantly change from day 2 to day 8. Cavin-1 and Cav1 appear coincidentally on day 2 followed by Cavin-2 on day 6. Glut4 appears on day 4, which serves as a control for the differentiation process and acquisition of insulin-sensitive glucose transport (Birnbaum, 1989). Therefore, the adipocyte differentiation model is not only useful for its classical role in determining the ontogeny of adipocyte-specific processes, but it also allows us to determine a possible assembly hierarchy for the proteins associated with and involved in caveola biogenesis. In agreement with these results, the immunogold electron microscopic (immuno-EM) analysis of adipocyte plasma membrane “sheets” (Vinten et al., 2001, 2005; Souto et al., 2003; Voldstedlund et al., 2003) documents the presence of Cavin-2 (Fig. 4 b) and Cavin-3 (Fig. 4 c) exclusively in or very near to caveola structures. Previous analysis by immuno-EM revealed Cav1 (Voldstedlund et al., 2003) and Cavin-1 (Vinten et al., 2001, 2005) to decorate these same structures in similar abundance, which is in accordance with a 1:1 ratio we have observed in fibroblasts (Hill et al., 2008). However, it is clear that Cavin-1 (Vinten et al., 2001, 2005) and Cavin-2 (Fig. 4 b) are present in multiple copies in all caveolae.
Figure 4.
Cavin-1 to -3 forms a complex at caveolae in adipocytes. (a) Cavin-1 to -3 and Cav1 are induced during 3T3-L1 adipocyte differentiation (note that Cavin-4 is not normally expressed in adipocytes). Total cell lysates were prepared from 3T3-L1 cells on the indicated day after induction of differentiation. Equal amounts of protein were analyzed by immunoblotting. Fodrin is shown as loading control. (b and c) Electron micrographs show a view of the cytoplasmic face of the adipocyte plasma membrane immunogold labeled for Cavin-2 (b) and Cavin-3 (c). The presence of Cavin-2 and -3 are primarily exclusively in or very near to clustered or single caveolar structures in multiple copies. Bars, 100 nm. (d) Cavin-1 to -3 forms a complex. 100 µg plasma membrane from primary adipocytes was solubilized in lysis buffer and immunoprecipitated with 3 µg anti–Cavin-1 or nonspecific IgG. The bound proteins were subjected to SDS-PAGE, silver stained as shown, and bands were identified as the indicated proteins (arrows) by mass spectrometry. (e) Similar immunoprecipitations were performed using anti–Cavin-1, -2, or -3 antibodies or nonspecific IgG. After SDS-PAGE, the bound proteins were analyzed by immunoblotting. IP, immunoprecipitation. Molecular masses (in parentheses) and markers are indicated in kilodaltons.
To determine how these proteins might interact at the plasma membrane of adipocytes, we performed immunoprecipitation from plasma membrane fractions with anti–Cavin-1 antibody followed by silver staining and mass spectrometry analysis of the stained bands. As shown in Fig. 4 d, all three proteins plus Cav1 and -2 were identified in this way. We often observe multiple Cavin-1 bands by immunoblotting (not depicted) as we see in Fig. 4 d, apparently a result of its susceptibility to proteolysis (also seen for Cavin-2; Fig. 2, a and b; and Fig. 4 e). It is notable that the only proteins stained in this gel aside from IgG are caveola components, and adipocyte caveolae-resident proteins such as CD36 (Souto et al., 2003) are absent. In confirmation of these data, we did reciprocal immunoprecipitations of Cavin-1, -2, and -3, and we show in Fig. 4 e that each antibody is able to bring down the complex consisting of the other species.
All members of the cavin family interact in a multimeric complex in the cytosol of fibroblasts
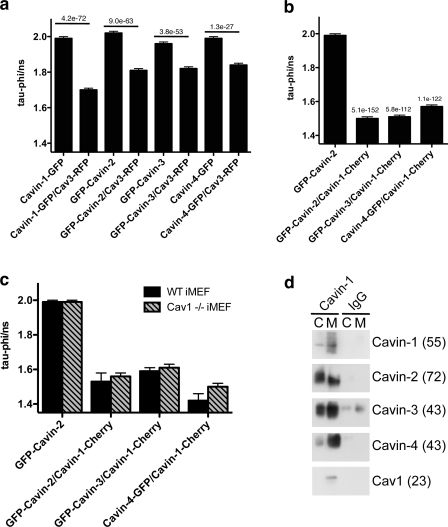
To further demonstrate the interaction of the cavins with caveolin, we chose fluorescence lifetime imaging microscopy (FLIM)/fluorescence resonance energy transfer (FRET). Using a cavin-EGFP construct as donor and Cav3–monomeric RFP as an acceptor, we observed a significant reduction in the EGFP fluorescence lifetime upon expression of all the cavin family members (Fig. 5 a), indicating that these proteins are within close proximity to caveolin. To evaluate the possibility that cavin family members form a multimeric complex, we used GFP-tagged SDR/Cavin-2, SRBC/Cavin-3, or MURC/Cavin-4 constructs as donors and PTRF/Cavin-1–Cherry as the acceptor. As shown by the very strong reduction in the donor lifetimes in Fig. 5 b, cavins are in close proximity to each other. Interestingly, as shown in Fig. 5 c, a similar interaction between the cavin proteins was observed in Cav1−/− iMEFs, suggesting that the cavin complex can be formed in the cytosol and is recruited to the forming caveolae.
Figure 5.
Cavin proteins form a multimeric complex and interact with caveolin. (a and b) BHK cells transiently expressing GFP-tagged cavin family members with or without Cav3-RFP (a) or expressing different combinations of fluorescently tagged cavin proteins (b) were analyzed using FLIM/FRET as detailed previously in Hill et al. (2008). In c, WT or Cav1−/− iMEFs expressing tagged cavin family members were analyzed by FLIM/FRET. Data represent mean GFP fluorescence lifetime ± SEM. All of the family members presented a significant reduction in GFP lifetime when coexpressed with Cav3-RFP (a) and Cavin-1–Cherry (b and c), indicating that these proteins are in close proximity. P-values of Student's t test are indicated; n = 80–150 cells. (d) Cavin-1–binding proteins were isolated from equivalent volumes of cytosol (C) and membrane (M) fractions prepared from WT iMEFs using Cavin-1 antibody or normal rabbit IgG as control. Binding to Cavin-2, -3, and -4 was observed in both cytosol and membrane fractions, whereas caveolin binding was only detected in the membrane fraction. Longer exposure of starting material detected cavin bands of corresponding sizes (not depicted). Result is representative of three independent experiments. Molecular masses (in parentheses) are indicated in kilodaltons.
We also examined the possible cytosolic interactions between cavins biochemically. Cavin-1–binding proteins were isolated from cytosol and membrane fractions prepared from WT iMEFs using an affinity-purified rabbit anti–Cavin-1 antibody. Analysis of bound proteins by immunoblotting showed an interaction between Cavin-1, -2, -3, and -4 in both the cytosol and membrane fractions, whereas interaction with caveolin was only observed in the membrane fraction (Fig. 5 d). Note that although Cavin-4 was present at very low levels in iMEFs, a significant coimmunoprecipitation could still be detected. Thus, both FLIM/FRET and coimmunoprecipitation experiments are consistent with the constitutive formation of a cavin complex in the cytosol, which is recruited to plasma membrane caveolae. To further confirm that these proteins interact in a multiprotein complex, we performed immunoblotting after native gel electrophoresis. A single band of around 230 kD was detected by the Cavin-1 and -4 antibodies in the cytosol of iMEFs (Fig. S1 a), which is consistent with a multiprotein complex formed between the cavin family members. This band was not observed in the membrane fraction, presumably because binding with caveolin multimers produces a complex that is too large to enter the gel. However, based on our tissue expression data, the exact components of the complex in specific cell types may well be determined by the tissue-specific expression of the family members, and we cannot at present rule out the existence of multiple cavin complexes containing two or more family members.
PTRF/Cavin-1 is required for recruitment of the cavin complex to caveolae
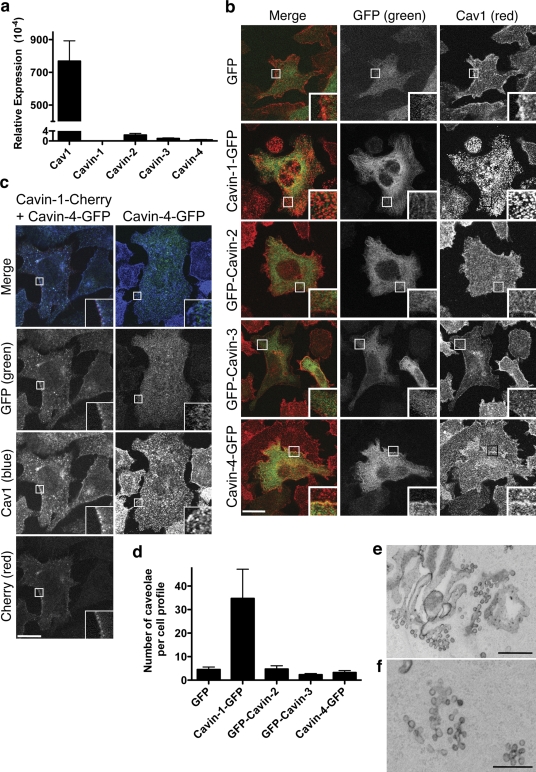
We have previously shown that prostate cancer PC3 cells express Cav1, but not PTRF/Cavin-1, and that expression of Cavin-1 alone is sufficient to generate caveolae (Hill et al., 2008). None of the four cavin proteins were detectable in PC3 cells (not depicted), and cavin transcripts were present at extremely low levels in PC3 cells in comparison with the mRNA for Cav1 (Fig. 6 a). We used this system to determine whether other cavins showed the same ability as Cavin-1 to induce caveola formation. PC3 cells stably expressing GFP-tagged cavins were generated and FACS sorted to obtain 95% pure populations of expressing cells. Western blotting showed that all four members were expressed as full-length fusion proteins with no significant degradation (Fig. S1 b). Examination of endogenous Cav1 by immunofluorescence suggested that only Cavin-1 caused a redistribution of Cav1 from a diffuse pattern over the cell surface to a punctate pattern (Fig. 6 b). In addition, Cavin-1, but not the other family members, colocalized with Cav1 at the plasma membrane and associated with the plasma membrane in a punctate pattern (note that the MeOH fixation used in Fig. 6 b clearly shows the redistribution of Cav1, but it does not preserve the colocalization between Cavin-1 and Cav1; Fig. S1 c). Consistent with this observation, EM analysis showed that only Cavin-1 induced abundant caveolae formation in >80% of the analyzed cells, whereas all the other family members showed several caveolae that were similar to that observed for GFP alone (Fig. 6, d–f). Note that in a very small proportion of SDR/Cavin-2–expressing cells, a very low number of morphological caveolae at the plasma membrane was observed, raising the possibility that Cavin-2 has some propensity to stabilize/generate caveolae, but this was orders of magnitude lower than for Cavin-1.
Figure 6.
Cavin-1, but not other cavins, generates caveolae in a heterologous system. (a) Relative expression of cavins and Cav1 mRNA in comparison with 18S RNA in PC3 cells was measured by RT-PCR. The y axis was divided to compare the cavins and Cav1 expression in the same scale. (b) PC3 cells stably expressing GFP-tagged cavins (green) were FACs sorted, methanol fixed, and immunolabeled for endogenous Cav1 (red). Only Cavin-1 caused Cav1 redistribution from a diffuse surface pattern to puncta consistent with caveolae formation (Hill et al., 2008). (c) PC3 cells transiently transfected with Cavin-4–GFP (green) alone or in combination with Cavin-1–Cherry (red) were methanol fixed and immunolabeled for endogenous Cav1 (blue). In cells expressing Cavin-4 plus Cavin-1, Cavin-4 colocalizes with Cav1 and Cavin-1 at the plasma membrane. Insets show a magnified view of the selected areas. (d) Quantitation of caveolae in stably transfected PC3 cell lines by EM expressed as mean ± SEM per cell. Caveolae were defined morphologically as plasma membrane (i.e., ruthenium red positive) pits/vesicular profiles of a diameter <100 nm. The number of such structures over the entire surface of at least 10 cell profiles per condition was determined and expressed as mean ± SEM per cell. Similar results were obtained in two independent experiments. (e and f) Representative micrograph of caveolae in PC3 cells stably expressing Cavin-1. Bars: (b) 10 µm; (e) 500 nm; (f) 200 nm.
Interestingly, when PTRF/Cavin-1 and MURC/Cavin-4 were coexpressed in PC3 cells, Cavin-4 was recruited to the plasma membrane where it colocalized with Cavin-1 and Cav1 (Fig. 6 c). These results suggest that Cavin-1 is required to recruit the cavin complex to caveolae where the other members may mediate distinct functions.
MURC/Cavin-4 localizes to sarcolemmal caveolae in muscle, and its distribution is perturbed in muscle disease
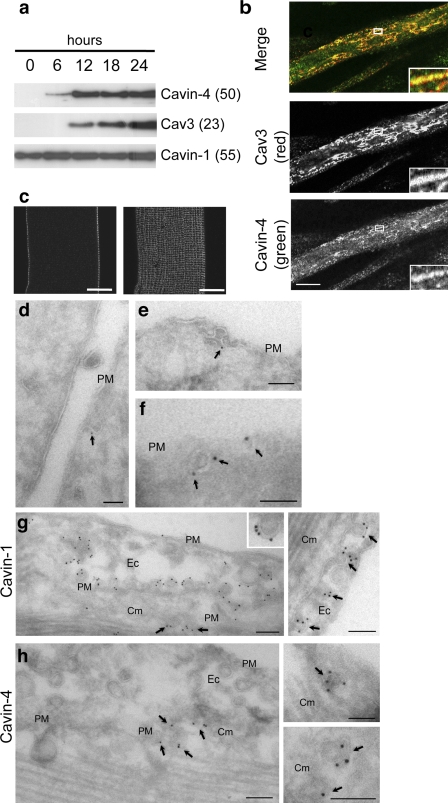
In contrast to our data showing association of cavins with caveolae, MURC/Cavin-4 was recently reported to be a cytosolic protein in muscle (Ogata et al., 2008; Tagawa et al., 2008). To further investigate the localization and function of Cavin-4 in muscle, we first determined the expression of Cavin-4 during C2C12 differentiation. As shown in Fig. 7 a, Cavin-4 expression is low in myoblasts but is increased upon differentiation, appearing after 6 h slightly before detection of Cav3 at 12 h (Fig. 7 a). Note that a very low level of Cavin-4 could be detected in myoblasts upon longer exposure times (not depicted) in concordance with the low expression level we detect in fibroblasts (Fig. 5). PTRF/Cavin-1 is expressed in myoblasts and myotubes (Fig. 7 a), which is consistent with its broader tissue distribution (Fig. 2). In C2C12 myotubes and adult mouse skeletal muscle tissue, Cavin-4 colocalizes with Cav3 at the light microscopic level (Fig. 7 b). In isolated muscle fibers, high resolution light microscopic analysis of Cavin-4 showed highly organized radial arrays of Cavin-4 labeling over the sarcolemma of the muscle (Fig. 7 c), which partially colocalized with Cav3 (not depicted). We next examined the distribution of Cavin-4 at the ultrastructural level in C2C12 myotubes and in skeletal muscle by immuno-EM on frozen sections. Specific labeling was observed in pits and vesicular profiles close to the plasma membrane of the C2C12 myotubes (Fig. 7, d–f) and skeletal muscle (not depicted) consistent with the association of Cavin-4 with caveolae. Myoblasts in the culture showed negligible labeling (unpublished data). Furthermore, labeling of PFA-fixed cardiac tissue showed labeling for Cavin-1 in both endothelial cells and cardiac muscle, whereas Cavin-4 was only detectable in the cardiac muscle (Fig. 7, g and h). In each case, labeling was observed in association with structures with the morphology of caveolae, i.e., ∼65-nm diameter pits or vesicular profiles generally close to the sarcolemma or endothelial cell plasma membrane. Although labeling per caveola was variable in thin sections, a labeling density of up to five golds per caveola profile for both Cavin-1 (Fig. 7 g) and Cavin-4 (Fig. 7 h) is consistent with a high number of cavin molecules per caveola in the same range as Cav1. This is consistent with labeling of Cavin-2 and -3 in plasma membrane lawns (Fig. 4) and with previous estimates of the stoichiometry of Cavin-1 and Cav1 (Hill et al., 2008). Collectively, these results show that Cavin-4 is a caveolar protein in skeletal and cardiac muscle, and together with the other members of the cavin family, it likely forms a structural complex at caveolae.
Figure 7.
Immunogold electron microscopic localization of endogenous Cavin-4 in C2C12 cells. (a) Expression of Cavin-1, -4, and Cav3 during C2C12 myotubes differentiation. Protein expression was analyzed by Western blotting at selected intervals after the initiation of differentiation. On longer exposures, low levels of Cavin-4 were also detected in myoblasts. Note that although in cardiac and skeletal muscle preparations Cavin-4 consistently migrates at 43 kD, in C2C12 myotubes, a major band is detected around 50 kD, possibly indicating a posttranslational modification in this cell type. (b) Myotubes (4 d) were methanol fixed, labeled for endogenous Cavin-4 (green) and Cav3 (red), and the top surface of the myotube was imaged using a confocal microscope. Insets show a magnified view of the selected areas. (c) Isolated muscle fibers were immunolabeled with anti–Cavin-4 antibody and analyzed by high resolution light microscopic analysis. Cavin-4 labeling was observed over the sarcolemma of the muscle with very low intracellular labeling. (d–f) C2C12 myotubes were immunolabeled for Cavin-4 using affinity-purified antibodies followed by 10 nm protein A gold. Specific labeling is associated with pits and vesicular profiles close to the plasma membrane (PM) with the typical morphology of caveolae. Arrows indicate gold particles labeling Cavin-4 at the plasma membrane. (g and h) Immuno-EM localization of Cavin-1 (g) and -4 (h) in cardiac muscle. A gallery of images showing regions of cardiac tissue containing both endothelial cells (Ec) and cardiac muscle (Cm). Cavin-1 is associated with caveolae of both endothelial cells and cardiac muscle (left, arrows). Note the high density of labeling associated with the cytoplasmic face of the caveolae (right, arrows). Cavin-4 is not detectable on endothelial caveolae but associates with vesicular profiles with caveolar morphology in cardiac muscle (arrows). Molecular masses (in parentheses) are indicated in kilodaltons. Bars: (b and c) 20 µm; (d–h) 100 nm.
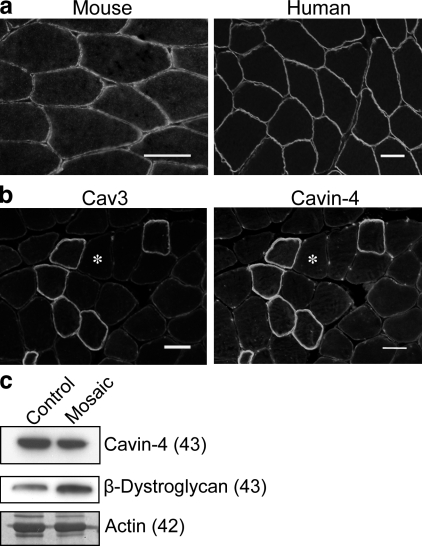
Changes in Cav3 are associated with muscle diseases termed caveolinopathies (for review see Woodman et al., 2004). We therefore investigated whether Cavin-4 is mislocalized in the muscle of these patients. In transverse sections of skeletal muscle from normal mouse and human samples, Cavin-4 localizes to the sarcolemmal membrane (Fig. 8 a). A patient with rippling muscle disease, who shows a mosaic pattern of Cav3 staining (unpublished data), shows a striking absence of Cavin-4 from the Cav3-negative fibers (Fig. 8 b). Cavin-1 showed a similar absence from Cav3-negative fibers (unpublished data). By Western blotting (Fig. 8 c), we observed a mild reduction (23%) in the levels of Cavin-4. We conclude that Cavin-4 is a new candidate protein for caveolin-associated muscle disease.
Figure 8.
Cavin-4 localization is perturbed in muscle disease. (a) Cryosections from mouse and human skeletal muscle were immunolabeled for Cavin-4, showing localization to sarcolemmal membrane. (b) Immunolabeling of Cav3 in a patient with rippling muscle disease (Cav3 mosaic) reveals markedly reduced Cav3 and Cavin-4 staining in a subpopulation of muscle fibers (indicated by asterisks). (c) Cavin-4 expression was reduced in muscle from Cav3 mosaic samples in comparison with control tissue. 10 µg muscle lysate was loaded. β-Dystroglycan expression and coomassie staining of actin demonstrate relative loading of muscle protein. Molecular masses (in parentheses) are indicated in kilodaltons. Bars, 50 µm.
Discussion
In this study, we provide the first description of a multimeric protein complex that associates with caveolae and regulates caveola formation. The cavin complex can be present in the cytosol, but it is recruited by caveolin to generate caveolae. PTRF/Cavin-1 is a crucial component for recruitment of this complex to caveolae, establishing a hierarchy of interactions required for caveola formation. Indeed, in cultured adipocytes, PTRF/Cavin-1 and Cav1 show identical temporal expression followed by that of SDR/Cavin-2, suggesting that the latter is recruited to caveolae after their formation at the cell surface. In addition, we have characterized a new member of the family, MURC/Cavin-4, and demonstrated both the caveolar localization of this protein and its redistribution in caveolin-associated muscle disease. The role of PTRF/Cavin-1 in caveola stability and/or formation and the density of the cavin proteins on the cytoplasmic face of caveolae, as shown by immuno-EM (Fig. 4 and Fig. 7), raises the possibility that the cavin complex acts as a coatlike structural component of caveolae.
This study has shown that cavins are dependent on caveolins for membrane association. Cavin family members show tissue-specific expression, pointing to regulation of the cavin complex at a transcriptional level, which in turn regulates caveola function. Strengthening the association and cofunction of the cavin proteins, we show that SDR/Cavin-2 and SRBC/Cavin-3 proteins are down-regulated in cavin-1 knockout mice and cell lines. Furthermore, in view of the differences in the expression of these proteins in different tissues, it is likely that the members of this family have functions outside the caveolar structure. Although cavins are dependent on caveolins for membrane association, we were able to detect similar levels of interaction between PTRF/Cavin-1 and the other cavin family members in the WT and Cav1−/− fibroblasts by FLIM/FRET, suggesting that the complex formation is not dependent on caveolins. By PTRF/Cavin-1 coimmunoprecipitation, we showed that the complex contained all the four cavin proteins in the cytosol with the additional binding of caveolin in the membrane fraction. This suggests a model in which cavin proteins constitutively associate in the cytosol and are recruited to membrane caveolae. Lipid binding is also likely to play a significant role in cavin association with caveolae through multiple PS-binding components with possible parallels to lipid (PIP2, in the AP2 complex) binding of adaptor complex components. The interaction of AP2 components with these lipids is of low affinity, but this weak binding is enhanced by other interactions including oligomerization to generate a complex with multiple lipid-interacting sites (Carlton and Cullen, 2005) as well as interactions with cargo proteins (Honing et al., 2005). These mutually stabilizing interactions cause the AP2 complex to act as a coincidence detector, relying on the multiple interactions to generate a high avidity interaction (Carlton and Cullen, 2005). In concordance with this hypothesis, MURC/Cavin-4 is also able to interact with PS in vitro (unpublished data) as previously shown for PTRF/Cavin-1, SDR/Cavin-2, and SRBC/Cavin-3 (Burgener et al., 1990; Izumi et al., 1997; Gustincich et al., 1999; Hill et al., 2008). Interestingly, caveolin peptides have been suggested to generate domains enriched in PS in a liposome-based system (Wanaski et al., 2003), a process that could further enhance the avidity of the cavin interaction with caveolin-rich domains. This model of coincidence detection also offers a mechanism to rapidly disassemble the caveolin–cavin complex upon modulation of one of the components, as suggested for phosphoinositide-based membrane complex formation (Carlton and Cullen, 2005). The association with surface caveolae is disrupted by cholesterol depletion, which perturbs caveolar structure (Hill et al., 2008). Collectively with the results of this study, these results suggest that the cavin complex can act to sense changes in caveolar structure or composition. The release of the cavin complex on caveolar disruption could then provide a signaling mechanism from the cell surface to intracellular compartments, a fascinating possibility in view of the strengthening links between caveolae and mechanosensation as well as potential nuclear functions (Ogata et al., 2008; Tagawa et al., 2008).
All four cavin proteins can be detected in skeletal muscle preparations (Fig. 2 a), but SRBC/Cavin-3 and MURC/Cavin-4 are highly expressed, suggesting a particularly important role in this tissue. MURC/Cavin-4 is evolutionarily conserved, and high throughput analysis of expression of proteins in the developing zebrafish embryo also suggests that a putative zebrafish orthologue is expressed predominantly in skeletal muscle (Thisse and Thisse, 2004). Recent studies provided the first insights into the function of MURC/Cavin-4 (Ogata et al., 2008; Tagawa et al., 2008). MURC/Cavin-4 was shown to activate the extracellular signal-regulated kinase pathway, influencing skeletal muscle differentiation, and to activate RhoA and regulate the Rho–ROCK pathway, modulating cardiac function. Overexpression of MURC/Cavin-4 in skeletal muscle and cardiac muscle promoted myogenesis and caused cardiac dysfunction and conduction disturbance, respectively. Consistent with this study, MURC/Cavin-4 was shown to interact with SDR/Cavin-2 (Ogata et al., 2008). MURC/Cavin-4 was suggested to be a purely cytosolic protein in muscle, but our results, using multiple independent techniques, including immunogold EM of the endogenous protein, show the caveolar localization of the protein. MURC/Cavin-4 is enriched in cardiac and skeletal muscle but is clearly present at low levels in other cell types, such as embryonic fibroblasts, as shown in this study. We have also shown that MURC/Cavin-4 is a new candidate protein for caveolin-related muscle disease. Muscle from a patient with mosaic expression of Cav3 shows a striking loss of sarcolemmal MURC/Cavin-4 in the Cav3-negative fibers. As a possible sensor of caveolar form, study of the cavin complex in muscle diseases of unknown etiology may provide insights into caveolinopathies where mutations in Cav3 are not the primary defect. In addition, these experiments raise the possibility, which is being tested at present, that mutations in MURC/Cavin-4 directly cause muscle disease.
The identification of a possible caveola coat complex, as shown in this study, will provide new insights into the generation and function of this newly discovered molecular assembly. Whether the cavin complex shows any similarity in basic design principles and in the mechanisms of assembly and disassembly to other well-characterized coat complexes is now amenable to analysis.
Materials and methods
Antibodies and reagents
Monoclonal antibodies recognizing Cavin-1 (Vinten et al., 2001), Cav1 (Souto et al., 2003), and Glut4 (James et al., 1988) have been previously described. Antibodies against Cav1-3 and Cavin-1 were obtained from BD. Antibodies against Cavin-1 to -4 were raised in rabbits using keyhole limpet hemocyanin–conjugated peptides and affinity purified. The following peptides from Cavin-1, -2, -3, and -4 were used, respectively: Ac-CTEESDAVLVDKSDSD, Ac-AEKFQHPNTDMLQEKPS, Ac-SDEEPVESRAQRLRRTGC (21st Century Biochemicals), and Ac-MEHNGSASNADKIHQGC (Auspep Pty. Ltd.). All other reagents were obtained from Sigma-Aldrich unless stated otherwise.
Sequence and phylogenetic analysis
Cavin protein sequences were gathered from the National Center for Biotechnology Information and aligned using MacVector 9.0.2 (MacVector, Inc.) using the entire sequence; phylogenetic trees were generated with the same program using the neighbor joining method. The tree was midpoint rooted, and numbers generated represent a percentage of 1,000 bootstrap trials that support the branch.
DNA constructs
Murine Cavin-1 constructs were described previously in Hill et al. (2008). Murine Cavin-2 (Fantom 2 clone 9530015P22), Cavin-3 (amplified from NIH-3T3 fibroblast cDNA), and Cavin-4 (Integrated Molecular Analysis of Genomes and their Expression clone MGC 107637) were cloned in pEGFP vectors. N- or C-terminal tagging gave similar results, and constructs showing the highest expression levels were chosen for the experiments.
Cell culture
Cell culture, transfection, and flow cytometry were performed as previously described (Carozzi et al., 2000; Hill et al., 2008; Kirkham et al., 2008; Liu and Pilch, 2008). PC3 cells stably expressing cavins were generated by transfecting GFP-tagged cavin constructs with Lipofectamine LTX (Invitrogen) and selected with G418 (Invitrogen). Muscle fibers were isolated as described previously (Hernandez-Deviez et al., 2006). Knockdown of Cavin-2 in 3T3-L1 fibroblasts was achieved using shRNA and lentivirally expressed RNA fragments against Cavin-2 or EGFP then selected with puromycin selection. Target sequences can be provided on request.
Immunofluorescence microscopy
Cultured cells were analyzed as described previously in Kirkham et al. (2008). Image acquisition of cells expressing GFP or Cherry fluorochromes or indirectly labeled with Alexa fluorophores (Invitrogen) was performed at room temperature in Acqua-Poly Mount medium (Polysciences, Inc.) using a confocal microscope with photomultiplier tube detectors (LSM 510 Meta; Carl Zeiss, Inc.) equipped with a 63x oil immersion objective (Carl Zeiss, Inc.). The data were captured using the LSM 510 Meta software (Carl Zeiss, Inc.), and unprocessed images were assembled using Photoshop (CS3; Adobe). Muscle specimens were snap frozen in isopentane-cooled liquid N2. Eight micron cryosections of muscle were fixed in 4% PFA followed by blocking in 2% BSA (in PBS). Sections were incubated with primary (overnight at 4°C) and secondary antibodies (1 h at room temperature) diluted in block solution. All staining in human samples was performed in comparison with normal age-matched muscle biopsies. Samples were obtained with approval from the Ethics Committee at The Children's Hospital at Westmead (Sydney, New South Wales, Australia). FLIM/FRET experiments were performed as described previously (Hill et al., 2008) with the indicated combinations of constructs transfected into BHK or iMEFs.
Biochemical methods
Tissue lysates from WT and Cavin-1−/− mouse tissues were prepared in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, and 0.2% sodium dodecyl sulfate) immediately supplemented with Laemmli's sample buffer.
Coimmunoprecipitation procedures were performed at 4°C. Whole cell lysates or plasma membrane fractions from primary adipocytes (Liu and Pilch, 2008) were solubilized in 1% Triton X-100, 150 mM NaCl, and 50 mM EDTA for 2 h with constant agitation. Insoluble material was removed by pelleting for 10 min in a microcentrifuge. Specific antibodies and nonspecific mouse or rabbit IgGs (5 µg) were incubated with the supernatant for 1 h, and 20 µl protein A beads (Santa Cruz Biotechnology, Inc.) was added for an additional hour. The beads were washed four times and eluted with SDS-PAGE loading buffer. The silver staining of the gels, the in-gel digestion of protein bands, and the identification of the proteins by mass spectrometry were performed as previously described (Pilch et al., 2007).
For experiments using WT or Cav1−/− iMEFs, cells harvested in buffer A (10 mM Tris, pH 7.4, 10 mM KCl, 10 mM NaF, 1 mM sodium pyrophosphate, 0.5 mM Na3VO4, 1 mg/ml leupeptin, and 1 mg/ml aprotinin) were disrupted using a dounce homogenizer and centrifuged at 2,000 g for 5 min. The supernatant was centrifuged at 100,000 g for 30 min to generate a cytosolic S100 fraction and a membrane P100 pellet. The fractions were adjusted to 50 mM NaCl and 0.1% NP-40, with the P100 being first solubilized in buffer A containing 250 mM NaCl and 0.5% NP-40. Fractions were incubated with protein A dynabeads precoupled with Cavin-1 antibody and rotated for 90 min. Beads were washed three times, and proteins bound were eluted using SDS-PAGE sample buffer without DTT and analyzed by Western blotting (Hill et al., 2008).
Quantitative RT-PCR
RNA was extracted using RNAeasy (QIAGEN), and 4–5 µg was reversely transcribed. Quantitative RT-PCR was performed in triplicate on three independent RNA preparations. cDNA levels were analyzed in PCR reactions with SYBR Green Technologies (Applied Biosystems), and the relative level of expression was normalized using 18S ribosomal RNA. Statistical analysis was performed on the average of three independent assays using Student's t test. Primer sequences can be provided on request.
EM analysis
Immuno-EM on adipocyte plasma membrane sheets was performed as described previously in Vinten et al. (2001). EM analysis of caveola formation and quantification in PC3 cells was performed as described previously in Hill et al. (2008). For localization of Cavin-4, C2C12 myotubes and mouse skeletal (quadriceps) or cardiac muscle was fixed with 4% PFA and processed for immuno-EM as described previously (Hill et al., 2008). Ultrathin sections were labeled using affinity-purified antibodies to Cavin-4 followed by 10 nm protein A gold.
Online supplemental material
Fig. S1 shows native gel immunoblotting for Cavin-1 and -4, immunoblotting of GFP-tagged cavins stably expressed in PC3 cells, and colocalization of Cavin-1 and Cav1 in PC3 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903053/DC1.
Acknowledgments
We are grateful to Charles Ferguson and Nicole Schieber for assistance with EM. Confocal microscopy was performed at the Australian Cancer Research Foundation (ACRF)/Institute for Molecular Bioscience (IMB) Dynamic Imaging Facility for Cancer Biology, established with funding from the ACRF. The authors acknowledge the use of the Australian Microscopy and Microanalysis Facility at the Center for Microscopy and Microanalysis at The University of Queensland. The IMB is a Special Research Center of the Australian Research Council.
This work was supported by grants from the National Health and Medical Research Council of Australia (to R.G. Parton and J.F. Hancock), the Australian Research Council (to R.G. Parton), and from the National Institutes of Health (DK30425 and DK56935 to P.F. Pilch; training grant DK07201 to M.R. Breen). L. Liu is supported by a Pilot and Feasibility award from the Boston Obesity Nutrition Research Center (DK046200). D. Abankwa (PAOOA-111446) and P.J. Walser (PAOOA-109094) were supported by the Swiss National Science Foundation.
Footnotes
Abbreviations used in this paper: Cav1, Caveolin-1; Cav3, Caveolin-3; FLIM, fluorescence lifetime imaging microscopy; FRET, fluorescence resonance energy transfer; iMEF, immortalized mouse embryonic fibroblast; MURC, muscle-restricted coiled-coil protein; PS, phosphatidylserine; PTRF, polymerase I and transcript release factor; SDR, serum deprivation response; shRNA, short hairpin RNA; SRBC, SDR-related gene product that binds to C kinase; WT, wild type.
References
- Aboulaich N., Vainonen J.P., Stralfors P., Vener A.V. 2004. Vectorial proteomics reveal targeting, phosphorylation and specific fragmentation of polymerase I and transcript release factor (PTRF) at the surface of caveolae in human adipocytes.Biochem. J. 383:237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum M.J. 1989. Identification of a novel gene encoding an insulin-responsive glucose transporter protein.Cell. 57:305–315 [DOI] [PubMed] [Google Scholar]
- Burgener R., Wolf M., Ganz T., Baggiolini M. 1990. Purification and characterization of a major phosphatidylserine-binding phosphoprotein from human platelets.Biochem. J. 269:729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozza F., Combs T.P., Cohen A.W., Cho Y.R., Park S.Y., Schubert W., Williams T.M., Brasaemle D.L., Jelicks L.A., Scherer P.E., et al. 2005. Caveolin-3 knockout mice show increased adiposity and whole body insulin resistance, with ligand-induced insulin receptor instability in skeletal muscle.Am. J. Physiol. Cell Physiol. 288:C1317–C1331 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Cullen P.J. 2005. Coincidence detection in phosphoinositide signaling.Trends Cell Biol. 15:540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. 2005. The transcriptional landscape of the mammalian genome.Science. 309:1559–1563 [DOI] [PubMed] [Google Scholar]
- Carozzi A.J., Ikonen E., Lindsay M.R., Parton R.G. 2000. Role of cholesterol in developing T-tubules: analogous mechanisms for T-tubule and caveolae biogenesis.Traffic. 1:326–341 [DOI] [PubMed] [Google Scholar]
- Cheng Z.J., Singh R.D., Marks D.L., Pagano R.E. 2006. Membrane microdomains, caveolae, and caveolar endocytosis of sphingolipids.Mol. Membr. Biol. 23:101–110 [DOI] [PubMed] [Google Scholar]
- Cohen A.W., Razani B., Schubert W., Williams T.M., Wang X.B., Iyengar P., Brasaemle D.L., Scherer P.E., Lisanti M.P. 2004. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation.Diabetes. 53:1261–1270 [DOI] [PubMed] [Google Scholar]
- Drab M., Verkade P., Elger M., Kasper M., Lohn M., Lauterbach B., Menne J., Lindschau C., Mende F., Luft F.C., et al. 2001. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice.Science. 293:2449–2452 [DOI] [PubMed] [Google Scholar]
- Fernandez M.A., Albor C., Ingelmo-Torres M., Nixon S.J., Ferguson C., Kurzchalia T., Tebar F., Enrich C., Parton R.G., Pol A. 2006. Caveolin-1 is essential for liver regeneration.Science. 313:1628–1632 [DOI] [PubMed] [Google Scholar]
- Fra A.M., Williamson E., Simons K., Parton R.G. 1995. De novo formation of caveolae in lymphocytes by expression of VIP21-caveolin.Proc. Natl. Acad. Sci. USA. 92:8655–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Agarwal A.K. 2008. Caveolin-1: a new locus for human lipodystrophy.J. Clin. Endocrinol. Metab. 93:1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Kehinde O. 1976. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells.Cell. 7:105–113 [DOI] [PubMed] [Google Scholar]
- Gustincich S., Schneider C. 1993. Serum deprivation response gene is induced by serum starvation but not by contact inhibition.Cell Growth Differ. 4:753–760 [PubMed] [Google Scholar]
- Gustincich S., Vatta P., Goruppi S., Wolf M., Saccone S., Della Valle G., Baggiolini M., Schneider C. 1999. The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein.Genomics. 57:120–129 [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez D.J., Martin S., Laval S.H., Lo H.P., Cooper S.T., North K.N., Bushby K., Parton R.G. 2006. Aberrant dysferlin trafficking in cells lacking caveolin or expressing dystrophy mutants of caveolin-3.Hum. Mol. Genet. 15:129–142 [DOI] [PubMed] [Google Scholar]
- Hill M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., Walser P., Abankwa D., Oorschot V.M., Martin S., et al. 2008. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function.Cell. 132:113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honing S., Ricotta D., Krauss M., Spate K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D.J. 2005. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2.Mol. Cell. 18:519–531 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Hirai S., Tamai Y., Fujise-Matsuoka A., Nishimura Y., Ohno S. 1997. A protein kinase Cdelta-binding protein SRBC whose expression is induced by serum starvation.J. Biol. Chem. 272:7381–7389 [DOI] [PubMed] [Google Scholar]
- James D.E., Brown R., Navarro J., Pilch P.F. 1988. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein.Nature. 333:183–185 [DOI] [PubMed] [Google Scholar]
- Jansa P., Mason S.W., Hoffmann-Rohrer U., Grummt I. 1998. Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes.EMBO J. 17:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa P., Burek C., Sander E.E., Grummt I. 2001. The transcript release factor PTRF augments ribosomal gene transcription by facilitating reinitiation of RNA polymerase I.Nucleic Acids Res. 29:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandror K.V., Stephens J.M., Pilch P.F. 1995. Expression and compartmentalization of caveolin in adipose cells: coordinate regulation with and structural segregation from GLUT4.J. Cell Biol. 129:999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.A., Delepine M., Boutet E., El Mourabit H., Le Lay S., Meier M., Nemani M., Bridel E., Leite C.C., Bertola D.R., et al. 2008. Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy.J. Clin. Endocrinol. Metab. 93:1129–1134 [DOI] [PubMed] [Google Scholar]
- Kirkham M., Nixon S.J., Howes M.T., Abi-Rached L., Wakeham D.E., Hanzal-Bayer M., Ferguson C., Hill M.M., Fernandez-Rojo M., Brown D.A., et al. 2008. Evolutionary analysis and molecular dissection of caveola biogenesis.J. Cell Sci. 121:2075–2086 [DOI] [PubMed] [Google Scholar]
- Le Lay S., Hajduch E., Lindsay M.R., Le Liepvre X., Thiele C., Ferre P., Parton R.G., Kurzchalia T., Simons K., Dugail I. 2006. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis.Traffic. 7:549–561 [DOI] [PubMed] [Google Scholar]
- Lee S.W., Reimer C.L., Oh P., Campbell D.B., Schnitzer J.E. 1998. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells.Oncogene. 16:1391–1397 [DOI] [PubMed] [Google Scholar]
- Liu L., Pilch P.F. 2008. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization.J. Biol. Chem. 283:4314–4322 [DOI] [PubMed] [Google Scholar]
- Liu L., Brown D., McKee M., Lebrasseur N.K., Yang D., Albrecht K.H., Ravid K., Pilch P.F. 2008. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance.Cell Metab. 8:310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S.W., Sander E.E., Grummt I. 1997. Identification of a transcript release activity acting on ternary transcription complexes containing murine RNA polymerase I.EMBO J. 16:163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo C., James G.L., Smart E.J., Anderson R.G. 1996. Localization of epidermal growth factor-stimulated Ras/Raf-1 interaction to caveolae membrane.J. Biol. Chem. 271:11930–11935 [DOI] [PubMed] [Google Scholar]
- Mineo C., Ying Y.S., Chapline C., Jaken S., Anderson R.G. 1998. Targeting of protein kinase Cα to caveolae.J. Cell Biol. 141:601–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S., Dietzen D.J., Hastings W.R., Lublin D.M., Kurzchalia T.V. 1996. Oligomerization of VIP21-caveolin in vitro is stabilized by long chain fatty acylation or cholesterol.FEBS Lett. 388:143–149 [DOI] [PubMed] [Google Scholar]
- Ogata T., Ueyama T., Isodono K., Tagawa M., Takehara N., Kawashima T., Harada K., Takahashi T., Shioi T., Matsubara H., Oh H. 2008. MURC, a muscle-restricted coiled-coil protein that modulates the Rho/ROCK pathway, induces cardiac dysfunction and conduction disturbance.Mol. Cell. Biol. 28:3424–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Simons K. 2007. The multiple faces of caveolae.Nat. Rev. Mol. Cell Biol. 8:185–194 [DOI] [PubMed] [Google Scholar]
- Parton R.G., Way M., Zorzi N., Stang E. 1997. Caveolin-3 associates with developing T-tubules during muscle differentiation.J. Cell Biol. 136:137–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L., Zerial M. 2005. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae.Nature. 436:128–133 [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Burli T., Zerial M., Helenius A. 2004. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic.Cell. 118:767–780 [DOI] [PubMed] [Google Scholar]
- Pilch P.F., Souto R.P., Liu L., Jedrychowski M.P., Berg E.A., Costello C.E., Gygi S.P. 2007. Cellular spelunking: exploring adipocyte caveolae.J. Lipid Res. 48:2103–2111 [DOI] [PubMed] [Google Scholar]
- Pol A., Martin S., Fernandez M.A., Ingelmo-Torres M., Ferguson C., Enrich C., Parton R.G. 2005. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies.Mol. Biol. Cell. 16:2091–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B., Combs T.P., Wang X.B., Frank P.G., Park D.S., Russell R.G., Li M., Tang B., Jelicks L.A., Scherer P.E., Lisanti M.P. 2002. Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hypertriglyceridemia with adipocyte abnormalities.J. Biol. Chem. 277:8635–8647 [DOI] [PubMed] [Google Scholar]
- Scherer P.E., Lisanti M.P., Baldini G., Sargiacomo M., Mastick C.C., Lodish H.F. 1994. Induction of caveolin during adipogenesis and association of GLUT4 with caveolin-rich vesicles.J. Cell Biol. 127:1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souto R.P., Vallega G., Wharton J., Vinten J., Tranum-Jensen J., Pilch P.F. 2003. Immunopurification and characterization of rat adipocyte caveolae suggest their dissociation from insulin signaling.J. Biol. Chem. 278:18321–18329 [DOI] [PubMed] [Google Scholar]
- Tagawa A., Mezzacasa A., Hayer A., Longatti A., Pelkmans L., Helenius A. 2005. Assembly and trafficking of caveolar domains in the cell: caveolae as stable, cargo-triggered, vesicular transporters.J. Cell Biol. 170:769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa M., Ueyama T., Ogata T., Takehara N., Nakajima N., Isodono K., Asada S., Takahashi T., Matsubara H., Oh H. 2008. MURC, a muscle-restricted coiled-coil protein, is involved in the regulation of skeletal myogenesis.Am. J. Physiol. Cell Physiol. 295:C490–C498 [DOI] [PubMed] [Google Scholar]
- Tahir S.A., Yang G., Goltsov A.A., Watanabe M., Tabata K., Addai J., Fattah el M.A., Kadmon D., Thompson T.C. 2008. Tumor cell-secreted caveolin-1 has proangiogenic activities in prostate cancer.Cancer Res. 68:731–739 [DOI] [PubMed] [Google Scholar]
- Thisse B., Thisse C. 2004. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. Available at: http://zfin.org (accessed August 25, 2008)
- Vinten J., Voldstedlund M., Clausen H., Christiansen K., Carlsen J., Tranum-Jensen J. 2001. A 60-kDa protein abundant in adipocyte caveolae.Cell Tissue Res. 305:99–106 [DOI] [PubMed] [Google Scholar]
- Vinten J., Johnsen A.H., Roepstorff P., Harpoth J., Tranum-Jensen J. 2005. Identification of a major protein on the cytosolic face of caveolae.Biochim. Biophys. Acta. 1717:34–40 [DOI] [PubMed] [Google Scholar]
- Voldstedlund M., Vinten J., Tranum-Jensen J. 2001. cav-p60 expression in rat muscle tissues. Distribution of caveolar proteins.Cell Tissue Res. 306:265–276 [DOI] [PubMed] [Google Scholar]
- Voldstedlund M., Thuneberg L., Tranum-Jensen J., Vinten J., Christensen E.I. 2003. Caveolae, caveolin and cav-p60 in smooth muscle and renin-producing cells in the rat kidney.Acta Physiol. Scand. 179:179–188 [DOI] [PubMed] [Google Scholar]
- Wanaski S.P., Ng B.K., Glaser M. 2003. Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains.Biochemistry. 42:42–56 [DOI] [PubMed] [Google Scholar]
- Way M., Parton R.G. 1995. M-caveolin, a muscle-specific caveolin-related protein.FEBS Lett. 376:108–112 [DOI] [PubMed] [Google Scholar]
- Woodman S.E., Sotgia F., Galbiati F., Minetti C., Lisanti M.P. 2004. Caveolinopathies: mutations in caveolin-3 cause four distinct autosomal dominant muscle diseases.Neurology. 62:538–543 [DOI] [PubMed] [Google Scholar]
- Xu X.L., Wu L.C., Du F., Davis A., Peyton M., Tomizawa Y., Maitra A., Tomlinson G., Gazdar A.F., Weissman B.E., et al. 2001. Inactivation of human SRBC, located within the 11p15.5-p15.4 tumor suppressor region, in breast and lung cancers.Cancer Res. 61:7943–7949 [PubMed] [Google Scholar]
- Yao Q., Chen J., Cao H., Orth J.D., McCaffery J.M., Stan R.V., McNiven M.A. 2005. Caveolin-1 interacts directly with dynamin-2.J. Mol. Biol. 348:491–501 [DOI] [PubMed] [Google Scholar]