Sculpting the Proteome with AAA+ Proteases and Disassembly Machines (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 28.
Abstract
Machines of protein destruction—including energy-dependent proteases and disassembly chaperones of the AAA+ ATPase family—function in all kingdoms of life to sculpt the cellular proteome, ensuring that unnecessary and dangerous proteins are eliminated and biological responses to environmental change are rapidly and properly regulated. Exciting progress has been made in understanding how AAA+ machines recognize specific proteins as targets and then carry out ATP-dependent dismantling of the tertiary and/or quaternary structure of these molecules during the processes of protein degradation and the disassembly of macromolecular complexes.
Proteases and disassembly chaperones are nature’s implements of protein destruction and recycling. Just as the creation of a metazoan body plan involves removing specific cells by apoptosis, cells use ATP-powered proteases and disassembly machines to remove unwanted proteins and to dismantle macromolecular complexes. Enzymes of the AAA+ ATPase family play critical roles in this sculpting of the proteome in all organisms. For example, the proteasome and related energy-dependent proteases, as well as NSF, p97, and Hsp104, all employ AAA+ ATPases for reactions that involve protein remodeling, unfolding, or disaggregation. AAA+ ATPases also mediate the mechanical activities of many different classes of enzymes including helicases, motor proteins, clamp loaders, protein and DNA translocases, and the rotary F1F0 ATP synthase.
This review will focus on the mechanism and regulation of ATP-dependent protein degradation, unfolding, and disassembly in bacteria, where these processes are currently best understood. There are at least five ATP-dependent proteases in bacteria (for review, see Gottesman, 2003). These proteases, which have orthologs in mitochondria and chloroplasts, function as large macromolecular machines, consisting of multimeric peptidases and AAA+ ATPases. The peptidase and ATPase activities can reside on separate polypeptides or a single chain depending on the enzyme. AAA+ ATPases can also function independently of peptidases as disassembly chaperones to dismantle protein complexes and solubilize aggregates (for reviews, see Hoskins et al., 2001; Glover and Tkach, 2001).
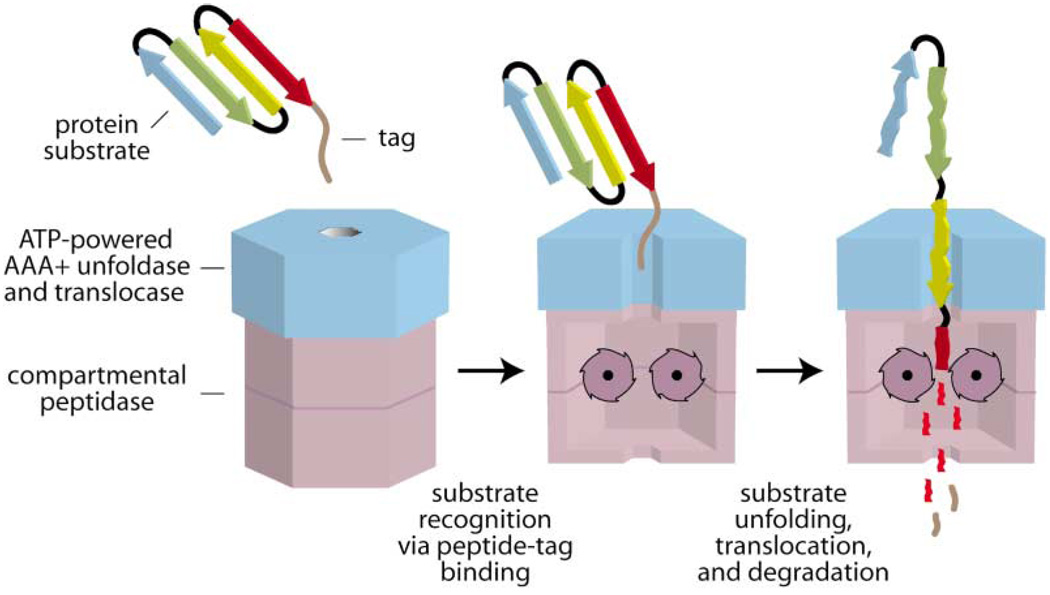
A bacterial cell, such as Escherichia coli, contains a total of about 3 million protein molecules representing the products of thousands of different genes. Because AAA+ proteases and disassembly chaperones destroy other proteins and protein complexes, the activities of these enzymes must be carefully controlled. As shown in the model of Figure 1, this control is largely accomplished at the level of substrate selection. Different AAA+ ATPases have distinct preferences for binding to short peptide sequences, called tags or recognition signals, often located near the N or C terminus of a target protein. Specialized adaptor or delivery proteins also modulate substrate specificity for these enzymes. For AAA+ proteases, native protein substrates are bound by the ATPase, which then catalyzes unfolding of this molecule and translocation of the denatured polypeptide into an associated peptidase for degradation (Figure 1). These protein unfoldase and protein translocation activities are key to the function of AAA+ proteases and are also involved in some peptidase-independent disassembly reactions. In the sections below, we first discuss the architectures of some of the best-characterized AAA+ proteases. We then return to questions of how substrates are recognized and then disassembled and/or degraded, emphasizing insights based on a few well-studied systems.
Figure 1. Cartoon Model of Substrate Recognition and Degradation by a AAA+ Protease.
The recognition step is mediated by binding of a peptide tag (brown) on the protein substrate to a AAA+ ATPase (blue). In subsequent steps, the protein is unfolded, translocated into a compartmental peptidase (magenta), and degraded. Peptide fragments are shown diffusing out of the peptidase, but active participation of the ATPase may be required for exit of large fragments.
Compartmental Proteases: Control of Substrate Access and Product Exit
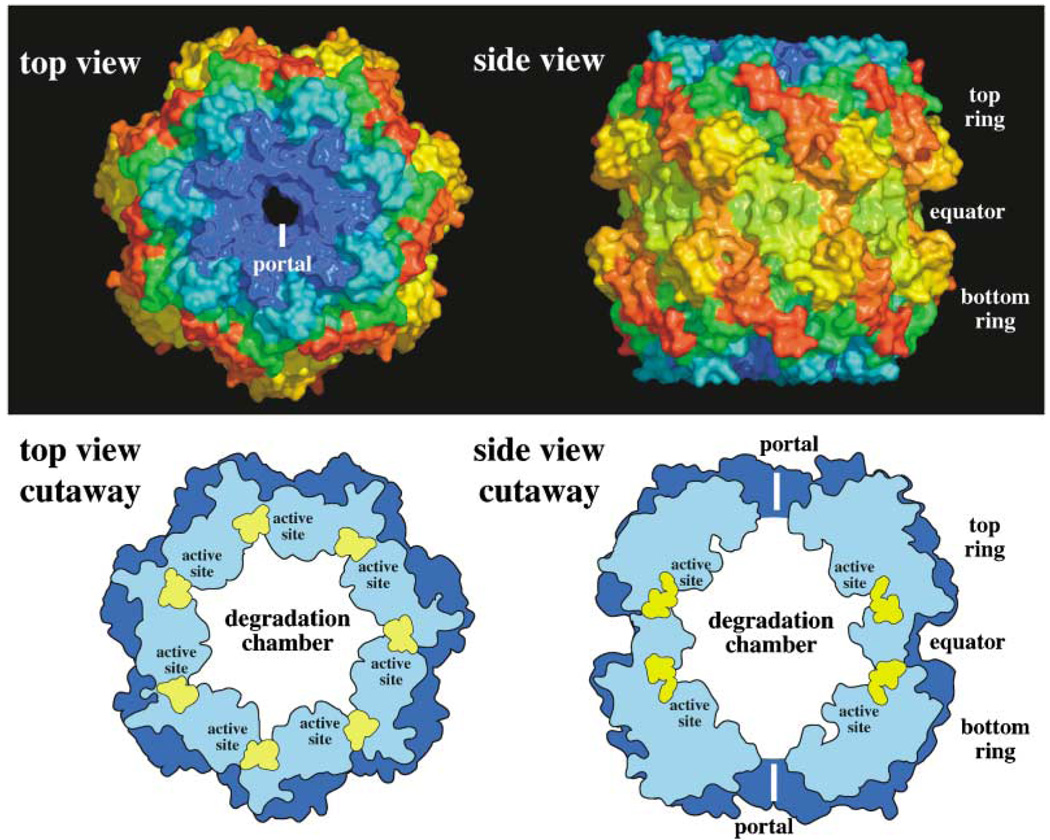
The face-to-face stacking of two radially symmetric rings creates compartmental peptidases (Wang et al., 1997; Bochtler et al., 1997). This architecture results in a hollow barrel-shaped multimer, with the proteolytic active sites in an internal chamber, as illustrated for ClpP and HslV in Figure 2 and Figure 3. At the symmetry axis of each ring, a narrow entry portal provides access to the proteolytic chamber. Even small native proteins have folded dimensions larger than the portal, and thus proteins in their native three-dimensional forms are excluded from inadvertent contact with the protease active sites. The 20S proteasome peptidases of archaea and eukaryotes are constructed in a similar fashion to the bacterial compartmental peptidases (Löwe et al., 1995; Groll et al., 1997). Each of these enzymes uses the same strategy of internal sequestration of its active sites, although differences exist in active-site chemistry, subunit structure, and ring architecture.
Figure 2. Structural Views of the ClpP Compartmental Peptidase.
The figure is based on the structure reported in Wang et al., (1997). In the top panel, each of the 14 identical subunits of ClpP is shown colored from blue to red from the N terminus to the C terminus. The 7-fold symmetry of a single ClpP ring is evident in the top view, which also shows the entry/exit portal. The side view shows the face-to-face stacking of both ClpP rings. The bottom panel shows cutaway diagrams that illustrate the positions of the active site residues (yellow) within the degradation chamber.
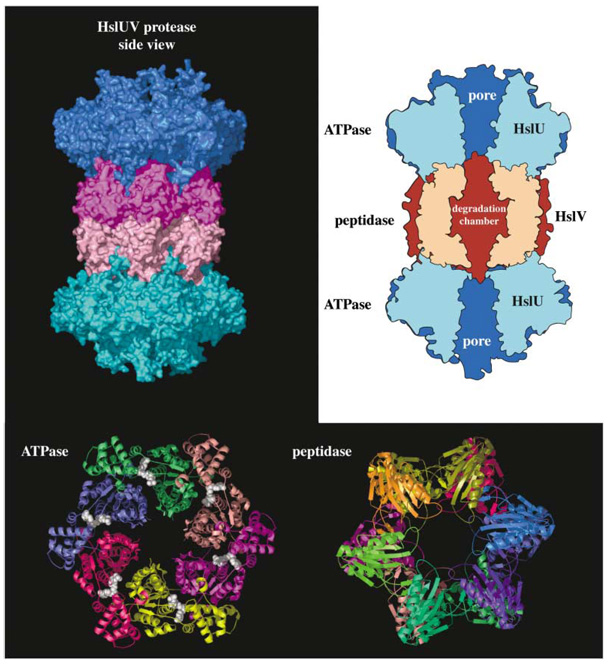
Figure 3. Structural Views of the HslUV Protease and Its Components.
The figure is based on the structure reported in Sousa et al. (2000). The top left panel shows a surface representation with individual hexameric ringsofthe HslU ATPase colored blue/cyan and the HslV peptidase colored magenta/pink. The top right panel is a cutaway diagram showing the positions of the pore through HslU and the degradation chamber within HslV. The bottom panel shows axial views of the ATPase and peptidase in ribbon representation with individual subunits in different colors. ATP molecules (white) are shown in CPK representation.
By themselves, the ClpP and HslV peptidases do not degrade native proteins or even unfolded peptides whose dimensions preclude diffusion through their entry portals (Thompson and Maurizi, 1994; Thompson et al., 1994; Seol et al., 1997). Degradation of these substrates requires collaboration with a AAA+ ATPase. For example, HslV partners with HslU (Figure 3), and ClpP works with either the ClpX or ClpA ATPases. These AAA+ ATPases bind appropriate substrates, denature them if necessary, and translocate the polypeptide into the peptidase (Figure 1). The portal in free ClpP is large enough to allow passage of only a single polypeptide (Wang et al., 1997), but several chains can be translocated concurrently during ClpXP degradation of proteins containing disulfide bonds (Burton et al., 2001a). Hence, the ATPase must increase the size of the peptidase portal in some fashion. Likewise, some activators of the proteasome increase the diameter of the portal to allow substrate entry or product exit (Whitby et al., 2000).
Once substrates are translocated into the degradation chamber of ClpP or HslV, the high concentration of active sites (hundreds of mM) ensures that most polypeptides are cleaved multiple times, typically generating peptide fragments from 5–20 residues in length (Thompson et al., 1994; Nishii and Takahashi, 2003). Very small cleavage products probably diffuse out of the degradation chamber, but efficient export of longer peptides requires the partner ATPase. For example, release of translocated polypeptides trapped in the degradation chamber of an inactive mutant ClpP depends on ATP hydrolysis by ClpX (Kim et al., 2000). Whether egress of larger cleavage fragments simply requires widening of the portal or active transport by the ATPase is an open question.
AAA+ ATPases Contain a Common Motor Module
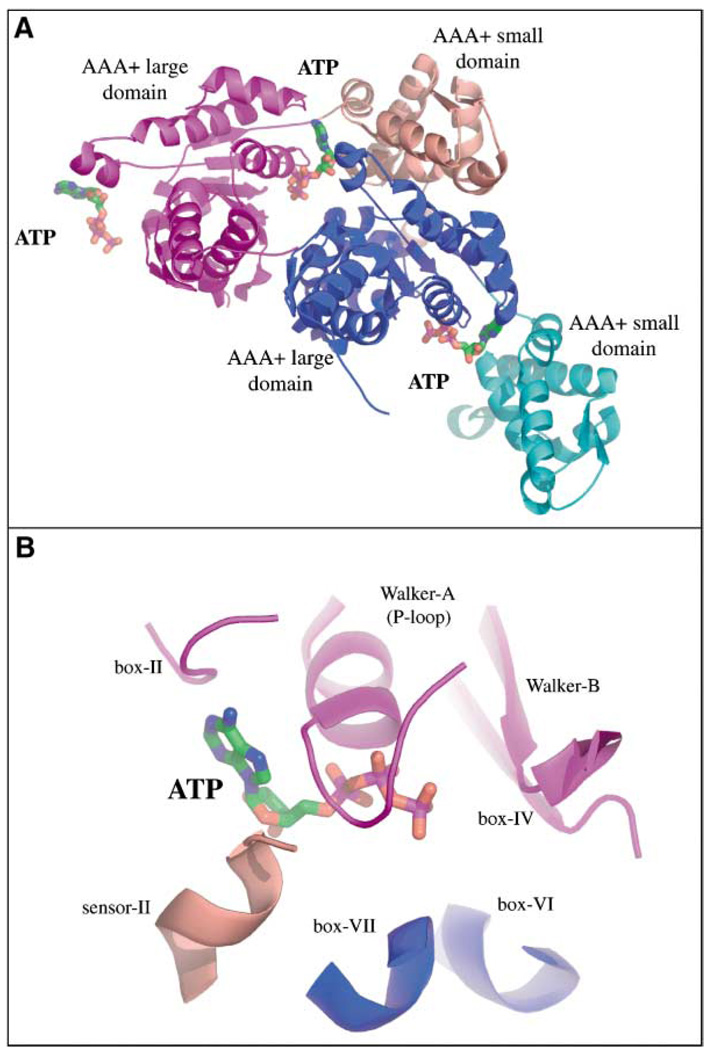
The bacterial AAA+ enzymes that function in proteolysis and disassembly are ring hexamers, containing one or two ATPase modules per polypeptide (Schirmer et al., 1996; Neuwald et al., 1999). These AAA+ ATPase modules are the motors that drive the mechanical processes of denaturation, disassembly, and translocation. Figure 3 and Figure 4 show the structure of the hexamer, individual subunits, and the nucleotide binding site for the HslU ATPase. Each AAA+ module consists of a large αβ domain followed by a smaller predominantly α-helical domain (Bochtler et al., 2000; Sousa et al., 2000; Wang et al., 2001). The ATP binding site, which is formed largely by sequence motifs that define the AAA+ family, is positioned both at an interface between the large and small AAA+ domains of a single subunit and at a subunit-subunit interface between the large domains (Figure 4). This intimate architecture allows ATP binding and/or hydrolysis to alter both the conformation of a single subunit and the overall conformation of the hexamer (see Wang et al., 2001).
Figure 4. Nucleotide Binding Sites in the HslU ATPase.
(A) Ribbon representation of two adjacent HslU subunits from a hexameric ring; stick representation of three ATP molecules. The large AAA+ domains are colored magenta and blue and the small AAA+ domains are colored pink and cyan. In this view, only the central ATP has a full complement of contacts, which are made from two large domains and one small domain.
(B) Closeup view of the central ATP binding site. The box-II, Walker-A, box-IV, Walker-B, box-VI, box-VII, and sensor-II sequence motifs are found in all AAA+ ATPases (Schirmer et al., 1996; Neuwald et al., 1999).
AAA+ ATPases that function in protein degradation and disassembly also contain distinct subfamily-specific domains (Schirmer et al., 1996). Some of these subfamily-specific domains contribute to substrate recognition by providing docking sites for adaptor or delivery proteins (see below). However, ATPase variants missing these specialized domains still recognize, denature, and translocate some substrates, showing that hexameric rings consisting of just the core AAA+ module catalyze all of its core processing activities.
For any machine, an energy source must be transformed into the mechanical movements that allow useful work to be performed. For AAA+ machines, the cycle of ATP binding, hydrolysis, and ADP/Pi release drives a coupled cycle of conformational changes in the enzyme. For example, nucleotide-dependent movements in the HslU ATPase involve flexing and rotating between the large and small AAA+ core domains with concomitant changes in the conformation of the HslU hexamer and its interactions with its peptidase partner (Sousa et al., 2000; Wang et al., 2001). There currently are no high-resolution structures of AAA+ ATPases with bound protein substrates, and it remains to be determined how changes in enzyme structure couple the ATP hydrolysis cycle to the mechanical processes of substrate disassembly, denaturation, and translocation.
ATPase-Peptidase Communication
Proteolytic AAA+ ATPases have shapes reminiscent of a hexagonal nut with the top and bottom surfaces connected by a central pore. One surface of the ATPase binds the protease, aligning the protease entry portal with the ATPase pore and providing a path for translocation of unfolded substrates through the ATPase and into the degradation chamber (Figure 1 and Figure 3; Ortega et al., 2000; Sousa et al., 2000). Active proteases can contain a single ATPase hexamer per protease molecule (as illustrated in Figure 1) or contain two hexamers, each docked with a portal on opposite sides of the peptidase (Figure 3). In the HslUV protease, both the ATPase and peptidase rings are hexameric and their interactions are symmetric (Figure 3). In ClpXP or ClpAP, by contrast, the ATPase is hexameric and the peptidase ring is heptameric, giving rise to an asymmetric interaction (Wang et al., 1997; Beuron et al., 1998).
Dynamic interactions between the ATPase and peptidase mediate functional communication during the ATPase cycle and substrate processing (Thompson et al., 1994; Yoo et al., 1996; Seol et al., 1997; Sousa et al., 2000, 2002; Kim et al., 2001; Wang et al., 2001; Ramachandran et al., 2002; Seong et al., 2002; Joshi et al., 2004). For instance, the interface between the ATPase and peptidase of HslUV changes depending on whether ATP or ADP is bound. Moreover, binding of HslU affects entry-portal gating and stimulates HslV’s peptidase activity. In ClpXP and ClpAP, flexible loops in the ATPase both connect to the nucleotide binding site and contact the peptidase. These interactions couple ATPase-pepti-dase affinityto changesinthe ATP hydrolysis rate during the processing of protein substrates. It is notable that many aspects of ATPase-peptidase communication are conserved between ClpXP and HslUV, despite dramatic structural differences in their peptidases and the interactions between their peptidases and ATPases. Communication between the ATPase and the peptidase is clearly an important common mechanism in the coordination of substrate import into and product export from the degradation chamber.
Peptide Signals Mediate Substrate Recognition and Tethering
The destructive activities of proteases and disassembly chaperones must be controlled to ensure that they engage the proper substrates. Bacteria have no ubiquitin system, and recognition of specific protein targets for degradation or disassembly is mediated by a diverse set of unstructured peptide signals displayed on otherwise native substrate proteins. Some of these peptide signals function directly as primary degradation tags, allowing recognition and engagement by the ATPase; others function indirectly to tether the substrate to the ATPase, increasing the local concentration and the probability of engagement for degradation. The importance of “peptide” recognition was initially suggested by studies showing that mutation of a few unstructured terminal residues could dramatically change the proteolytic susceptibility of an attached protein (Bowie and Sauer, 1989; Parsell et al., 1990). Subsequent studies revealed that peptide signals in many substrates were responsible for their recognition by bacterial proteases or disassembly chaperones (see, for example, Keiler et al., 1996; Laachouch et al., 1996; Levchenko et al., 1997; Gonciarz-Swiatek et al., 1999; Wang et al., 1999; Hoskins et al., 2000; Gonzalez et al., 2000).
The importance of short peptide sequences in bacterial protein destruction is exemplified by the SsrA quality control system, which adds a degradation tag to nascent proteins on stalled ribosomes (Keiler et al., 1996). When recruited to a distressed ribosome, SsrA acts as tmRNA to direct addition of the tag sequence AANDENYALAA to the C terminus of the nascent polypeptide. This ssrA tag, in turn, targets the attached protein for degradation by ClpXP, ClpAP, and other proteases (Keiler et al., 1996; Gottesman et al., 1998; Herman et al., 1998). No additional substrate information is required for degradation, and thus the ssrA tag functions as a “strong” primary degradation signal.
In terms of protease specificity, it is natural to think about a single class of substrates being cleaved by a given enzyme, but this model is inadequate for bacterial AAA+ proteases. ClpXP, for example, degrades hundreds of different E. coli proteins in addition to ssrA-tagged substrates, and many of these substrates interact with ClpX using different types of peptide signals (Flynn et al., 2003). Indeed, at least five distinct classes of naturally encoded peptide motifs target proteins for degradation by ClpXP. This result can be rationalized from a biological perspective, as the existence of multiple classes of degradation signals would allow differential regulation of the degradation of disparate classes of substrates depending upon the demands of cell physiology. In fact, the intracellular levels of many ClpXP substrates change in response to oxygen levels, starvation, DNA damage, heat and cold shock, etc. (Flynn et al., 2003). Thus, one role of AAA+ proteases may be to readjust the composition of the proteome following the global changes in gene expression that accompany responses to stress.
The existence of multiple classes of degradation signals for a single AAA+ protease raises obvious biochemical questions about how these sequences function and are recognized. Some peptide signals represent primary degradation signals, whereas others function in tethering roles. Indeed, multiple peptide signals can be present in a single substrate and be required for efficient degradation. For example, one peptide signal in the UmuD/D′ heterodimer tethers it to ClpXP, allowing efficient recognition of a second “weak” degradation signal (Neher et al., 2003b). In a conceptual sense, the distinction between primary degradation or disassembly signals and secondary tethering signals is clear. Primary signals mediate engagement and subsequent denaturation/translocation by the AAA+ ATPase, whereas tethering signals simply increase binding affinity and the probability of engagement. Experimentally, however, determining the type of signal may not be straightforward. Eliminating either class of signal from a substrate could prevent degradation. Moreover, transplanting either type of signal to a new protein could potentially lead to its degradation. For example, fusing a tethering tag to a protein with a weak primary signal, which might comprise almost any region of unstructured polypeptide, could make the fusion protein an efficient substrate. For some substrates and AAA+ enzymes, tethering signals bind to accessory domains of the ATPase, whereas primary signals bind directly to the pore of the AAA+ ATPase (see below), but it is not yet clear whether this will be a general rule.
Degradation signals are present near the N or C terminus of a wide variety of substrates for ClpXP (Flynn et al., 2003), and terminal recognition signals seem to be common in substrates of other proteases as well. The a-carboxylate of the ssrA tag is critical for ClpXP recognition, and signals of this type would therefore only be efficiently recognized at the C terminus of a protein (Kim et al., 2000; Neher et al., 2003a). Placement of signals at the beginning or end of protein sequences may also be common because these regions are often unstructured, making a degradation tag in the native protein accessible to AAA+ proteases. Some degradation tags function at either protein terminus and even at internal sites (Hoskins et al., 2002). Hence, the key features of functional recognition signals are accessibility and the presence of apposite binding determinants.
EM (electron microscopy) studies show native substrates bound at or near the “protein-processing” pore on the protease-distal face of the ClpX ATPase (Ortega et al., 2000, 2002). This observation plus the fact that degradation substrates must ultimately translocate through the ATPase pore raises the possibility that the pore contains the binding and engagement sites for the recognition tags of some substrates. Indeed, in some instances, ATPase pore residues have been directly implicated in substrate recognition by mutational or cross-linking studies (Song et al., 2000; Siddiqui et al., 2004; Schlieker et al., 2004).
Cryptic Recognition Tags Allow Conditional Degradation
Some proteins only become substrates for degradation by AAA+ proteases following prior cleavage by another protease. During the SOS response to DNA damage, for example, the LexA repressor, which regulates this response, undergoes autocleavage to produce an N-and C-terminal fragment. ClpXP degrades both LexA fragments but does not degrade intact LexA (Flynn et al., 2003; Neher et al., 2003a). Latent signals for ClpXP degradation in LexA must be masked or hidden in the intact, native protein. For the N-terminal LexA fragment, the new C-terminal carboxylate is part of the ClpXP recognition signal; for the C-terminal fragment, autocleavage destabilizes a region of protein structure making a recognition signal, which is normally buried, accessible to ClpX. In this way, damage-induced autocleavage of LexA—an environmentally sensitive event—is coupled to a processive proteolysis reaction that efficiently destroys the resulting protein fragments. Uncovering of cryptic signals by unfolding, subunit dissociation, or cleavage by another protease may be a common mechanism for regulation of degradation by AAA+ proteases.
Adaptors Regulate Degradation and Disassembly
Adaptor proteins can be required for efficient substrate degradation or disassembly by AAA+ enzymes. Adaptors can also inhibit recognition of specific substrates. Although we are only beginning to learn how adaptors and related molecules are used to control intracellular degradation, these factors clearly play a central role in regulating proteolysis.
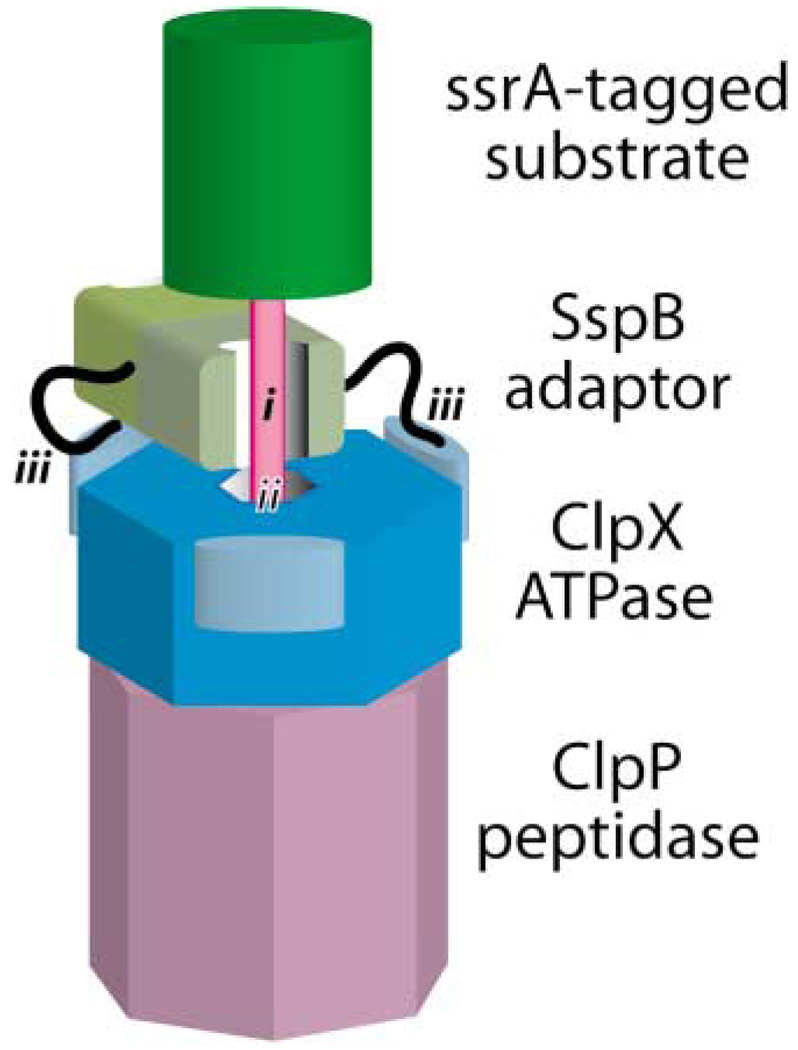
Adaptors that activate or enhance degradation help to bring the enzyme and substrate together. For example, the SspB adaptor binds both to ssrA-tagged substrates and to the ClpX ATPase, forming a delivery complex that permits efficient degradation at low substrate concentrations (Levchenko et al., 2000; Flynn et al., 2001; Wah et al., 2002). Importantly, only the tagged substrate and not SspB is degraded in this reaction. Three distinct sets of peptide-protein interactions stabilize delivery complexes (Figure 5): (1) a core dimeric domain of SspB binds part of the ssrA degradation tag of a substrate; (2) another part of the tag contacts the AAA+ portion of ClpX; and (3) the N-terminal domain of ClpX binds to the flexible tails of SspB, tethering the adaptor and enzyme (Levchenko et al., 2003; Song and Eck, 2003; Wah et al., 2003; Dougan et al., 2003; Bolon et al., 2004a; Hersch et al., 2004).
Figure 5. Model for Adaptor-Mediated Delivery of a Tagged Substrate to a AAA+ Protease.
The protein substrate (green) has an ssrA-degradation tag (pink). One part of the tag (i) is bound by the SspB adaptor (light brown); another part (ii) is bound by the ClpX ATPase (blue). Flexible tails (black) of the SspB adaptor make tethering interactions (iii) with the N-terminal domains (light blue) of ClpX.
Like SspB, the UmuD/D′ heterodimer and the adaptor/substrate pairs MecA/ComK and RssB/σs all employ tethering interactions to stabilize substrate binding to a AAA+ ATPase (Turgay et al., 1998; Zhou et al., 2001; Neher et al., 2003b). In fact, the UmuD subunit of UmuD/D′ acts like an adaptor. It delivers the truncated UmuD′ subunit to ClpXP for degradation but is not itself degraded despite containing a degradation signal and a tethering signal (Gonzalez et al., 2000; Neher et al., 2003b). These peptide signals in the UmuD structure appear to be too close or arranged improperly to interact simultaneously with the tethering site and protein-processing pore on the AAA+ protease. This observation illustrates a general principle. In cases where multiple degradation signals in a substrate need to be integrated by a protease, proper recognition will depend on the way in which these signals are displayed in the three-dimensional structure.
The tethering signals present in SspB, UmuD, and RssB share sequence homology and bind to a common site on the subfamily-specific N-terminal domain of the ClpX ATPase (Wah et al., 2003; Dougan et al., 2003; Neher et al., 2003b; Siddiqui, 2004). This binding ensures that these adaptors deliver substrates to the ClpXP protease but do not interact with other proteases, which lack the necessary tethering sites. This common mode of tethering also ensures that this set of adaptors compete with each other for delivery of substrates to ClpXP, allowing changes in the priority of substrate degradation depending on the relative cellular levels of each adaptor.
Many adaptors inhibit degradation or disassembly of specific substrates by masking their recognition signals. The MuB regulatory protein binds to the recognition tag of MuA transposase, for example, and inhibits ClpX disassembly of transposase-DNA complexes (Levchenko et al., 1997). SspB inhibits ClpAP degradation of ssrA-tagged substrates by masking recognition determinants in the tag (Flynn et al., 2001). SspB also inhibits ClpXP degradation of ssrA-tagged substrates when its tethering interactions with ClpX are blocked (Wah et al., 2003). This inhibition occurs because concurrent binding to the ssrA tag results in modest clashes between the ClpX ATPase and the SspB adaptor (Hersch et al., 2004). Inhibition can also occur by binding of an adaptor to the AAA+ ATPase rather than to the substrate. For example, ClpS binds to the ClpA ATPase and inhibits binding of ssrA-tagged substrates (Dougan et al., 2002). It may seem counterintuitive that cells would want to inhibit degradation of aberrant translation products bearing ssrA tags. However, under adverse conditions where translational mistakes and ssrA tagging occur frequently, inhibiting degradation of ssrA-tagged substrates could free AAA+ proteases to deal with more critical substrates.
Degradation can also be controlled by the opposing activities of different adaptors. In Bacillus subtilis, ComK regulates genetic competence. During growth at low cell densities, the MecA adaptor binds ComK and targets it for degradation by the ClpCP protease (Turgay et al., 1997, 1998). At high cell densities, however, another adaptor (ComS) is synthesized, binds to MecA, and displaces ComK. This prevents ComK degradation and allows it to activate expression of genes required for competence. Interestingly, many ClpCP substrates appear to use the MecA adaptor, and thus competition for MecA may be a common regulatory mechanism (Schlothauer et al., 2003). Furthermore, a second MecA-like protein appears to function to deliver a separate group of substrates to ClpCP, providing potential for additional regulation (Persuh et al., 2002).
Common Recognition Strategies in All Kingdoms
Protein substrates of the 26S proteasome of eukaryotic cells are marked for destruction by covalent attachment of polyubiquitin chains, via the sequential actions of the E1, E2, and E3 enzymes (Hershko and Ciechanover, 1998). Different E2 and E3 enzymes modify distinct sets of substrates, conferring specificity and allowing individual proteins to be modified and degraded at appropriate times. At first glance, this approach seems very different from the bacterial systems described above. However, recent work has shown that substrates of the proteasome must also have unfolded peptide elements to allow enzymatic engagement and efficient degradation (Prakash et al., 2004). These engagement signals clearly resemble the primary degradation signals of bacterial substrates. Moreover, polyubiquitin would then function as a covalent adaptor to tether the attached substrate to the proteasome. As a result, bipartite recognition of distinct tethering and engagement signals appears to be a common mechanism for substrate selection by ATP-powered proteases in all organisms.
Why does degradation by AAA+ proteases frequently involve recognition of multiple signals, which individually are insufficient for degradation and often bind with relatively weak µM affinity to the protease? Coupling weak interactions can create stronger binding through increased local concentrations but has additional biological consequences. First, recognition motifs may not need to meet strict sequence requirements, making them easier to evolve, as a wide range of peptide sequences can mediate the modest binding affinities required. This latitude also makes it easier to create signals that can be masked by protein folding or binding and/or signals compatible with other protein functions. Second, if efficient binding requires several recognition events, then numerous possibilities for combinatorial regulation emerge. For example, two weak signals might only be efficiently recognized following assembly of a specific macromolecular complex or following protein processing or modification. Third, use of weak signals in combination allows highly dynamic recognition because individual contacts can be broken relatively easily, allowing susceptibility to degradation or disassembly to re-equilibrate rapidly in response to environmental changes or cell cycle cues.
AAA+ ATPases Catalyze Mechanical Protein Unfolding
Unfolding of native proteins by AAA+ ATPases is an essential step in degradation and represents one way to disassemble macromolecular complexes. Very stable proteins—those for which spontaneous unfolding can take months or even years—are unfolded in a few minutes or less in the enzymatic reaction (Weber-Ban et al., 1999; Kim et al., 2000; Singh et al., 2000; Burton et al., 2001a; Kenniston et al., 2003). Denaturation and translocation of substrates begins with the primary degradation signal (Lee et al., 2001). The ATPase then appears to unfold structure adjacent to the degradation tag, with global denaturation of single-domain proteins occurring rapidly thereafter in a spontaneous reaction because of the cooperativity of protein unfolding. In support of this model, it has been shown in several cases that the degradation tags of substrates enter the peptidase first, and altering the position of the tag or destabilizing adjacent structural elements can alter degradation rates dramatically for variants of the same protein (Lee et al., 2001; Reid et al. 2001; Ortega et al., 2002; Kenniston et al., 2003).
How does a AAA+ ATPase unfold a protein substrate? Current evidence suggests that, following binding of the peptide tag, the enzyme begins to translocate this tag through its protein-processing pore (Figure 1 and Figure 6). Attempting to pull the attached native protein through this small aperture would generate a denaturation force. This pulling model makes structural sense and is attractive for several additional reasons. First, the same ATP-dependent conformational changes that drive translocation would also cause denaturation. Second, for a large number of different proteins, a single “strong” degradation signal like the ssrA tag is sufficient for efficient denaturation by AAA+ ATPases. If a second signal is required for unfolding of these substrates, then it must be generic and present in almost all proteins. The “pulling” model of denaturation requires just one point of contact between the ATPase and the substrate and is therefore compatible with unfolding mediated by a single signal. Most other models of mechanical deformation, by contrast, require two contact points between the enzyme and substrate.
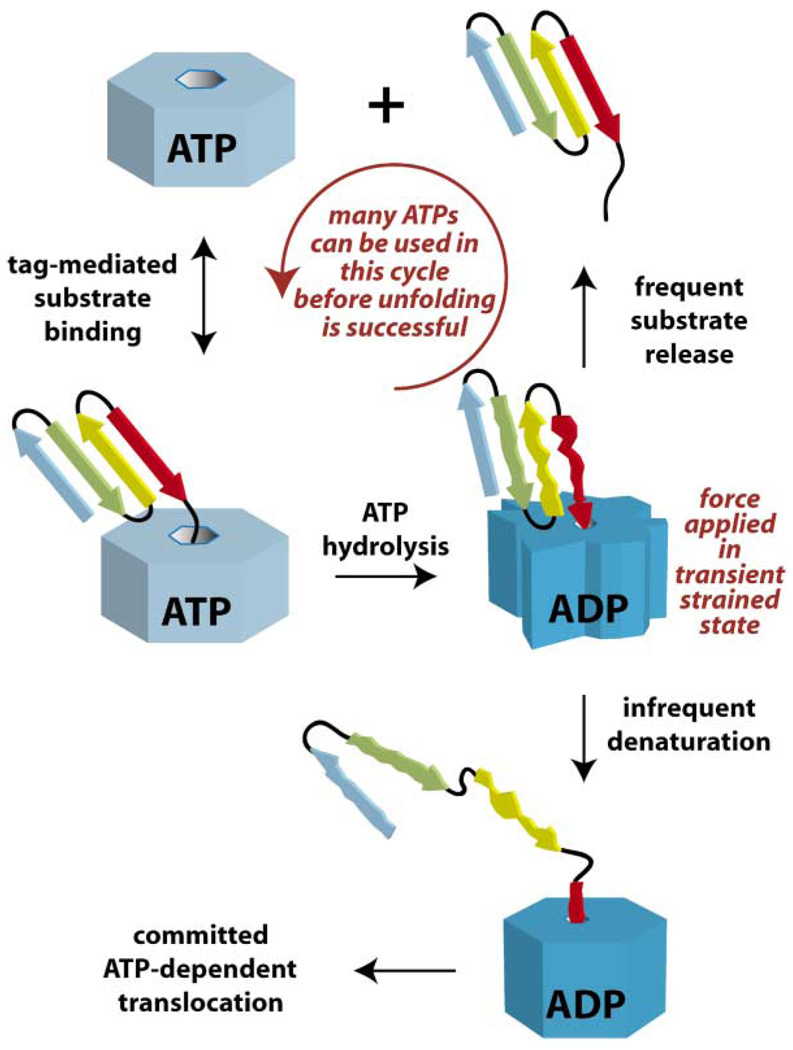
Figure 6. Model for Enzyme-Catalyzed Protein Denaturation.
A tagged native protein binds to a AAA+ ATPase in its ATP bound state (light blue). A conformational change, which accompanies ATP hydrolysis, pulls the native protein into the central pore, creating an unfolding force and a highly strained enzyme (dark blue). At this point, the substrate can either dissociate or unfold. The probability of either event depends upon the stability of the structural elements in the substrate adjacent to the recognition tag. For very stable substrates, hundreds of cycles of protein binding and release may be required before denaturation is successful (Kenniston et al., 2003). Once denaturation occurs, additional rounds of ATP hydrolysis drive processive protein translocation. If a compartmental protease is bound to the ATPase, then the translocated protein is degraded; otherwise, it is released in a denatured state and has the opportunity to refold.
The stability of the three-dimensional structure of substrates influences their rates of unfolding by AAA+ ATPases and can determine whether a protein with an appropriate recognition tag is degraded and at what rate. For example, FtsH, a membrane bound AAA+ protease, can degrade proteins of moderate stability but is incapable of degrading very stable proteins (Herman et al., 2003). By contrast, ClpXP can unfold and degrade hyperstable substrates, albeit more slowly than it processes less stable variants (Kenniston et al., 2003, 2004). These results suggest that some AAA+ enzymes generate larger denaturation forces and are therefore more powerful than others in terms of substrate unfolding. Interestingly, ClpXP loses the ability to degrade hyperstable proteins when it uses a slowly hydrolyzed ATP analog as the energy source (Burton et al., 2003). Similarly, mutations that interfere with communication between nucleotide binding sites in adjacent subunits diminish the unfoldase activity of ClpX (Joshi et al., 2003). Thus, the intrinsic resistance of a substrate to enzymatic unfolding and the rate and coordination of enzymatic ATP hydrolysis are both important in determining whether a given substrate can be denatured and thus degraded.
ATP Costs Are High and Substrate Dependent
ATP hydrolysis is required for the denaturation and translocation steps of protein degradation. The energetic costs of these processes for ClpXP degradation have been determined for several proteins (Kenniston et al., 2003, 2004). Depending on the substrate, ClpXP hydrolyzes an average of 30 to 80 ATPs for each 100 residues that are translocated and degraded. Protein denaturation shows even larger substrate-dependent variations. For example, ClpXP denaturation of a hyperstable substrate required hydrolysis of an average of 550 ATPs, whereas unfolding of a variant with a destabilizing mutation near the degradation tag used only 12 ATPs. Protein degradation by AAA+ proteases can clearly be very costly, often using as much or even more energy than was expended for biosynthesis of the protein.
Variability in the number of ATPs required for the denaturation of different substrates emphasizes the importance of local structure in resisting denaturation and provides additional clues about the protein-unfolding mechanism. ATP hydrolysis drives repetitive cycles of conformational change in the AAA+ ATPase. The enzyme has no way of storing energy from one hydrolysis cycle to the next, and denaturation of single-domain substrates occurs in an all-or-none fashion and not, for example, by taking the protein apart one structural piece at a time. Hence, each cycle of ATP hydrolysis must represent a new attempt to denature the native protein, which either resists or succumbs to unfolding with a probability that depends upon its structural stability (Figure 6). Stable native proteins appear to require hundreds of denaturation attempts by the ATPase before the probability of unfolding is high (Kenniston et al., 2003). Nevertheless, there must be the same chance that denaturation will occur on the first attempt as on any subsequent attempt. The stochastic nature of this process indicates that some protein molecules in a population must be more susceptible than others to enzyme-mediated denaturation, presumably because of structural distortions induced by thermal motions in which a few noncovalent interactions near the primary degradation tag are broken transiently. For substrates with very stable structural elements neighboring the degradation tag, the probability of such distortions and thus the chance of unfolding per cycle is low. As a result, hydrolysis of many ATPs is needed on average to ensure denaturation. For substrates with less stable local structure, the unfolding probability per cycle is high and only a small number of enzyme turnovers are required on average to denature most molecules.
In the model of Figure 6, the partitioning between substrate release and denaturation acts as a biochemical “clutch” that allows the AAA+ motor to disengage rather than stall when denaturation fails (Kenniston et al., 2003). Releasing “hard to denature” substrates in this fashion would prevent AAA+ proteases from becoming jammed but would also require rebinding of substrates after each enzymatic cycle.
If substrates are normally released after each futile ATPase cycle, adaptors may keep the substrate tethered to the enzyme, thus making rebinding a unimolecular process (Bolon et al., 2004b). Moreover, flexibility in the tethering linkage between the adaptor and enzyme could help maintain the integrity of the delivery complex during the conformational excursions of enzyme during the ATPase cycle.
Energy-Dependent Demolition Allows Escape from Deep Energetic Wells
The assembly of more-and-more stable protein/protein or protein/nucleic-acid complexes often drives biological reactions forward. Although this strategy provides directionality by proceeding downhill to a thermodynamic minimum, it creates problems if the structure assembled is not the final product of the pathway. In such cases, AAA+ ATPases are frequently recruited as “remodeling” or disassembly chaperones to carry out energy-dependent dismantling of a macromolecular complex that would otherwise persist. These destructive activities can allow progress to a desired biological end point, recycle the molecular components, or remove unneeded and potentially harmful complexes. Aggregates of misfolded, misassembled, and damaged proteins represent a second class of stable complexes that need to be dismantled to return proteins to soluble functional forms or to prevent the toxic effects of accumulation. Many AAA+ ATPases can solubilize aggregated proteins, although the participation of other molecular chaperones often appears to be required for efficient reactions (see Glover and Tkach, 2001).
How do AAA+ ATPases disassemble stable protein complexes and promote disaggregation? One simple idea, namely that the same global protein-unfolding activity used for degradation is also responsible for disassembly, is supported by studies of ClpX-mediated demolition of complexes of the tetrameric MuA transposase with recombined DNA (Burton et al., 2001b; Burton and Baker, 2003). The recombination steps catalyzed by MuA are driven by the increasing stability of the protein-DNA complexes. Once recombination is complete, however, the protein-DNA complex must be dismantled to allow subsequent DNA replication and repair steps. ClpX promotes this disassembly reaction by unfolding the transposase. Importantly, only one subunit of the MuA tetramer needs to bear a recognition signal for the disassembly reaction to proceed. This and other results suggest that unfolding of just one subunit of the MuA tetramer is sufficient to destabilize the entire complex. In the same way that removing one block from an arch could lead to its collapse, unfolding and removing one subunit from a macromolecular complex can create a fragile structure that will dissociate spontaneously.
Global unfolding is not required for all remodeling reactions carried out by AAA+ ATPases. During DNA replication, for example, the AAA+ clamp loader opens one of the subunit-subunit interfaces of the processivity clamp, allowing this ring-like molecule to encircle DNA (Davey et al., 2002). Moreover, AAA+ ATPases may break up large protein aggregates by acting like molecular “crowbars” to pry apart protein-protein interfaces (Glover and Tkach, 2001). The key to understanding these less dramatic remodeling reactions will be to determine how AAA+ ATPases recognize specific substrates and then use ATP-driven conformational changes to apply an appropriate molecular force.
Summary
The pace of advances in our understanding of the biological roles and mechanisms of the AAA+ ATPases involved in protein degradation and disassembly has been dramatic. This progress has depended on the availability of information about the three-dimensional conformations of these enzymes, on the discovery and characterization of new substrates and adaptors, and on the availability of substrates and adaptors that are well characterized in terms of both recognition determinants and structural properties. Numerous important questions, at both the biological and biochemical levels, remain unanswered but now seem within reach.
Acknowledgments
We thank members of the Baker and Sauer labs for helpful discussions. This work was supported in part by NIH grants AI-15706, AI-16892, and GM-499224 and the Howard Hughes Medical Institute. T.A.B., J.M.F., and I.L are employees of HHMI.
References
- Beuron F, Maurizi MR, Belnap DM, Kocsis E, Booy FP, Kessel M, Steven AC. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J. Struct. Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Huber R. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1997;10:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent tethering of the SspB adaptor to the AAA+ protease ClpXP is required for efficient substrate delivery: a protein-design study. Mol. Cell. 2004a;13:443–449. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004b doi: 10.1016/j.molcel.2004.10.001. in press. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Sauer RT. Identification of C-terminal extensions that protect proteins from intracellular proteolysis. J. Biol. Chem. 1989;264:7596–7602. [PubMed] [Google Scholar]
- Burton BM, Baker TA. Mu transpososome architecture ensures that unfolding by ClpX or proteolysis by ClpXP remodels but does not destroy the complex. Chem. Biol. 2003;10:463–472. doi: 10.1016/s1074-5521(03)00102-9. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001a;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BM, Williams TL, Baker TA. ClpX-mediated remodeling of Mu transpososomes: selective unfolding of subunits destabilizes the entire complex. Mol. Cell. 2001b;8:449–454. doi: 10.1016/s1097-2765(01)00307-0. [DOI] [PubMed] [Google Scholar]
- Burton RE, Baker TA, Sauer RT. Energy-dependent degradation: Linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Sci. 2003;12:893–902. doi: 10.1110/ps.0237603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Weber-Ban E, Bukau B. Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol. Cell. 2003;12:373–380. doi: 10.1016/j.molcel.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA. 2001;11:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Glover JR, Tkach JM. Crowbars and ratchets: Hsp100 chaperones as tools in reversing protein aggregation. Biochem. Cell Biol. 2001;79:557–568. [PubMed] [Google Scholar]
- Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA, McMacken R, Kelley WL, Georgopoulos C, Sliekers O, Zylicz M. Recognition, targeting, and hydrolysis of the λ O replication protein by the ClpP/ClpX protease. J. Biol. Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, Rasulova F, Maurizi MR, Woodgate R. Subunit-specific degradation of the UmuD/D′ heterodimer by the ClpXP protease: the role of trans recognition in UmuD′ stability. EMBO J. 2000;19:5251–5258. doi: 10.1093/emboj/19.19.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Baker TA, Sauer RT. SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc. Natl. Acad. Sci. USA. 2004;101:12136–12141. doi: 10.1073/pnas.0404733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Kim SY, Wickner S. Substrate recognition by the ClpA chaperone component of ClpAP protease. J. Biol. Chem. 2000;275:35361–35367. doi: 10.1074/jbc.M006288200. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Sharma S, Sathyanarayana BK, Wickner S. Clp ATPases and their role in protein unfolding and degradation. Adv. Protein Chem. 2001;59:413–429. doi: 10.1016/s0065-3233(01)59013-0. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Yanagihara K, Mizuuchi K, Wickner S. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. USA. 2002;99:11037–11042. doi: 10.1073/pnas.172378899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SA, Baker TA, Sauer RT. C-terminal domain mutations in ClpX uncouple substrate binding from an engagement step required for unfolding. Mol. Microbiol. 2003;48:67–76. doi: 10.1046/j.1365-2958.2003.03424.x. [DOI] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of a AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J. Struct. Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Laachouch JE, Desmet L, Geuskens V, Grimaud R, Toussaint A. Bacteriophage Mu repressor as a target for the Escherichia coli ATP-dependent Clp Protease. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Yamauchi M, Baker TA. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Wah DA, Sauer RT, Baker TA. Structure of a delivery protein for a AAA+ protease in complex with a peptide degradation tag. Mol. Cell. 2003;12:365–372. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Neher SB, Flynn JM, Sauer RT, Baker TA. Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev. 2003a;17:1084–1089. doi: 10.1101/gad.1078003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher SB, Sauer RT, Baker TA. Distinct peptide signals in the UmuD and UmuD′ subunits of UmuD/D′ mediate tethering and substrate-processing by the ClpXP protease. Proc. Natl. Acad. Sci. USA. 2003b;100:13219–13224. doi: 10.1073/pnas.2235804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Nishii W, Takahashi K. Determination of the cleavage sites in SulA, a cell division inhibitor, by the ATP-dependent HslVU protease from Escherichia coli. FEBS Lett. 2003;553:351–354. doi: 10.1016/s0014-5793(03)01044-5. [DOI] [PubMed] [Google Scholar]
- Ortega J, Singh SK, Ishikawa T, Maurizi MR, Steven AC. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Ortega J, Lee HS, Maurizi MR, Steven AC. Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J. 2002;21:4938–4949. doi: 10.1093/emboj/cdf483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Silber KR, Sauer RT. Carboxy-terminal determinants of intracellular protein degradation. Genes Dev. 1990;4:277–286. doi: 10.1101/gad.4.2.277. [DOI] [PubMed] [Google Scholar]
- Persuh M, Mandic-Mulec I, Dubnau D. A MecA paralog, YpbH, binds ClpC, affecting both competence and sporulation. J. Bacteriol. 2002;184:2310–2313. doi: 10.1128/JB.184.8.2310-2313.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004 doi: 10.1038/nsmb814. in press. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Hartmann C, Song HK, Huber R, Bochtler M. Functional interactions of HslV (ClpQ) with the ATPase HslU (ClpY) Proc. Natl. Acad. Sci. USA. 2002;99:7396–7401. doi: 10.1073/pnas.102188799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BG, Fenton WA, Horwich AL, Weber-Ban EU. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc. Natl. Acad. Sci. USA. 2001;98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- Schlothauer T, Mogk A, Dougan DA, Bukau B, Turgay K. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. USA. 2003;100:2306–2311. doi: 10.1073/pnas.0535717100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Yoo SJ, Shin DH, Shim YK, Kang MS, Goldberg AL, Chung CH. The heat-shock protein HslVU from Escherichia coli is a protein-activated ATPase as well as an ATP-dependent proteinase. Eur. J. Biochem. 1997;247:1143–1150. doi: 10.1111/j.1432-1033.1997.01143.x. [DOI] [PubMed] [Google Scholar]
- Seong IS, Kang MS, Choi MK, Lee JW, Koh OJ, Wang J, Eom SH, Chung CH. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J. Biol. Chem. 2002;277:25976–25982. doi: 10.1074/jbc.M202793200. [DOI] [PubMed] [Google Scholar]
- Siddiqui SM. Dissecting the steps of substrate processing by the energy-dependent protease ClpXP. PhD thesis. Cambridge, Massachusetts: Massachusetts Institute of Technology; 2004. [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the protein-processing pore of ClpX, a AAA+ ATPase, in recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HK, Eck MJ. Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine. Mol. Cell. 2003;12:75–86. doi: 10.1016/s1097-2765(03)00271-5. [DOI] [PubMed] [Google Scholar]
- Song HK, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Kessler BM, Overkleeft HS, McKay DB. Crystal structure of HslUV complexed with a vinyl sulfone inhibitor: corroboration of a proposed mechanism of allosteric activation of HslV by HslU. J. Mol. Biol. 2002;318:779–785. doi: 10.1016/S0022-2836(02)00145-6. [DOI] [PubMed] [Google Scholar]
- Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J. Biol. Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- Thompson MW, Singh SK, Maurizi MR. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- Turgay K, Hamoen LW, Venema G, Dubnau D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;7:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a specificity factor for a AAA+ ATPase: Assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem. Biol. 2002;9:1237–1245. doi: 10.1016/s1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Flexible linkers leash the substrate-binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol. Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Wang L, Elliott M, Elliott T. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J. Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of the 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Seol JH, Kang M-S, Chung CH. Poly-L-lysine activates both peptide and ATP hydrolysis by the ATP-dependent HslUV protease in Escherichia coli. Biochem. Biophys. Res. Commun. 1996;229:531–535. doi: 10.1006/bbrc.1996.1838. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets σs for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]