Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin (original) (raw)
Abstract
Peripheral membrane proteins of the Bin/amphiphysin/Rvs (BAR) and Fer-CIP4 homology-BAR (F-BAR) family participate in cellular membrane trafficking and have been shown to generate membrane tubules. The degree of membrane bending appears to be encoded in the structure and immanent curvature of the particular protein domains, with BAR and F-BAR domains inducing high- and low-curvature tubules, respectively. In addition, oligomerization and the formation of ordered arrays influences tubule stabilization. Here, the F-BAR domain-containing protein Pacsin was found to possess a unique activity, creating small tubules and tubule constrictions, in addition to the wide tubules characteristic for this subfamily. Based on crystal structures of the F-BAR domain of Pacsin and mutagenesis studies, vesiculation could be linked to the presence of unique structural features distinguishing it from other F-BAR proteins. Tubulation was suppressed in the context of the full-length protein, suggesting that Pacsin is autoinhibited in solution. The regulated deformation of membranes and promotion of tubule constrictions by Pacsin suggests a more versatile function of these proteins in vesiculation and endocytosis beyond their role as scaffold proteins.
Keywords: endocytosis, membrane trafficking, protein structure
Pacsins (also known as Syndapins or FAP52) comprise a conserved family of peripheral membrane proteins in eukaryotes that play a central role in synaptic vesicle recycling and receptor-mediated endocytosis, ultimately controlling the cell surface expression of transmembrane proteins (1–4). The 3 isoforms of Pacsin in higher eukaryotes differ in their expression pattern, cargo selectivity, and recruitment of other proteins involved in endocytosis. Pacsin 1 is a brain-specific isoform and has been implicated in neuronal transmission and the neuropathology of Huntington's disease, whereas Pacsin 2 and 3 are more widely expressed (2, 5, 6). On a molecular level, Pacsins bind to Dynamin and Eps15 homology domain (EHD)-containing proteins, nucleotide-dependent enzymes with membrane remodeling and fission activities, as well as the actin cytoskeleton via neural Wiskott-Aldrich syndrome protein (N-WASP), linking membrane trafficking and cytoskeletal rearrangements (4, 7, 8). Although Pacsin isoforms bind a specific subset of proteins via the central linker region and their SH3 domain, they all share a highly conserved Fer-CIP4 homology-BAR (F-BAR) domain at their N terminus (9).
F-BAR domains constitute a subfamily of Bin/amphiphysin/Rvs (BAR) domains, protein modules that stabilize or induce membrane curvature (9–13). In contrast to N-BAR domains that prefer highly curved membrane tubules, F-BAR domains usually stabilize membrane structures with larger diameter (9, 14, 15). Such curvature preference can be attributed in part to the structures of the particular proteins (11, 14–16). Both BAR and F-BAR domains consist of a 3-helix bundle and form extended, crescent-shaped dimers. The concave surface of the arc presents an overall positive electrostatic potential that allows for interactions with the plasma membrane. To some degree, the intrinsic curvature of the dimer correlates with the preference for a particular membrane tubule or vesicle size (9, 11, 14, 15). N-terminal, amphipathic helices in N-BAR domains contribute to the driving force required to deform membranes, an energetically expensive process (17–19). The amphipathic helix in F-BAR domain-containing proteins appears to be dispensable for their tubulation activity (14). Instead, F-BAR dimers can form ordered arrays on membranes, predominantly driven by electrostatic and intermolecular interactions (15, 20).
BAR and F-BAR domain-containing proteins encoded in eukaryotic genomes cluster into distinct branches, but little is known with regard to the functional relevance of such complexity (9). Elucidating the morphogenic potential and molecular mechanism of these proteins will provide insight into their role in membrane trafficking and endocytosis. Here, we show that Pacsin proteins stabilize tubules of both large and narrow diameter, but also facilitate membrane constriction in vitro, suggesting unanticipated diversity in the family of F-BAR domain-containing proteins. Structural studies elucidate the molecular mechanism and unique motifs in Pacsin that contribute to its distinct membrane remodeling potential.
Results and Discussion
Pacsin-Mediated Membrane Deformation.
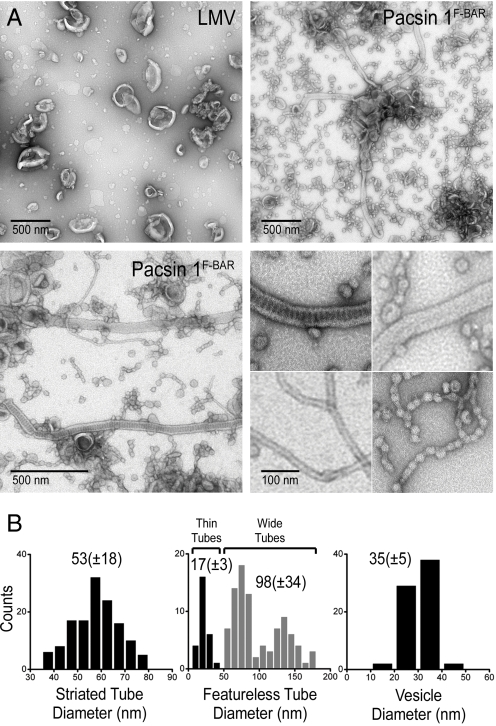
The F-BAR domain of human Pacsin 1 and 2 were overexpressed in Escherichia coli and purified to homogeneity. Analytical ultracentrifugation revealed a predominantly dimeric state of the isolated domain (Fig. S1_A_). Using the purified proteins, we analyzed their morphogenic potential in a membrane tubulation assay using negative stain electron microscopy. Large multilamellar vesicles (LMVs) generated from isolated brain lipids (Folch fraction I) were incubated with the F-BAR domain of Pacsin 1. Unlike the homogeneous and distinctive tubule morphology induced by other BAR and other F-BAR proteins (Endophilin, Sorting nexin 9, and Toca-1), the F-BAR domain of Pacsin 1 generated a broad spectrum of membrane structures (Fig. 1A and Fig. S2).
Fig. 1.
Pacsin-mediated liposome tubulation. (A) Negative-stain EM. LMVs from Folch fraction I lipids were incubated with the isolated F-BAR domain of human Pacsin 1 (residues 1–325). Two representative micrographs are shown in low magnification. The bottom-right image shows higher magnifications of the 4 main membrane morphotypes induced by Pacsin. (B) Statistical analysis. Diameters of the different morphotypes shown in A were quantified by determining their size on at least 3 independently prepared EM grids. Diameters (especially for wider tubules) may appear larger because of flattening effects during negative staining.
We observed 2 classes of tubules with shallow curvature, striated and featureless, with a mean diameter of 53 ± 18 nm and 98 ± 34 nm, respectively, and average length of 1.3–1.4 μM (Fig. 1 and Fig. S2 B and C). Given the size of LMVs, the observed tubules were likely to originate from a single vesicle, not requiring fusion events. Occasionally, tubes were associated with vesicular structures (Fig. S2 B and C, arrows). Featureless tubules were comparable with structures observed with other F-BAR proteins and showed a wider distribution with regard to diameter, both across an individual tubule, as well as between tubules, which in part may be attributed to flattening effects during negative staining (Fig. 1B Middle) (9). The striated tubules induced by Pacsin had an invariable diameter and regular staining pattern, suggesting that these structures were more rigid and ordered (Fig. 1B Left and Fig. S2_B_). Striation has been reported for Dynamin assemblies on membranes, as well as for the BAR domain-containing proteins endophilin and amphiphysin, but the staining pattern generated by Pacsin was distinct in that its repeat units were perpendicular to the long tube axis (10, 12, 21). A similar pattern has been observed on 40-nm tubules formed with Sorting Nexin 9 (22), but the significance and molecular basis for such patterns awaits further elucidation.
In contrast to the morphologies observed with the majority of F-BAR domain-containing proteins, the F-BAR domain of Pacsin also induced narrow tubules with high curvature (d = 17 ± 3 nm) (Fig. 1 A and B Middle). Such structures were comparable with the ones observed with Endophilin or Amphiphysin and were slightly narrower than those obtained with the F-BAR domain of FCHo2 (11, 14). With both Pacsin and FCHo2, narrow tubules occurred at the same conditions as broader tubes. Variations in the higher-order assemblies of F-BAR domains on the membranes might contribute to the stabilization of different degrees of curvature (14, 20). In addition, narrower and broader tubes may originate from liposomes with different properties, such as in bilayer tension, membrane stiffness, or intrinsic curvature.
Most strikingly though, the background of the electron micrographs was dominated by small vesicle structures (d = 35 ± 5 nm) that often were connected and clustered together resembling beads on a string (Fig. 1 and Fig. S2_D_). These structures occurred at protein concentrations at which tubulation was observed, suggesting that this activity was not because of excess F-BAR domains, unlike the vesiculation activity described for samples containing an excess of BAR domains (11). Both narrow tubules and tube constrictions were absent in the LMV preparations in the absence of protein and significantly less abundant in samples containing other BAR and F-BAR domain, indicating a Pacsin-specific effect (Fig. 1A and Fig. S2_A_). In contrast to isolated vesicles with irregular diameters found in LMV preparations, the Pacsin-induced morphologies were regular and significantly more abundant. The tight clustering of vesicular structures and their occasional association with narrow tubes suggested a pathway where constrictions emerge on highly curved tube structures (Fig. 1A and Fig. S2_D_).
Comparable results regarding the occurrence of the various membrane morphologies were obtained by using large vesicles that were generated by extrusion through a 400-nm filter (Fig. S3_A_). Usage of smaller vesicles with diameters between 50 and 200 nm abolished the formation of wider tubules, but vesiculation remained apparent. Formation of tubules and vesicles was rapid, being visible and fully developed after short incubation times (Fig. S3_B_). Incubation at 4 °C as opposed to 25 °C or 37 °C prevented formation of tubules with large diameters, and an increased abundance of high-curvature tubules was apparent, suggesting that bilayer deformability and lipid order may play a role for the membrane sculpting potential (Fig. S3_B_). In addition, Pacsin-mediated vesiculation was observed with inner-leaflet lipid mixtures comprising phosphatidylserine, phosphatidylcholine, and phosphatidylethanolamine instead of brain lipid extracts (Fig. S3_C_).
Structures of the F-BAR Domain of Pacsin 1 and 2.
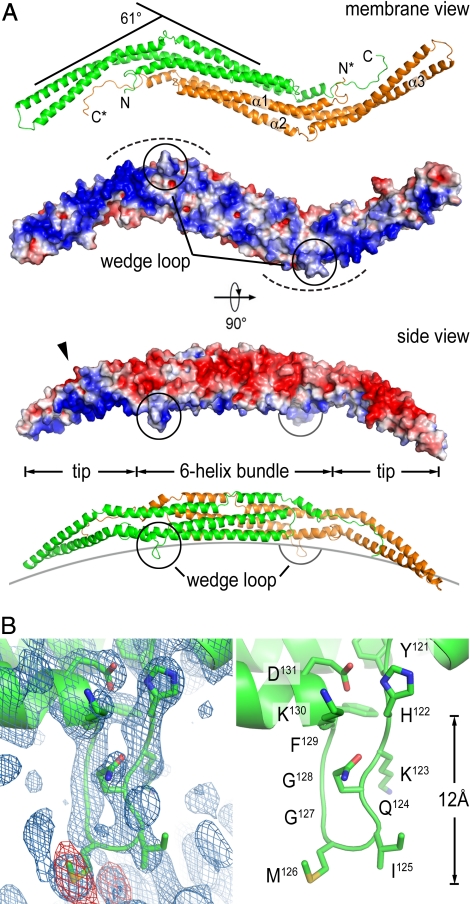
To elucidate the molecular basis for the generation of Pacsin-specific membrane morphologies, we determined the crystal structures of the F-BAR domain of Pacsin 1 and 2 (see SI Appendix and Table S1 for details). Screens using the full-length proteins yielded crystals of the F-BAR domains because of limited proteolysis over the course of the crystallization. The structure of the F-BAR domain from human Pacsin 1 was solved by single anomalous dispersion (SAD) phasing, using data collected on a crystal grown from selenomethionine-derivatized protein. The model was refined to a maximum resolution of 2.9 Å in the space group P21 (Fig. 2A). Final refinement steps were aided by determining the structure of the isolated F-BAR domain (residues 1–325 of human Pacsin 1) at 2.8 Å in space group C2 using SAD phasing. The structure of the F-BAR domain of Pacsin 2 was solved by molecular replacement in space group P212121 and was refined to a maximum resolution of 2.8 Å. The F-BAR domains of Pacsin 1 and 2 are very similar (rms deviation of 1.4 Å between all main chain atoms) and resemble the structure of F-BAR domains from CIP4, FBP17, and FCHo2 regarding overall fold and dimeric state, with a central 6-helix bundle and distal tips composed of a helix-turn-helix motif (Fig. 2A and Fig. S4_A_) (14, 15). All F-BAR domains display a rather shallow concave surface that is enriched in positively charged residues that form the major membrane interaction interface (Fig. 2A).
Fig. 2.
Structure of Pacsin's F-BAR domain. (A) Ribbon and surface presentation of the F-BAR domain dimer of Pacsin 1. Two perpendicular views are shown. Only 2 protomers are shown and are colored in green and orange, respectively. Asterisks denote the termini of the second protomer. The electrostatic potential of the F-BAR domain was mapped onto its molecular surface, with red representing negative potential and blue representing positive potential (−4 to +4 kBT). Electrostatic potentials were calculated by using the program adaptive Poisson-Boltzmann solver (APBS). (B) Close-up view of the wedge loop. The blue and red maps in the left image show a (2Fo-Fc) omit map (contoured at 1.6σ) and an anomalous map (contoured at 3σ) around the wedge loop, respectively. The data collected on a selenomethionine-derivatized Pacsin 1 crystal (space group P21) were used. The right image shows the same view with the wedge-loop residues presented as sticks.
Distinct Features of the F-BAR Domain of Pacsin.
A striking difference to previously determined structures of F-BAR proteins is the extreme degree of lateral kinking at the tip regions relative to the central dimerization region in Pacsin. Whereas the Toca-related F-BAR domains (from CIP4 and FBP17) are rather straight molecules and the one from FCHo2 has moderately kinked tips (14, 15), Pacsin's tips are bent away from its central body in a ≈61° angle, giving the dimeric molecule a pronounced twisted S-shape (Fig. 2A and Fig. S4_A_). Although the curvature of the concave surface remains shallow similar to other F-BAR domains, it adds a second degree of curvature to the Pacsin F-BAR dimer. Interaction with the membrane will require that both principle curvatures of the protein will be satisfied, indicating that Pacsin may have the tendency to bind to or stabilize membranes with higher curvature or vesicular structure. In contrast, other BAR and F-BAR domain-containing proteins may predominantly stabilize radial curvature, e.g., that of a tube (see SI Appendix).
In this conformation, positively charged residues that mediate interactions with the negatively charged lipid head groups are concentrated at the concave surface but are arranged in a slight pitch lining the bending point and the tip regions, which results in the display of a twisted, nonflat surface with positive electrostatic potential (Fig. 2A). The pitch and presence of both curvatures suggests a mechanism by which Pacsin not only tubulates bilayers, but also may introduce or stabilize vesicular structures and potentially saddle points. Such regions of negative curvature occur during fission at the bud-neck and the plasma membrane-neck junctions during vesicle fission, and the interconnected, Pacsin-induced vesicles observed in vitro resemble such topologies (Fig. 1A and Fig. S2_D_).
Another feature that distinguishes Pacsin's F-BAR domain from that of other F-BAR domain-containing proteins is a unique, 8-residues-long insertion in helix 2 of Pacsin that forms a flexible loop in the crystal structures. The loops are located at the end of the central 6-helix bundle close to the bending points of the tips and protrude from the membrane interaction surface (Fig. 2B and Fig. S4_B_). At the base of the loop, there are 2 bulky hydrophobic residues, isoleucine125-methionine126 (I125-M126) or a double-methione123–124 (M123-M124) motif in Pacsin 1 and 2, respectively. The loop is flanked by lysines on either side (arginines in Pacsin 3) and contains a double-glycine motif next to the central hydrophobic residues. Given the nature, position, and protrusion depth of the loop (≈12 Å), we proposed a function equivalent to an amphipathic helix that would intercalate into a bilayer leaflet and would work as a wedge for membrane bending. We will refer to this motif as a wedge loop. Similar motifs are of functional importance in proteins involved in fusion and fission.
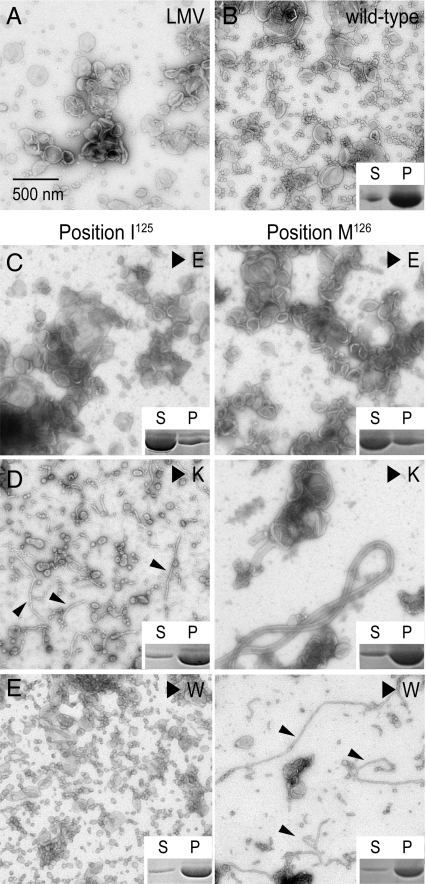
Wild-type protein and site-directed mutants were analyzed with regard to their tubulation activity and membrane binding affinity. Protein binding to liposomes was assessed in a pelleting assay by detection of protein in the pellet or supernatant fraction after high-speed centrifugation. Replacing either isoleucine or methionine in the wedge loop of Pacsin 1's F-BAR domain with a negatively charged residue (I125E or M126E) decreased the overall affinity of the protein for membranes accompanied by the loss of its membrane sculpting potential (Fig. 3 A–C). In contrast, introduction of positively charged or tryptophan residues at those positions (I125K, M126K, I125W, or M126W) maintained near wild-type membrane affinity of the mutant proteins (Figs. 3 D and E Inset). Although tryptophan is hydrophobic, its indole nitrogen may prevent it from deeply inserting into the membrane. In all cases except for the mutation I125W, vesiculation was largely suppressed (Fig. 3 D and E). Instead, predominantly wide tubes (M126K) or thin tubules (I125K, M126W), similar to morphologies that have been observed at low temperature, formed when LMVs were incubated with the mutant proteins. In samples prepared with proteins containing a tryptophan instead of the wedge loop isoleucine (I125W), vesiculation dominated, coinciding with a loss of tubular structures (Fig. 3E). The results are consistent with a model in which the methionine functions as a wedge that acts close to the acyl chains of a membrane leaflet and drives vesiculation. The adjacent isoleucine serves a supporting role, but may be closer to the lipid head group-acyl chain junction, explaining the bilateral sensitivity to tryptophan or lysine mutations at that site. Decreased salt-sensitivity of the wild-type protein compared with the lysine mutants (I125K, M126K) in binding and tubulation assays support a wedge-loop-insertion mechanism (Fig. S5).
Fig. 3.
Mutational analysis of the wedge-loop motif. (A) Negative-stained micrograph of LMVs. (B–E) Tubulation activity and membrane binding of wild-type and mutant Pacsin 1F-BAR. LMVs were incubated with wild-type Pacsin 1F-BAR (B) or proteins with glutamate (C), lysine (D), or tryptophan (E) substitutions at positions 125 and 126 located at the center of the wedge loop. Small arrows point to thin tubular structures. Insets show results from vesicle copelleting assays assessing the binding of proteins to the liposomes. Supernatant (S) and pellet (P) fractions were analyzed by SDS/PAGE followed by Coomassie staining. (Scale bars, 500 nm.)
Taken together, the structural and mutational analyses identified the wedge loop as a motif that contributes to membrane affinity and determines Pacsin's unique vesiculation activity. Also, our data suggest that vesicles originated from tubules, which can form in the absence of loop insertions.
Higher-Order Arrangements Observed in Crystals of the F-BAR Domain of Pacsin.
Previous studies revealed that the F-BAR domains of CIP4 and FBP17 could form ordered arrays that cover the membrane tube and that distinct membrane curvatures can be attributed to different lattice arrangements (15, 20). Both tip-to-tip and lateral intermolecular interactions are responsible for the assembly of protein lattices (20). Given the laterally curved state and distinct structural motifs of Pacsin, membrane engagement, lattice properties, and spacing will be different from those of the more straight F-BAR domains studied thus far. In our crystallographic studies of Pacsin 1 and 2, the F-BAR domains pack in unique, primitive lattices that may provide insight into the Pacsin-specific membrane sculpting activities. A detailed description of these lattices is provided in the SI Appendix and Fig. S6. The 2 different types of higher-order arrangements observed with Pacsin 1 and 2, respectively, suggest a loose packing mode with lower density of F-BAR domains (Pacsin 1, space group P21) (Fig. S6 A and B) that may be responsible for the stabilization of wide tubes and a tight spiral-like assembly (Pacsin 2, space group P212121) (Fig. S6_C_) that at high protein density may induce narrow tubule formation and possibly vesiculation.
Suppressed Activities in Full-Length Pacsin.
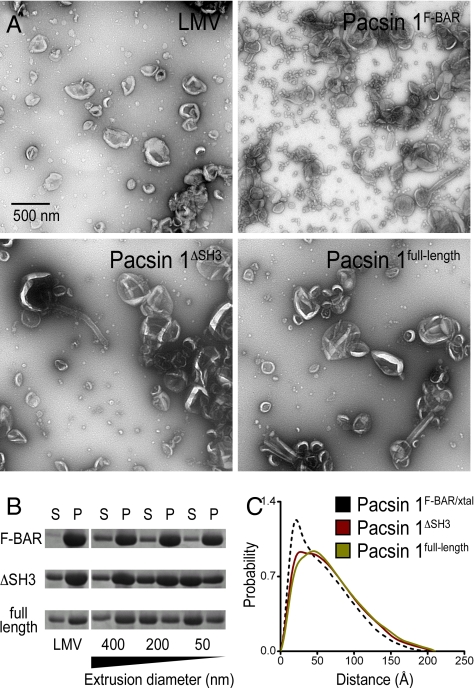
Experiments so far were carried out with the isolated F-BAR domain. Its domain boundaries were determined based on limited proteolysis and the available crystal structures. We next determined how the central linker and C-terminal SH3 domain affect Pacsin's membrane sculpting activity. When the linker (Pacsin 1ΔSH3) or linker-SH3 module (Pacsin 1full-length) was included in the construct, Pacsin's membrane deformation activity was significantly impaired, suggesting an autoinhibitory role of these units in Pacsin (Fig. 4A and Fig. S7_A_). In the case of the protein containing the linker region, full activity comparable with that of the isolated F-BAR domain could be achieved at a higher protein concentration used in the tubulation assays. In contrast, the autoinhibition in the full-length protein including the SH3 domain appeared stronger and could not be compensated for by high protein concentration (Fig. S7_B_).
Fig. 4.
Autoinhibition of human Pacsin 1. (A) Negative-stain EM. LMVs were incubated with Pacsin 1F-BAR, Pacsin 1ΔSH3, or Pacsin 1full-length. Samples were analyzed by negative-stain EM. (B) Membrane binding of Pacsin to liposomes of different size. LMVs or LUVs that were extruded through filters with pore sizes of 400, 200, or 50 nm were incubated with the 3 different Pacsin 1 construct. Samples were subjected to high-speed centrifugation. Pellet (P) and supernatant (S) fraction were analyzed on a Coomassie-stained SDS/PAGE. (C) Solution conformation of Pacsin 1ΔSH3 and Pacsin 1full-length. SAXS data were collected on homogeneous solutions of Pacsin 1ΔSH3 and Pacsin 1full-length. Based on the scattering profile in solution, the distance distribution functions, P(R), were computed and compared to the P(R) function calculated from the crystal structure of Pacsin 1F-BAR. (Scale bars, 500 nm.)
In addition, we analyzed the curvature preference of the different constructs. Folch liposomes were extruded using filters with pore sizes raging from 50–400 nm, and protein binding was determined by using a vesicle pelleting assay (Fig. 4B). The isolated F-BAR domain showed no curvature preferences, binding equally well to vesicles of all sizes and comparable with LMVs. In contrast, the full-length protein bound preferentially to LMVs and liposomes extruded through a 400-nm filter. Pacsin 1ΔSH3 yielded intermediate results. It still bound fairly well to liposomes with an average diameter of 200 nm, but binding to smaller vesicles was weaker. For the F-BAR domain of FCHo2, curvature-insensitive binding has been linked to the presence of an intact N-terminal amphipathic helix (14). In Pacsin, domains located at the C terminus of the protein altered its curvature preference and overall membrane deformation potential.
The dissociation constant for the F-BAR domain and full-length dimer in solution were comparable as determined by analytical ultracentrifugation (2.1 μM and 1.2 μM, respectively), indicating that different dimerization propensities or additional intermolecular contacts via the additional domains are unlikely to contribute to the attenuated activity of full-length Pacsin (Fig. S1). Small angle X-ray scattering (SAXS) studies revealed that the overall size (maximum diameter and radius of gyration) of the F-BAR domain dimer was comparable with the dimensions of the full-length protein, suggesting that the SH3 domains were positioned in close proximity to the F-BAR domains in solution (Fig. 4C). Modeling of SAXS data has been used successfully to determine the position of the SH3 domain of Endophilin in the context of the full-length protein and to reconstruct the PX-BAR domain module of Sorting Nexin 9 (23). Yet, a similar approach indicated that the SH3 domains of Pacsin may be linked flexibly to a rigid F-BAR domain dimer in solution, because their position relative to the F-BAR domain could not be determined unambiguously (Fig. S8). The shape of the F-BAR domain in full-length Pacsin modeled from the SAXS data were comparable with the structure obtained by X-ray crystallography, indicating that the domain is indeed rigid in solution. We speculate that flexibility between the F-BAR and SH3 domains may interfere with efficient lattice or oligomer formation. Although intramolecular occlusion of certain motifs cannot be ruled out, this model would suggest that membrane deformation may not be determined by a single F-BAR domain, but may require F-BAR domain multimers. Activation may be achieved by the binding of protein ligands to the SH3 or linker domains of Pacsin, or by posttranslational modifications.
Energetic and Geometric Considerations for Pacsin-Mediated Membrane Deformation.
Calculations estimating the energy density requirements and geometrical constraints for Pacsin-mediated membrane deformation provided a quantitative assessment of the factors that contribute to the various liposome morphologies (see SI Appendix for a detailed description of the calculations and Fig. S9). The energy required to bend a membrane is in part dependent on the degree of curvature. Although the stabilization of wide tubes (d = 60 nm) requires only a small energy density (≈0.011 kBT/nm2), the generation of narrow tubules (d = 18 nm) or vesicles (d = 30 nm) is energetically expensive (≈0.125 kBT/nm2 or ≈0.18 kBT/nm2, respectively) (Fig. S9_A_). Considering the electrostatic interactions between proteins and the membrane, a single F-BAR domain could contribute ≈0.133 kBT/nm2 at most. Based on these estimations, the generation of wide tubes may only call for 10%–20% of membrane coverage by protein. In contrast, the generation of thin tubules would require densely packed F-BAR domains on the membrane. The narrow tubules observed in the tubulation assays may be the highest degree of curvature that can be achieved by F-BAR domains (Fig. S9_E_). In addition, vesiculation cannot be rationalized solely by protein packing and is likely to involve other mechanisms such as the insertion of a wedge to increase local curvature (19, 24).
Pacsin's unique S-shaped conformation is likely to play a pivotal role in determining the total (Gaussian) membrane curvature preferred by this F-BAR domain. According to our estimations, rather straight F-BAR domains with mainly one-dimensional intrinsic curvature may preferably orient on a membrane tube (close to) vertical to the long tube axis and appear optimized to maintain tubes with radii close to the radius imposed by the curvature of an F-BAR dimer (Fig. S9_C_). In contrast, the F-BAR domain of Pacsin constitutes a scaffold that is optimized for the stabilization of a high degree of membrane curvature because of its two-dimensional curvature profile (Fig. S9_D_). The Pacsin fold may also accommodate vesicular structures better than the F-BAR domains of CIP4, FBP17, and FCHo2.
Conclusions
The generation and stabilization of high membrane curvature is of fundamental biological importance for cellular membrane trafficking and maintenance of organellar structures, for example the endoplasmic reticulum (ER) (19). In the tubular ER, Reticulons, membrane-associated proteins, maintain such high curvature by the stable insertion of helical elements into a leaflet of the bilayer, providing a more permanent anchorage (25, 26). In contrast, endocytosis and membrane trafficking relies on the reversible association of proteins at the lipid bilayer, resulting in its deformation, fission, or fusion.
From our structural and functional data, we propose a model in which tubulation and Pacsin-specific membrane bulging activities rely on the distinct curvatures and particular distribution of positively charged residues within a Pacsin dimer, in addition to the insertion of the wedge loops into the membrane. Similar to other F-BAR domain-containing proteins, Pacsin's full deformation potential is likely to rely on higher-order packing on the membrane, tunable and alternative lattices stabilizing different degrees of curvature. At the same time, the association of Pacsin with itself and the membrane is rather labile, based on the weak interactions observed crystallographically and its salt and dilution sensitivity in tubulation assays. The wedge loop is crucial for the vesiculation activity, and we speculate that Pacsin may contribute more directly to fission by the insertion of this motif that resembles fusion and fission loops in other proteins.
Recently, it has been shown that Dynamin-mediated fission proceeds through a hemi-fission event (27, 28). Scission succeeded spontaneously on GTPase-triggered disassembly of Dynamin coats that initially introduced an unstable intermediate. Pacsin may facilitate initial steps that will aid Dynamin-mediated vesicle scission or may play a regulatory role by forming metastable lattices on the membrane while stressing the bilayer via wedge-loop insertion. It has been shown that perturbing Pacsin function under intense action potential stimulation decreases the pool of endocytic vesicles and increases the number of cisternae at the synapse (29). Although Pacsin's role in recruiting Dynamin may explain such an effect on vesicle recycling, it may also suggest a more central role for Pacsin under these conditions. In Drosophila, the F-BAR domain of Pacsin was recently shown to be required for the expansion of the postsynaptic membrane system, whereas the SH3 domain serves both an inhibitory as well as a targeting role (30). These results are consistent with our findings and support our model. It remains to be investigated whether there is an apparent redundancy in the activities of Pacsin and other factors involved in endocytosis such as BAR domain-containing proteins, Dynamin, and EHD-type ATPases and how their activities are coordinated and used in the cell to drive vesicle fission.
Materials and Methods
Protein Expression and Purification.
Human full-length Pacsin 1 and 2, Pacsin 1F-BAR (residues 1–325), Pacsin 1ΔSH3 (residues 1–381), and Pacsin 2F-BAR (residues 1–324) were produced following standard molecular biology and liquid chromatography techniques. A detailed protocol is provided in the SI Appendix.
Crystallization, X-Ray Data Collection, and Structure Solution.
Crystallization conditions, crystal morphologies, and structure solution are described in detail in the SI Appendix. Datasets were collected using synchrotron radiation at the Cornell High Energy Synchrotron Source (CHESS) (beamline A1) and Argonne Photon Source.
Liposome Preparation.
Folch fraction 1 lipids (Sigma Aldrich) were dissolved in chloroform and stored at −20 °C. For liposome preparations, chloroform was evaporated under a nitrogen atmosphere for 15 min, and the samples were subjected to vacuum desiccation for 2 h at 60 mtorr. Dry films were hydrated in low salt buffer [25 mM Tris-HCl (pH 7.4) and 50 mM NaCl] to a concentration of 10 mg/mL.
To prepare LMVs, hydrated liposomes were sonicated for 15 min, followed by 8 freeze–thaw cycles using liquid nitrogen. For most applications, liposomes were diluted to a concentration of 2 mg/mL in low-salt buffer and were incubated in a heat block at 30 °C for 1 h before exposure to protein solutions. This method yields LMVs with an average diameter of 300–600 nm and the occasional occurrence of larger vesicles (≥1 μm). For the preparation of large unilamellar vesicles (LUVs), hydrated liposomes were subjected to 6 freeze–thaw cycles, followed by extrusion through filters with pore sizes ranging from 50 nm to 400 nm. LUV preparations were diluted to a concentration of 2 mg/mL and used immediately after extrusion.
Liposome Copelleting Assay.
Liposome samples (1 mg/mL) were incubated in the presence or absence of protein (10 μM) in 40 μL of low-salt buffer (corresponding to a 1:1 lipid–protein ratio) for 20 min at room temperature. Samples were centrifuged in a Optima MAX-E ultracentrifuge (Beckman) equipped with a TLA-100 rotor at 329,200 × g at 4 °C for 45 min. Pellets and supernatants were separated, pellets were resuspended in 40 μL of low-salt buffer, and both fractions were analyzed by SDS/PAGE and Coomassie Blue staining.
Negative-Stain Electron Microscopy (EM).
Liposome samples (2 mg/mL) were incubated in the presence or absence of protein (0.2–1 mg/mL or 4–20 μM) in low-salt buffer for 1 min at 25 °C, if not indicated otherwise. The sample (8 μL) was applied to a Carbon-formvar-coated copper grid (EMS) and incubated for 2 min. Excess liquid was carefully removed by using a wet absorbent tissue (Kimwipes; Kimberly Clark). The grids were incubated 3 times with 2% filtered uranyl acetate solution for 5 sec and air-dried. Negative staining was performed at 25 °C (except for samples prepared at 4 °C, which were stained in the cold room). Membrane morphologies were examined on a FEI Morgagni Transmission Electron Microscope (TEM) and a FEI Tecnai G2 F20 TEM/STEM with the electron energy set to 80 kV and 200 kV, respectively. Representative images were taken on an AMT camera with a direct magnification of 11kx–36kx. Distance and size measurements were taken by using the program ImageJ (National Institutes of Health).
SAXS.
SAXS data were collected at the Advanced Photon Source (APS), beamline 12-IDC at the electron energy of 12 KeV and 4 °C on homogeneous samples of Pacsin 1ΔSH3 and Pacsin 1full-length. Samples of Pacsin 1F-BAR showed a high sensitivity to radiation damage and were not considered in this study. Data reduction, analysis, and free-atom modeling of the SAXS data were carried out by using the program package ATSAS (31). A detailed description is provided in the SI Appendix.
Supplementary Material
Supporting Information
Acknowledgments.
We thank John Grazul and Huolin Xin at the Cornell Center for Materials Research for their assistance with scanning transmission electron microscopy and Jim Sethna, Ben Machta, Scott Emr, and Rick Cerione for helpful discussion. This work is based on research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source and at the Cornell High Energy Synchrotron Source. The facilities are supported by National Center for Research Resources at the National Institutes of Health Award RR-15301 (to Advanced Photon Source), National Science Foundation Award DMR-0225180 (to Cornell High Energy Synchrotron Source), and National Institutes of Health Award RR-01646 (to Cornell High Energy Synchrotron Source). This work was supported by a PEW Scholar award in Biomedical Sciences (to H.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the RCSB Data Bank, www.rcsb.org (PDB ID codes 3HAH, 3HAI, and 3HAJ).
References
- 1.Anggono V, et al. Syndapin I is the phosphorylation-regulated dynamin I partner in synaptic vesicle endocytosis. Nat Neurosci. 2006;9:752–760. doi: 10.1038/nn1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci. 2000;113:4511–4521. doi: 10.1242/jcs.113.24.4511. [DOI] [PubMed] [Google Scholar]
- 3.Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiProspero NA, et al. Early changes in Huntington's disease patient brains involve alterations in cytoskeletal and synaptic elements. J Neurocytol. 2004;33:517–533. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- 6.Modregger J, DiProspero NA, Charles V, Tagle DA, Plomann M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington's disease brains. Hum Mol Genet. 2002;11:2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- 7.Braun A, et al. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessels MM, Qualmann B. The syndapin protein family: Linking membrane trafficking with the cytoskeleton. J Cell Sci. 2004;117:3077–3086. doi: 10.1242/jcs.01290. [DOI] [PubMed] [Google Scholar]
- 9.Itoh T, et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Farsad K, et al. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 12.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 13.Tsujita K, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henne WM, et al. Structure and analysis of FCHo2 F-BAR domain: A dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Shimada A, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 16.Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J Mol Biol. 2005;351:653–661. doi: 10.1016/j.jmb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Gallop J, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda M, et al. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerberg J, Kozlov M. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 20.Frost A, et al. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 22.Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Kaan HY, Hooda RN, Goh SL, Sondermann H. Structure and plasticity of Endophilin and Sorting Nexin 9. Structure. 2008;16:1574–1587. doi: 10.1016/j.str.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 26.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 27.Bashkirov PV, et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson F, Jakobsson J, Löw P, Shupliakov O, Brodin L. Perturbation of syndapin/PACSIN impairs synaptic vesicle recycling evoked by intense stimulation. J Neurosci. 2008;28:3925–3933. doi: 10.1523/JNEUROSCI.1754-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar V, et al. Syndapin promotes formation of a postsynaptic membrane system in Drosophila. Mol Biol Cell. 2009;20:2254–2264. doi: 10.1091/mbc.E08-10-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J Appl Cryst. 2007;40:S223–S228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information