Mapping Anatomical Connectivity Patterns of Human Cerebral Cortex Using In Vivo Diffusion Tensor Imaging Tractography (original) (raw)
Abstract
The characterization of the topological architecture of complex networks underlying the structural and functional organization of the brain is a basic challenge in neuroscience. However, direct evidence for anatomical connectivity networks in the human brain remains scarce. Here, we utilized diffusion tensor imaging deterministic tractography to construct a macroscale anatomical network capturing the underlying common connectivity pattern of human cerebral cortex in a large sample of subjects (80 young adults) and further quantitatively analyzed its topological properties with graph theoretical approaches. The cerebral cortex was divided into 78 cortical regions, each representing a network node, and 2 cortical regions were considered connected if the probability of fiber connections exceeded a statistical criterion. The topological parameters of the established cortical network (binarized) resemble that of a “small-world” architecture characterized by an exponentially truncated power-law distribution. These characteristics imply high resilience to localized damage. Furthermore, this cortical network was characterized by major hub regions in association cortices that were connected by bridge connections following long-range white matter pathways. Our results are compatible with previous structural and functional brain networks studies and provide insight into the organizational principles of human brain anatomical networks that underlie functional states.
Keywords: anatomical connectivity, betweenness centrality, DTI tractography, network, small world
Introduction
The human brain is a complex system that is capable of generating and integrating information from multiple sources with high efficiency (Sporns et al. 2004). Characterization of the global architecture of the anatomical connectivity patterns in the human brain is therefore crucial because it could increase our understanding of how functional brain states emerge from their underlying structural substrates and provide new insights into the association of brain function deficits with underlying structural disruption in brain disorders (Sporns et al. 2005).
Although the single neuron is the basic element of the brain, constructing and analyzing anatomical networks at the level of the neuron are unrealistic, given the huge amount of neurons (∼1011) in the human brain. Currently, anatomically segregated brain regions containing large population of neurons with similar cytoarchitecture or functional involvement and interregional pathways possibly represent the most appropriate organizational level for the brain network analyses (Sporns et al. 2005). At this level, several anatomical networks have been established using chemical tract-tracing methods but are limited to the brain of mammalia such as the cat and primate (Felleman and van Essen 1991; Scannell and Young 1993; Young 1993). Further network analyses have revealed that these anatomical networks contain many nontrivial topological properties such as the existence of clusters of brain regions (Hilgetag, Burns, et al. 2000; Honey et al. 2007) and hierarchical organization (Hilgetag et al. 1996; Hilgetag, O'Neill, et al. 2000). It has been also demonstrated (Sporns and Zwi 2004) that these mammalian cortical networks have a “small-world” topology that is characterized by greater local interconnectivity or cliquishness as compared with a “random” network and smaller characteristic path length linking individual nodes as compared with a “regular” network (Watts and Strogatz 1998). However, the direct evidence for anatomical connectivity networks in the human brain remains scarce, even at a macroscale, mainly due to the fact that most invasive experimental methods (e.g., chemical tracing) used in the animal brain cannot be directly applied to the human brain (Crick and Jones 1993). Recently, Sporns et al. (2005) have referred to the comprehensive, detailed structural description of the network with elements and connections forming the human brain as the “human connectome” and advocated urgent research efforts in this area.
Recent advances in modern neuroimaging techniques have allowed for noninvasive investigation of human brain networks. Using neurophysiological data (e.g., functional magnetic resonance imaging [fMRI], electroencephalography [EEG], magnetoencephalography [MEG]), several research groups have established the functional brain networks in humans and further reported important characteristics of these networks, such as small-world attributes (Stam 2004; Stam et al. 2007; Eguiluz et al. 2005; Salvador et al. 2005a; Achard et al. 2006; Micheloyannis et al. 2006). Recently, He et al. (2007) established a human brain morphological network with cortical thickness measurement as a proxy for connectivity and observed network topology compatible with the functional brain networks. Considerable progress has been made in looking into the brain anatomical circuitry with the development of diffusion MRI that can characterize the orientation of white matter (WM) fiber bundles by detecting underlying water molecule diffusion (for a review, see Le Bihan 2003). Specifically, diffusion tractography methods (also called fiber tracking) were developed to investigate the brain anatomical connectivity in vivo. Deterministic “streamline” tractography using diffusion tensor imaging (DTI) infers the continuity of fiber bundles from voxel to voxel (Mori and van Zijl 2002). Along with multiple manual/automatic regions of interest (ROIs) selection, DTI deterministic tractography is capable of noninvasive visualization of major WM tracts faithful to the known WM anatomy (Catani et al. 2002; Wakana et al. 2004). Recently, probabilistic diffusion tractography methods, which focus on the connectivity probabilities rather than the actual WM pathways between voxels, have also been developed (Behrens et al. 2003; Parker and Alexander 2005).
To our knowledge, there are only 2 recent studies using diffusion MRI to investigate human brain anatomical networks. The first effort was made by Hagmann et al. (2007), in which small-world topology was first confirmed in anatomical networks of individual brains. This initial study included 2 subjects and defined network nodes subject-specifically at a voxel population level (i.e., thousands of small ROIs) rather than a regional level, which makes the brain anatomical network less comparable across subjects. The second study, from Iturria-Medina et al. (2008), established a weighted anatomical network for individual brains using diffusion MRI-based “anatomical connection probabilities” in a group of subjects (n = 20) and further reported a broad range of the network characteristics, such as small-world properties and efficiency. Under this probabilistic scheme, however, a nonzero connection probability value was assigned to many brain region pairs, even those that other technologies suggest are unlikely to be connected (e.g., left frontal and right occipital cortex). Given the relatively low sensitivity of diffusion techniques, additional large data set studies are necessary.
The objective of this study was to utilize DTI on a large sample of healthy subjects (80 young adults) to 1) construct a macroscale anatomical connectivity network in human cerebral cortex and 2) further analyze its underlying topological properties. Notably, we aimed to establish a population-based anatomical network capturing the underlying common connectivity pattern of the cerebral cortex (i.e., backbone) across young healthy adults, rather than a subject-specific and very detailed network for an entire individual brain. To do this, the entire cerebral cortex was first parcellated into multiple cortical regions (each region was defined as a network node) using the automated anatomical labeling (AAL) (Tzourio-Mazoyer et al. 2002), and the resulting regions can be classified as the primary, association, paralimbic, or limbic cortex (Mesulam 2000). The network connections were then inferred from DTI tractography. With the constructed cortical network, graph theoretical approaches were further employed to examine the topological characteristics of the network. Specifically, the present study would determine: 1) whether this population-based cortical network has a small-world architecture and a specific connectivity distribution, 2) whether the cortical network includes vital hub nodes/bridge edges, and 3) whether the topological characteristics of the network are compatible with those of previous non-DTI brain networks studies.
Materials and Methods
Subjects
The present study included 80 right-handed young adults (males/females: 38/42; age: 18–31 years) who were selected from a large developmental data set (Lebel et al. 2008). The subjects were recruited through advertising on campus and in local communities, most of them are Caucasian (>75%). All subjects have no history of neurological or psychiatric disorders. Informed consent was obtained from each subject, and our protocol was approved by the University of Alberta Health Research Ethics Board.
MRI Acquisition
All scans were performed on the same Siemens Sonata 1.5T MRI scanner (Siemens Medical Systems, Erlangen, Germany). Diffusion tensor images were acquired by using a twice-refocused single-shot Echo-Planar Imaging-based sequence: coverage of the whole brain, 3-mm slice thickness with no interslice gap, 40 axial slices, time repetition (TR) = 6400 ms, echo time (TE) = 88 ms, 6 diffusion directions with b = 1000 s/mm2, number of excitations (NEX) = 8, in-plane acquisition matrix = 128 × 128 with 75% phase partial Fourier (zero filled and interpolated to 256 × 256), field of view (FOV) = 220 × 220 mm2, and scan time = 6:06 min. The voxel size of diffusion-weighted images was trilinearly interpolated to 1-mm isotropic dimension during postprocessing. Three-dimensional (3D) _T_1-weighted images with high resolution were obtained by a magnetization prepared rapid acquisition gradient-echo (MPRAGE) sequence with the following parameters: 1-mm slice thickness with no interslice gap, 144 axial slices, TR = 1890 ms, TE = 4.38 ms, time to inversion = 1100 ms, NEX = 1, in-plane acquisition matrix = 256 × 192, FOV = 256 × 192 mm2, and scan time = 6:03min.
The Construction of the Anatomical Connectivity Network
Network Node Definition
Node definition is important in the brain network construction as the node is the most basic element of a network (Sporns et al. 2005). In this study, we employed the AAL template (Tzourio-Mazoyer et al. 2002) to parcellate the cerebral cortex into 78 cortical regions (39 for each hemisphere, see Supplementary Table 1), each representing a node of the cortical network. For each subject, the parcellation process was conducted in the DTI native space. To do this, each individual structural image (i.e., _T_1-weighted MP–RAGE image) was first coregistered to the b0 image in the DTI space using a linear transformation (Fig. 1). The transformed structural image was then mapped to the _T_1 template of ICBM152 in the Montreal Neurological Institute (MNI) space using a nonlinear transformation. The resulting inverse transformation was then used to warp the AAL mask from the MNI space to the DTI native space in which the discrete labeling values were preserved by using a nearest neighbor interpolation method (Fig. 1). Both the linear and nonlinear mappings were implemented in the SPM5 package (http://www.fil.ion.ucl.ac.uk/spm/software/spm5/).
Figure 1.

A flowchart for the construction of the cortical anatomical network in the human brain using DTI tractography. (1) Rigid coregistration from _T_1-weighted structural MRI (a) to DTI native space (c, DTI color-coded map; red: left to right; green: anterior to posterior; blue: inferior to superior) for each subject. (2) Nonlinear registration from the resultant structural MRI to _T_1 template of ICBM152 in the MNI space (b), resulting in a nonlinear transformation (T). (3) Applying the inverse transformation (_T_−1) to the AAL template in the MNI space (d), resulting in the subject-specific AAL mask in the DTI native space (f). All registrations were implemented in the SPM5 package. (4) Reconstructing all the WM fibers (e) in the whole brain by using DTI deterministic tractography. (5) Determining the WM fibers connecting every pair of cortical regions for each subject. (6) Identifying the population-based cortical network matrix (g, blue: 1; blank: 0) by applying nonparametric sign test to every pair of cortical regions (P < 0.05, Bonferroni corrected). For more details, see Materials and Methods. R, right; L, left. The abbreviations of the cortical regions were established by Achard et al. (2006) and are included as the Supplementary Table 1.
Interregional Anatomical Connections by DTI Tractography
The distortion of diffusion-weighted images due to eddy currents was first corrected using an affine registration (Woods et al. 1998). The diffusion tensor matrix was then calculated voxel-by-voxel and diagonalization was performed to yield 3 eigenvalues and eigenvectors (Basser and Pierpaoli 1996). The DTI tractography was further implemented using a continuous streamline-tracking algorithm as follows (Mori et al. 1999). Briefly, the structural _T_1-weighted image was first classified into gray matter (GM), WM, and cerebrospinal fluid (CSF) in SPM5. Then all fiber bundles of the brain were reconstructed with DTI studio-2.5 (Johns Hopkins University, Baltimore, MD) by selecting all WM voxels as seed voxels for fiber tracking. In each voxel, the orientation of the largest component of the diagonalized diffusion tensor was assumed to represent the orientation of the dominant fiber bundles. The tracking was initiated from the center of the seed voxel and proceeded along its fiber bundle orientation. When the track left the current voxel and entered that one, the proceeding direction was changed to the fiber bundle orientation of the neighboring voxel. This tracking procedure continued until a voxel classified as GM or CSF was reached or the turning angle between adjacent voxels was greater than 45 degrees. Notably, we did not adopt the so-called “single tracking approach”, which initiates fiber tracking only from specific ROIs. Instead, we chose the so-called “exhaustive search approach” in which fiber tracking is first performed from all voxels and then fiber bundles crossing (or ending in) specific ROIs are selected. This approach has been demonstrated to be more practical to handle the branching problem of WM tracts (Mori and van Zijl 2002).
Under the tracking criterion above, the trajectories of all fiber bundles were restricted to WM voxels. Fiber bundles were considered linked to a cortical region if one of the fiber bundle's end points was adjacent to the cortical region. Notably, each cortical mask of the AAL template is not a pure cortical GM mask but includes tissues from both cortical GM and subcortical WM (Tzourio-Mazoyer et al. 2002). The inclusion of subcortical WM allowed us to determine whether fiber bundles were linked to a cortical region using the corresponding AAL mask directly. But fiber bundles may be chosen erroneously if an AAL mask contained too many WM voxels that are not truly adjacent to the cortex. To address this issue, WM voxels in the raw AAL cortical mask were removed if no cortical voxels existed within their 2-mm cubic neighborhood. Using the refined cortical masks, 2 cortical regions were considered as “anatomically connected” if fiber bundles with 2 end points located in their respective masks were present. For each subject, the number of existing fiber bundles connecting every pair of regions was counted. It should be noted that the number of fiber bundles was used only to indicate the existence/absence of fiber bundles, and not to represent the connectivity strength or probability, as the fiber bundle numbers are strongly dependent on the tracking algorithm, image resolution, etc. Notably, probabilistic tractography can also be used to identify the cortical connections but was not an option for our current study as our data were only acquired with 6 diffusion directions.
Anatomical Connectivity Network Across Population
Given the individual variability of brain anatomy (Thompson et al. 1996; Amunts et al.1999; Westbury et al. 1999), it is not surprising that anatomical connectivity between regions differs across subjects. In this study, we focused on the connections that were most consistent across subjects, that is, the backbone network. To identify the highly consistent cortical connections, a nonparametric one-tailed sign test was applied. For each pair of cortical regions, the sign test was performed with the null hypothesis that there is no existing connection, that is, “fiber bundle number = 0.” The Bonferroni method was used to correct for multiple comparisons (i.e., 78 × 77/2 = 3003 pairs of regions) at P < 0.05 (Fig. 1). The sign test was chosen because of doubts about the validity of absolute fiber bundle number estimates and to minimize inclusion of false positives. The use of this conservative statistical criterion generated a symmetric binarized matrix that captured underlying anatomical connectivity patterns in the human cerebral cortex. In the current study, we focused on a simpler on–off connectivity pattern. This binarized network could potentially be further developed to a weighted one in the future.
The Cortical Network Topological Analysis
Evaluation of the Small-World Property
Small-world measures of a network (clustering coefficient, Cp, and characteristic path length, Lp) were originally proposed by Watts and Strogatz (1998). Briefly, the Cp is the average of the clustering coefficients over all nodes in a network, where the clustering coefficient Ci of a node i is defined as the number of existing connections among the node's neighbors divided by all their possible connections. Cp quantifies the extent of local cliquishness or local efficiency of information transfer of a network (Watts and Strogatz 1998; Latora and Marchiori 2001). The Lp of a network is measured here by using a “harmonic mean” distance between pairs proposed by Newman (2003), that is, the reciprocal of the average of the reciprocals. Lp quantifies the ability of parallel information propagation or global efficiency (in terms of 1/Lp) of a network (Latora and Marchiori 2001). A real network would be considered small world if it meets the following criteria: γ=Cpreal/Cprand≫1 and λ=Lpreal/Lprand≈1 (Watts and Strogatz 1998), where Cprand and Lprand are the mean clustering coefficient and characteristic path length of 1000 matched random networks that preserve the same number of nodes, edges, and degree distribution as the real network (Maslov and Sneppen 2002; Sporns and Zwi 2004). Of note, these topological parameters may change with the selection of statistical threshold. When the statistical criterion is stiffened, fewer connections will survive, leading to a sparser network. To test the effect of thresholding, we repeatedly calculated the topological parameters as a function of the statistical thresholds (P value range: 1–10−6).
Betweenness Centrality
The betweenness of a node Binode or an edge Biedge is defined as the number of shortest paths between pairs of other nodes that pass through the node or the edge (Freeman 1977; Girvan and Newman 2002). Binode and Biedge are global centrality measures that capture the influence of a node or edge over information flow between other nodes in the network. Binode and Biedge were first calculated using the MatlabBGL package (http://www.stanford.edu/∼dgleich/programs/matlab_bgl/). The normalized betweenness was then calculated as binode=Binode/<Bnode> or biedge=Biedge/<Bedge>, where <_B_node> and <_B_edge> were the average node or edge betweenness of the network, respectively. The nodes or edges with the largest normalized betweenness values were considered pivotal nodes (i.e., hubs) or edges (i.e., bridges) in the network. Specifically, nodes or edges were identified as the hubs or bridges in the cortical network if their betweenness values were at least one standard deviation (SD) greater than the average betweenness of the network (i.e., binode>mean+SD or biedge>mean+SD).
Vulnerability
The vulnerability is widely used to quantitatively measure the damage on the network performance caused by the simulated failure of its elements (Costa et al. 2007). To calculate the vulnerability of an individual node or edge in the cortical network, we removed the nodes or edges one by one from the network and calculated the changes in the mean shortest path length (i.e., global efficiency) of resulting network by Vi=100×(Lp−L′p)/Lp%, where Lp is the shortest path length of the real brain network and L′p is the shortest path length of the brain network after removing the node or edge. To test the effects of hubs/bridges and non-hubs/non-bridges on the network performance, we compared the vulnerability values of these 2 groups using a 2-sample _t_-test. Notably, vulnerability and betweenness centrality are 2 important measures for characterizing the influence of nodes/edges in a network. In many cases, a node/edge with high betweenness has more vulnerability. However, the nodes/edges with higher betweenness centrality are not necessarily more vulnerable because vulnerability quantifies the node/edge influence from a distinct aspect as compared with betweenness.
Topological Distribution
Small-world networks can be classified into different categories (e.g., power-law, exponential, and exponentially truncated power-law) according to their node degree (degree of a node is the number of connections linking the node) distribution (Amaral et al. 2000), each showing different network behavior such as degree of resilience to targeted attacks (Albert et al. 2000; Achard et al. 2006). Here, we examined the node degree distribution of the human cortical anatomical network. Additionally, we also investigated the node and edge betweenness distribution of the cortical network. These features have not been examined in previous brain network studies. Three possible forms of distribution were fitted to the probability of degree, node, and edge betweenness: a power-law, p(x) ∼ _x_α−1; an exponential, p(x) ∼ exp(x/xc); and an exponentially truncated power-law, p(x) ∼ _x_α−1exp(x/xc). Here a cumulative distribution was used to reduce the effects of noise on this smaller data set (Strogatz 2001), and goodness-of-fit was tested using _R_2 values (a value closer to 1 indicates a better fitting).
Results
Connections of Human Cortical Network via Fiber Bundles
Under our statistical criterion (P < 0.05, Bonferroni corrected), 329 pairs of cortical regions showed significant anatomical connectivity in our study population of young adults. The connection indicates the existence of DTI deterministic tractography-derived fiber bundles between the cortical regions. The cortical network WM is mostly comprised of association (intrahemispheric) and commissural fibers (interhemispheric), in terms of classic WM anatomy (Nolte 1993). The projection fibers connecting the cortex with noncortical structures (e.g., brainstem and thalamus, etc.) are not involved in our cortical network.
Figure 2 illustrates some examples of interregional cortical connections, involving 4 short WM tracts (Fig. 2_a_–d) and 9 major WM tracts (Fig. 2_e_–m, the genu of corpus callosum [CC], body of CC, splenium of CC, inferior longitudinal fasciculus [ILF], arcuate fasciculus [AF], superior longitudinal fasciculus [SLF], uncinate fasciculus [UF], cingulum, and inferior frontooccipital fasciculus [IFO]). The cortical regions linked by these tracts in the cortical network are listed in Figure 2. For the major WM tracts (Fig. 2_e_–m), their 3D trajectory and linked cortical regions are faithful to the postmortem WM anatomy (Crosby et al. 1962) as well as the human WM anatomy from previous DTI studies (Wakana et al. 2004).
Figure 2.
Examples of cortical connections and their corresponding WM fibers in one subject. The 13 selected cortical connection examples included 4 short WM tracts (a_–_d) and 9 well-known major WM tracts (e_–_g, CC; h, ILF; i, AF; j, SLF; k, UF; l, cingulum; m, IFO) that are well identified with DTI deterministic tractography. As well, the linked cortical regions for each selected connection are listed with the abbreviations in the Supplementary Table 1. It should be noted that the fiber bundles shown here are only a part of a specific major WM tract, rather than the entire tract.
Human Cortical Network and Its Topological Property
Seventy-eight cortical regions and 329 identified interconnections constitute a binarized cortical network with a sparsity of ∼11% (329 interconnections of 3003 potential between-region connections), in which there are no isolated nodes (i.e., cortical regions).
Is the Human Cortical Network Small World?
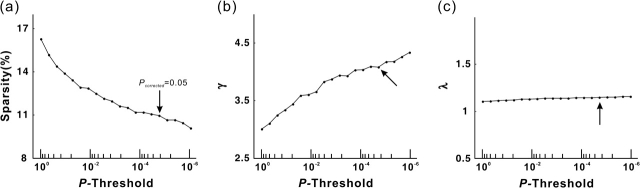
The clustering coefficient of the cortical network (Cpcortex=0.49) is approximately 4 times that of a comparable random network (Cprand=0.12), whereas the path length (Lpcortex=2.32) is approximately equivalent to the random network (Lprand=2.02). The relationship between the Cp and Lp (γ = 4.07; λ = 1.15) fits the definition of a small-world network (Watts and Strogatz 1998). To test for robustness, we divided all 80 subjects into 2 groups (40 subjects for each group, age matched) and calculated the split-half reliability. The cortical networks were constructed with the same criterion listed in the Materials and Methods for the 2 groups. The resulting networks of the 2 subgroups showed high similarities (group 1: λ = 4.63, γ = 1.16; group 2: λ = 4.69, γ = 1.17; node-betweenness correlation: R = 0.90, P < 10−29), suggesting the reliability of our method. In Table 1, we listed the topological parameters across studies in which small-world properties have been consistently demonstrated in the human brain networks at a regional level. To determine whether the human anatomical cortical networks had the small-world properties at other statistical thresholds, we further plotted the topological parameters as a function of P value threshold (Fig. 3). As the threshold becomes lower (i.e., more conservative statistical criterion), the clustering coefficient increases rapidly, whereas the average path length changes little (Fig. 3). Overall, the small-worldness of the cortical anatomical network tends to increase as the P value threshold decreases. However, it is currently difficult to determine a definitive statistical criterion during the construction of human brain networks (Achard et al. 2006; He et al. 2007). The small-world characteristic shown here indicates that cortical anatomical networks of the human brain have greater local interconnectivity or cliquishness and short mean distance between cortical regions.
Table 1.
Topological parameters of human brain networks at a macroscale level
| Human brain network (regional level) | N | Cp | Lp | γ | λ | Topological distribution |
|---|---|---|---|---|---|---|
| Anatomical network (the present study) | 78 | 0.49 | 2.32 | 4.07 | 1.15 | Exponentially truncated power-law distribution (degree and betweenness) |
| Anatomical network (Iturria-Medina et al. 2008) | 90 | Not reported | Not reported | 1.85 | 1.12 | Exponentially truncated power-law distribution (degree) |
| Morphological network (He et al. 2007) | 54 | 0.30 | 3.05 | 2.36 | 1.15 | Exponentially truncated power-law distribution (degree) |
| Functional network (Salvador et al. 2005a) | 45 | 0.25 | 2.82 | 2.08 | 1.09 | Not reported |
| Functional network (Achard et al. 2006) | 90 | 0.53 | 2.49 | 2.37 | 1.09 | Exponentially truncated power-law distribution (degree) |
Figure 3.
The topological parameters as a function of the statistical threshold. (a) The sparsity of the cortical anatomical networks decreases as the P value threshold lowers (i.e., more conservative statistical criterion). (b) The clustering coefficient ratio (gamma) increases as the P value threshold lowers. (c) The path length ratio (lambda) shows little change as the P value threshold lowers. Overall, the small-worldness of the cortical anatomical network tends to increase as a function of lowering the P value threshold. The cortical anatomical network also exhibits small-world attributes even under very relaxing statistical criterion. The black arrow indicates the values of topological parameter of the human cortical network under our conservative statistical criterion (P < 0.05, Bonferroni corrected, which corresponds to 0.05/(78 × 77/2) = 0.05/3003 ∼1 × 10−5 without correction).
Betweenness Centrality of Node/Edge in the Network
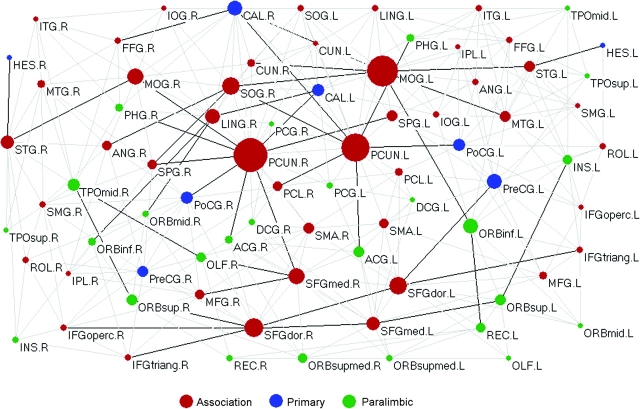
Betweenness centrality is an important metric that can be used to determine the relative importance of a node or edge within a network and identify the pivotal nodes/edges in the complex network. Figure 4 illustrates the topological map of the human cortical network in conjunction with node/edge betweenness (for a full list of parameters for all nodes, see Supplementary Table 2). We found that node betweenness of left hemisphere is linearly correlated with that of right hemisphere (t = 6.2, P < 10−6) although with a rightward asymmetry on average (Fig. 5). The absolute connectivity pattern and the betweenness centrality showed large hemispheric differences for some individual regions (e.g., middle occipital gyrus [MOG] and superior occipital gyrus [SOG]) (Figs 1_g_ and 5). Further, nodes or edges are identified as the hubs or bridges in the cortical network if their betweenness values are at least one SD greater than the average betweenness of the network (i.e., binode>mean+SD orbiedge>mean+SD). Tables 2 and 3 summarize the hub nodes and bridge edges, respectively. The identified hub nodes (9 in total, Figs 4 and 6 and Table 2) include 8 regions of the heteromodal or unimodal association cortex (bilateral precuneus [PCUN], bilateral MOG, bilateral dorsolateral superior frontal gyrus [SFGdor], right SOG, and right medial superior frontal gyrus [SFGmed]) and 1 region of the primary cortex (right calcarine cortex). The identified bridge edges (43 in total, Fig. 4 and Table 3) include 11 interhemispheric, 17 interlobe, and 15 intralobe connections that are mainly associated with several major WM tracts (e.g., CC, IFO, ILF, SLF, and cingulum). Moreover, most of the bridge edges are linked to association cortex regions with high node-betweenness centrality (i.e., hub regions) (Fig. 4).
Figure 4.
The topological map of human cortical network. In the map, nodes represent brain cortical regions and lines represent the DTI tractography–derived anatomical connections between regions. Circle size (i.e., diameter) represents the magnitude of normalized node-betweenness centrality (Table 2 and Supplementary Table 2). Association, primary, and paralimbic cortex regions are marked as red, blue, and green, respectively. Dark solid lines represent bridge connections with high normalized edge-betweenness values (Table 3). The geometric distance between regions on the drawing space approximately corresponds to the shortest path length between them. The network was visualized with the Pajek software with slight manual adjustment for the locations of brain regions (Batagelj and Mrvar 1998). For the abbreviations of the regions, see Supplementary Table 1.
Figure 5.
The relation between the node betweenness of left and right hemispheres. Each circle represents one cortical region (39 in total). The black line indicates the linear-fitted curve and the dash lines indicate 95% confidence interval. The node betweenness of left hemisphere is linearly correlated with that of right hemisphere (t = 6.2, P < 10−6). Of note, the absolute betweenness centrality of some individual cortical regions (e.g., MOG and SOG) demonstrates large hemispheric asymmetry.
Table 2.
Cortical regions identified as hub nodes in the human cortical network and their properties
| Hub regions | Class | binode | ki | Ci | Li | Vinode | Identified as a hub in previous human brain networks studies |
|---|---|---|---|---|---|---|---|
| PCUN.R | Association | 6.19 | 20 | 0.27 | 1.74 | 2.47 | Anatomicala and functionalb networks |
| MOG.L | Association | 5.56 | 17 | 0.34 | 1.88 | 1.97 | Functional networksb |
| PCUN.L | Association | 4.97 | 19 | 0.29 | 1.78 | 2.09 | Anatomicalaand functionalb networks |
| SFGdor.R | Association | 2.90 | 11 | 0.35 | 2.13 | 0.78 | Anatomicala, functionalb and morphologicalc networks |
| SFGdor.L | Association | 2.84 | 11 | 0.38 | 2.08 | 0.77 | Anatomicala, functionalband morphologicalc networks |
| SOG.R | Association | 2.73 | 13 | 0.54 | 1.93 | 0.85 | Functionalb and morphologicalc networks |
| SFGmed.R | Association | 2.53 | 13 | 0.35 | 1.96 | 0.76 | Morphologicalc networks |
| MOG.R | Association | 2.31 | 12 | 0.44 | 2.10 | 0.44 | Functionalbnetworks |
| CAL.R | Primary | 2.25 | 13 | 0.56 | 1.95 | 0.77 | Functionalbnetworks |
Table 3.
Cortical connections identified as bridges in the human cortical network and their properties
| Region A | Region B | Class | Adjacent | biedge | Viedge | Potentially involved major WM tracts |
|---|---|---|---|---|---|---|
| SOG.R | MOG.L | Inter-H | N | 5.16 | 0.357 | CC |
| SFGdor.L | SFGdor.R | Inter-H | N | 4.79 | 0.332 | CC |
| MOG.L | CAL.R | Inter-H | N | 4.29 | 0.331 | CC |
| ORBinf.L | MOG.L | Inter-L | N | 3.96 | 0.428 | IFO |
| SPG.L | PCUN.R | Inter-H | N | 3.83 | 0.376 | CC |
| PoCG.L | PCUN.L | Intra-L | Y | 3.81 | 0.290 | SLF I |
| MOG.L | STG.L | Inter-L | N | 3.79 | 0.336 | ILF |
| MOG.R | STG.R | Inter-L | N | 3.67 | 0.202 | ILF |
| PoCG.R | PCUN.R | Intra-L | Y | 3.64 | 0.326 | SLF I |
| SFGmed.R | PCUN.R | Inter-L | N | 3.37 | 0.232 | Cingulum |
| PCUN.L | ACG.L | Inter-L | N | 3.37 | 0.403 | Cingulum |
| SFGdor.L | PreCG.L | Intra-L | Y | 3.21 | 0.277 | SLF I |
| ANG.R | SOG.R | Inter-L | Y | 3.16 | 0.222 | SLF II |
| ORBinf.R | LING.R | Inter-L | N | 3.08 | 0.368 | IFO |
| HES.R | STG.R | Intra-L | Y | 3.01 | 1.702 | N/A |
| PCUN.R | MOG.R | Inter-L | N | 2.99 | 0.218 | N/A |
| REC.L | ORBinf.L | Intra-L | Y | 2.74 | 0.255 | N/A |
| ORBsup.L | INS.L | Inter-L | Y | 2.46 | 0.272 | N/A |
| PCUN.R | ACG.R | Inter-L | N | 2.44 | 0.206 | Cingulum |
| MOG.L | MTG.L | Inter-L | Y | 2.41 | 0.209 | N/A |
| OLF.R | TPOmid.R | Inter-L | N | 2.41 | 0.208 | UF |
| PCUN.L | PHG.L | Inter-L | N | 2.40 | 0.223 | Cingulum |
| ORBsup.R | SFGdor.R | Intra-L | Y | 2.40 | 0.235 | N/A |
| OLF.R | SFGmed.R | Intra-L | N | 2.29 | 0.166 | N/A |
| PCUN.R | PHG.R | Inter-L | N | 2.29 | 0.232 | Cingulum |
| CAL.L | LING.R | Inter-H | N | 2.28 | 0.201 | CC |
| CAL.R | FFG.R | Intra-L | N | 2.24 | 0.210 | N/A |
| SPG.R | PCUN.R | Intra-L | Y | 2.11 | 0.148 | SLF I |
| SFGdor.R | SFGmed.L | Inter-H | N | 2.11 | 0.146 | CC |
| PCL.R | PCUN.L | Inter-H | N | 2.08 | 0.165 | CC |
| ORBmid.R | LING.R | Inter-L | N | 2.00 | 0.281 | IFO |
| SPG.R | SOG.R | Inter-L | Y | 1.98 | 0.096 | N/A |
| MFG.R | SFGmed.R | Intra-L | N | 1.98 | 0.169 | N/A |
| PCUN.L | CAL.R | Inter-H | N | 1.97 | 0.169 | CC |
| ORBsup.R | TPOmid.R | Inter-L | N | 1.97 | 0.187 | UF |
| PCUN.L | SOG.R | Inter-H | N | 1.97 | 0.152 | CC |
| SFGdor.R | IFGoperc.R | Intra-L | N | 1.96 | 0.164 | N/A |
| MOG.L | CUN.R | Inter-H | N | 1.93 | 0.168 | CC |
| HES.L | STG.L | Intra-L | Y | 1.92 | 0.187 | N/A |
| ORBsup.L | SFGmed.L | Intra-L | Y | 1.86 | 0.171 | N/A |
| SFGdor.L | IFGtriang.L | Intra-L | N | 1.85 | 0.152 | N/A |
| PCUN.R | CAL.L | Inter-H | N | 1.85 | 0.188 | CC |
| SFGdor.R | IFGtriang.R | Intra-L | N | 1.84 | 0.152 | N/A |
Figure 6.
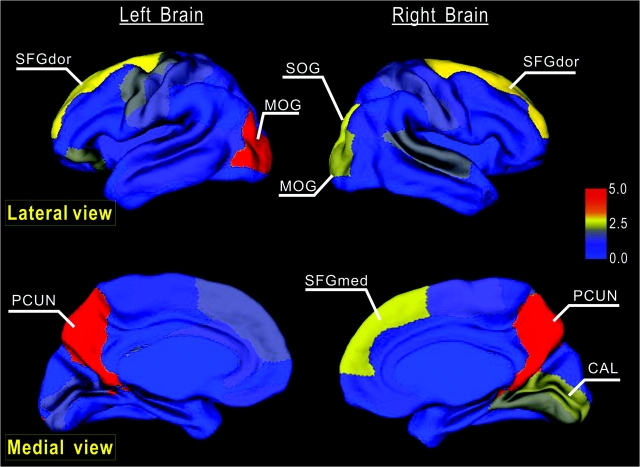
Node betweenness centrality map on the human cerebral cortex. According to the AAL template (Tzourio-Mazoyer et al. 2002), the cerebral cortex was parcellated into 78 regions (39 per hemisphere), each representing a node in the anatomical cortical network. Regions were mapped into an average cortical surface obtained from ICBM152 according to their normalized betweenness centrality values. The color bar indicating the range of normalized node betweenness is shown on the right. Hub regions identified in this study are marked on the map. Note that several hubs (PCUN, SFGdor, and MOG) appear in a bilaterally symmetric fashion (for details, see Table 2).
Vulnerability
To simulate the effects of individual node or edge “lesions” on the performance of the cortical network, we calculated the vulnerability values (_V_i) of each node and edge. We found that eliminating the hubs/bridges resulted in significantly higher vulnerability than eliminating non-hubs/non-bridges (hubs vs. non-hubs, t(76) = 7.13, P < 10−9; bridges vs. non-bridges, t(327) = 12.5, P < 10−28), which highlights the importance of these hubs/bridges in transferring information flow of the human cortical network.
Topological Distribution
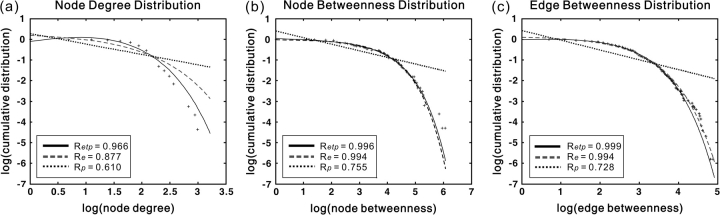
In the present study, we found that node degree distributions of the cortical network in the human brain, as well as the node/edge–betweenness distribution, were best fitted by the exponentially truncated power-law form (i.e., p(x) ∼ _x_α−1exp(x/xc)) (Fig. 7). The estimated parameters are as follows: node degree, estimated exponent α = 1.66, and cutoff degree kcnode=3.65 (Fig. 7_a_); node betweenness, estimated exponent α = 0.96, and cutoff betweenness Bcnode=76.93 (Fig. 7_b_); edge betweenness, estimated exponent α = 1.10, and cutoff betweenness Bcedge=18.95 (Fig. 7_c_). These exponentially truncated power-law distribution models indicate that the cortical network includes some pivotal nodes (i.e., hubs) and edges (i.e., bridges) but prevents the existence of huge hubs or bridges with too much load.
Figure 7.
The degree and betweenness distributions of the human cortical network. (a) Log-log plot of the cumulative node degree distribution; (b) log-log plot of the cumulative node-betweenness distribution; (c) log-log plot of the cumulative edge-betweenness distribution. The plus sign represents observed data, the solid line is the fit of the exponentially truncated power-law (p(x) ∼ _x_α−1exp(x/xc)), the dashed line is an exponential (p(x) ∼ exp(x/xc)), and the dotted line is a power-law (p(x) ∼ _x_α−1). _R_2 was calculated to assess the goodness-of-fit (a larger value indicates a better fitting; _R_etp, _R_2 for exponentially truncated power-law fit; Re, _R_2 for exponential fit; Rp, _R_2 for power-law fit). The exponentially truncated power-law is the best fitting for all the 3 distributions (a, estimated exponent α = 1.66 and cutoff degree _kc_node = 3.65; b, estimated exponent α = 0.96 and cutoff betweenness _B_cnode = 76.93; c, estimated exponent α = 1.10 and cutoff betweenness _B_cedge = 18.95).
Discussion
In this study, DTI tractography was employed to construct a macroscale anatomical network that captures the underlying common connectivity pattern of human cerebral cortex (i.e., backbone) across a large population (N = 80) of healthy young adults. The constructed cortical network had a prominent small-world topological organization and an exponentially truncated power-law topological distribution. Also, this cortical network was characterized by pivotal regions (i.e., hubs) predominantly in association cortex that were connected by pivotal connections (i.e., bridges) mainly following long-range WM tracts. These results further our understanding of the large-scale topological organization of the human anatomical cortical network.
Node Definition of the Human Anatomical Cortical Network
The term “connectome” was proposed recently by Sporns et al. (2005), referring to the comprehensive, detailed anatomical description of the network with elements and connections forming the human brain. The key issue for compiling the connectome is how to define basic structural elements of the human brain in terms of network nodes and edges. The cortical network nodes in our study were defined using the AAL template, which divides each cerebral hemisphere into 45 regions according to functional and anatomical criteria (Tzourio-Mazoyer et al. 2002), and has been used in previous human brain functional (Salvador et al. 2005a, 2005b; Achard et al. 2006) and anatomical (Iturria-Medina et al. 2008) network studies. Although the AAL template includes both cortical and deep GM structures, such as the thalamus and the amygdala, our current network was confined to the cortical system (i.e., 39 regions per hemisphere) because of the relatively large errors associated with defining basal structures during mapping AAL mask from the MNI space to native space and the difficulty of defining the adjacent WM to basal structures that is required in our method. In contrast, Hagmann et al. (2007) proposed a data-driven method to partition the WM–GM interface, resulting in thousands of small ROIs as the network nodes. This scheme potentially avoids grouping together pieces of GM that are functionally different but makes it hard to compare the network across subjects because the ROIs are subject specific. In future studies, it might be more meaningful to define the cortical nodes based on a finer myeloarchitectonic feature.
Edge Definition of the Human Anatomical Cortical Network
The organization of WM tracts has been previously investigated using invasive techniques such as dissection, histological staining, and axonal tracing (Kobbert et al. 2000). The existing mammalian (e.g., cat and primate) large-scale connectivity networks are mainly based on these invasive techniques (Felleman and van Essen 1991; Scannell and Young 1993; Young 1993). Recently, noninvasive DTI has been developed, which is capable of providing 2 types of information: the extent of water diffusion anisotropy and its orientation (Basser and Pierpaoli 1996). The former is widely used to evaluate the integrity of underlying brain tissue (for a review, see Beaulieu 2002) and the latter can be indirectly utilized to reconstruct WM tracts, referred to as DTI tractography (Conturo et al. 1999; Jones et al. 1999; Mori et al. 1999; Basser et al. 2000). It has been well demonstrated that many WM tracts derived from DTI deterministic tractography follow known WM anatomy as shown in previous studies (Catani et al. 2002, 2003; Wakana et al. 2004). However, previous DTI deterministic tractography studies have mainly focused on several specific WM tracts such as CC, cingulum, and fornix (Xu et al. 2002; Concha et al. 2005; Gong et al. 2005). Rather than a local focus, we applied DTI deterministic tractography globally to identify the most common cortical connections in a large sample. Notably, in addition to the specifics of the tractography algorithm, the resulting connections here depend on the selection of statistical criterion, as well as the sample size. With more conservative criterion, fewer connections will survive, leading to greater sparsity of the network (Fig. 3_a_). On the other hand, a smaller sample size may yield fewer connections even under the same statistical criterion due to the reduction of the statistical power.
As shown in Figure 2, major WM tracts were successfully reconstructed, supporting the validity of DTI deterministic tractography. Of note, although WM tracts are the basis of the network connections, a 1-to-1 mapping relationship between each WM tract and each cortical connection is unlikely because 1) the anatomical definition and description of short-range WM tracts (e.g., U-fiber bundles) and even the major WM tracts are limited, 2) a named major tract (e.g., CC, SLF, etc.) generally links multiple cortical regions, and 3) the involved fiber bundles of a specific cortical connection may belong to multiple WM tracts. Consequently, the fiber bundles shown in Figure 2 are only a part a specific major WM tract, rather than the entire tract.
Small-World Cortical Anatomical Networks in Humans
The small-world network introduced by Watts and Strogatz (1998) has made a tremendous impact on the studies of numerous complex networks, from social, economic to biological networks (for a review, see Strogatz 2001). The existing mammalian cortical networks derived from chemical tracing methods (Sporns and Zwi 2004) along with the recent human structural networks derived from diffusion MRI and MRI-based cortical thickness consistently exhibited small-world attributes (Hagmann et al. 2007; He et al. 2007; Iturria-Medina et al. 2008). Also, recent studies demonstrated small-world properties in human brain functional networks using neurophysiological data, for example, fMRI (Eguiluz et al. 2005; Salvador et al. 2005a; Achard et al. 2006), EEG (Micheloyannis et al. 2006; Stam et al. 2007), and MEG (Stam 2004). In agreement with these previous studies, small-world properties were observed in our cortical network using DTI tractography of an adult population of 80 healthy subjects. Although the topological parameters are quantitatively incomparable across various brain network studies due to the diversity of species (e.g., cat, primate, and human) and network construction approaches (e.g., different node/edge definition criterion), these common findings suggest that the small-world topology is a fundamental principle of structural and functional organization of complex brain networks.
The small-world topology has high clustering coefficient and short path length, indicating the local clustering or cliquishness of the connectivity network and the small number of connections between any pair of regions. Previous computational simulation studies have demonstrated that small-world topologies emerge when networks are evolved for high complexity (Sporns et al. 2000). Therefore small-world architecture of our cortical network represents an optimal organizational pattern according to evolution and development. In terms of information flow, high clustering allows modularized information processing, which is functionally segregated from one area to another, and short paths allow effective interactions or rapid transfer of information between regions, which is essential for functional integration. The coexistence of functional segregation and functional integration ensures the effective integration of multiple segregated sources of information in the brain (Tononi et al. 1994; Sporns and Zwi 2004; Sporns et al. 2004). The small-world properties in our cortical anatomical network might provide the underlying structural substrates of such functional coexistence in the human brain. Future systematic studies exploring both anatomical and functional brain network in the same subjects would provide more direct evidence for the associations of structural and functional network properties.
Exponentially Truncated Power-Law Topological Distribution
In the present investigation, we demonstrated that both the node- and edge-betweenness centrality of the human cortical network followed exponentially truncated power-law distribution (Fig. 7). From the information flow perspective, betweenness represents the communication “load” of a node or edge within the entire network and, therefore, indicates the node/edge relative importance (Goh et al. 2001). The observed distribution model suggests that the cortical network has some “core” regions and connections but prevents the appearance of huge hubs or bridges with too much “load.” Previous studies have demonstrated that networks with truncated power-law distribution are highly resilient to random errors and targeted attacks in comparison to those with scale free (i.e., power-law) distribution (Albert et al. 2000; Achard et al. 2006). In this study, we also investigated the node degree distribution that was commonly explored in the mammalian cortical anatomical networks (Sporns and Zwi 2004), human brain structural (He et al. 2007; Iturria-Medina et al. 2008), and functional networks (Achard et al. 2006). Consistent with these previous studies, we demonstrated that the node degree distribution of the cortical network also showed an exponentially truncated power-law pattern. Nonetheless, there are inconsistent findings. For example, Kaiser et al. (2007) recently reported a scale-free (i.e., power-law degree distribution) cortical network at the regional level in cat and primate. The discrepancies in the topological distribution could be attributed to different data types and analysis method applied to these studies. Hagmann et al. (2007), however, reported an exponential distribution of node degree in the human brain anatomical networks at a voxel population level, whereas Eguiluz et al. (2005) showed a scale-free degree distribution in the human brain functional networks at a voxel level. The discrepancy among the topological distributions could be associated with the different spatial scale analysis applied in these studies.
Hub Regions and Bridge Connections in Human Cortical Network
Based on the node betweenness, 9 hub regions were identified in the cortical network (Table 2 and Fig. 6), which are predominately involved in the recently evolved heteromodal [PCUN, SFGdor, and SFGmed] or unimodal (MOG and SOG) association cortex that plays a central role receiving convergent inputs from multiple cortical regions (Mesulam 2000). Three cortical regions (PCUN, SFGdor, and MOG) appear as hubs in a bilaterally symmetric fashion. As shown in Table 2, all hub regions here were previously identified as hub regions in human brain anatomical (Iturria-Medina et al. 2008), morphological (He et al. 2007), or functional (Achard et al. 2006) networks. Of note, PCUN and SFGdor were significant in various human brain networks across studies, and their approximately equivalent regions (i.e., areas 7 and 46, respectively) have also been reported as pivotal nodes in the macaque cortical network (Honey et al. 2007; Sporns et al. 2007). Previous studies exploring the neuroanatomy of cognitive functions have demonstrated the important roles of these hub regions in multiple highly integrated functional systems. For example, PCUN has shown significant involvement in visuospatial imagery, episodic memory retrieval, self-processing, and consciousness (for a review, see Cavanna and Trimble 2006); SFGdor fundamentally contributes to diverse functional systems, such as working memory and attention (Fox et al. 2006; for a review, see Petrides 2005). There are also inconsistent hub regions across brain networks. For example, the middle frontal gyrus was previously identified as a hub in the human brain functional networks (Achard et al. 2006) but was not a hub in our cortical network. The methodological differences (e.g., network construction, hub definition criterion, etc.) and diversity of the targeted brain system (e.g., with/without subcortical regions) could be responsible for the discrepancy. As the hub regions are believed to handle multimodal or integrative function, their damage could dramatically affect the stability and efficiency of the network (Sporns and Zwi 2004; Achard et al. 2006). In accordance with this view, our vulnerability results confirmed that “lesions” in the hubs lead to significantly higher changes of the path lengths as compared with “lesions” in nonhubs.
Based on the edge betweenness, we identified 43 pivotal connections (i.e., bridges), which are mainly associated with major WM tracts connecting the 2 hemispheres/different lobes within 1 hemisphere (11/17 of 43) or connecting nonadjacent cortical regions (30 of 43). These long-range connections might play roles as shortcuts to ensure short mean path lengths in the small-world networks (Kaiser and Hilgetag 2004). It should be noted that most of the bridge edges (33 of 43) are linked with the identified hub nodes, suggesting their possible involvement in the multimodal or integrative brain function that is related to the association cortex regions. Similarly, the vulnerability results also revealed that the “lesion” of bridge connections affected the performance of this cortical network more strongly than “lesions” in nonbridge connections.
Methodological Issues
Several methodological issues in the present study need to be addressed. First, DTI deterministic tractography was employed to construct the cortical network of the human brain. Despite being widely used, this method has a limited capacity for resolving crossing fiber bundles (Mori and van Zijl 2002), which may result in the loss of some existing fiber bundles and hence miss some cortical–cortical connections (i.e., false negative). For example, those fiber bundles that are involved in interhemispheric connections of lateral cortical regions are frequently missed by this type of tractography. Also some long-distance fiber bundles within a hemisphere may be missed due to this methodological inability. Recently developed probabilistic diffusion tractography methods have the advantage of overcoming fiber crossings, as well as robustness to the image noise (Behrens et al. 2003; Parker and Alexander 2005), and thus are potentially capable of identifying those connections missed by DTI deterministic tractography. This kind of method, however, may yield spurious connections (i.e., false positive) that have not been described with other methods (Parker and Alexander 2005). In the present, we aimed to construct a population-based backbone network, the strategy was therefore to control the “false-positive” connections as minimal (e.g., using conservative statistical criterion) in the network but at the expense of “false-negative” connections. This strategy will inevitably lead to an incomplete representation of the individual cortical network. Although the compatibility of our network properties with previous findings across a wide range of studies (e.g., using fMRI, cortical thickness, etc.) suggests the validity of the constructed network, those “false negative” caused by consistent loss across individuals due to our tractography limitations may induce systematic bias to the network properties. Future studies with more sophisticated tractography methods (Behrens et al. 2007) or diffusion imaging techniques (Tuch et al. 2003), as well as finer imaging resolution or quality, could be conducted to yield a more complete representation of the anatomical brain networks.
Second, an automatic registration of individual brain to a common template was applied in the present study to parcellate the entire cerebral cortex into different cortical regions. Notably, current automatic registration techniques are difficult to guarantee the exact match of every gyrus/sulcus across subjects, especially given the intersubject variability in the anatomical boundaries of regions (Amunts et al. 1999). The limitation of current registration techniques may cause location errors of the network nodes across subjects and therefore induce bias into the network properties, for example, the hub region locations. Another concern is that we used the AAL template to parcellate cerebral cortex into 78 cortical regions. A different cortical parcellation scheme has been applied in recent human brain networks studies (He et al. 2007, 2008). The resulting networks with different parcellation schemes may therefore have different network properties. Future studies could be conducted to explore the influence of different parcellation schemes, as well as the impact of the spatial scales (e.g., region level/voxel level), on the network architectures.
Third, we observed that node betweenness of left hemisphere is linearly correlated with that of right hemisphere (t = 6.2, P < 10−6, Fig. 5). We also noted that there were obvious differences in connectivity patterns of the cortical regions between hemispheres (Fig. 1_g_). The node betweenness showed large hemispheric difference in some cortical regions (e.g., MOG and SOG) and have a rightward asymmetry on average (Fig. 5). The observed hemispheric difference is compatible with previous reports of structural and functional asymmetries (for a review, see Toga and Thompson 2003). The hemispheric differences in the connectivity patterns found in the present study were also supported from the WM tract asymmetries reported recently in DTI studies (Highley et al. 2002; Nucifora et al. 2005; Barrick et al. 2007; Vernooij et al. 2007). In future, it would be interesting to systematically examine the asymmetry of the cortical network and even its relationship with brain function.
Finally, in this study, the topological organization of the cortical network was studied in a young adult population. Previous studies have demonstrated the alteration of topological properties of functional or morphological brain networks in normal aging (Achard and Bullmore 2007) and brain disorders such as Alzheimer's disease (Stam et al. 2007; He et al. 2008). Therefore, we suspect that the topological organization of the human brain anatomical networks may be altered during normal development and aging as well as under specific brain disorders, which could be examined in future studies.
Conclusion
Using DTI tractography, we established a macroscale anatomical network capturing the underlying common connectivity pattern of the cerebral cortex in a healthy young adult population. This cortical network exhibits a prominent small-world attribute and an exponentially truncated power-law topological distribution, with the embedded pivotal regions and connections mainly involving the association cortex regions and long-range WM tracts, respectively. Our findings are largely compatible with previous human brain functional network studies using neurophysiological data and structural network studies using anatomical and diffusion MRI data, thus providing insights into our understanding of how the architecture of anatomical connection network in the human brain underlies functional and morphological organization.
Supplementary Material
Supplementary Tables 1 and 2 can be found at: http://www.cercor.oxfordjournals.org/.
[Supplementary Data]
Funding
The Canadian Institutes of Health Research (to D.W.G. and C.B.); the Canadian Language and Literacy Research Network of Centres of Excellence (to C.B.); the Alberta Heritage Foundation for Medical Research (to C.B.); Natural Sciences and Engineering Research Council (to C.L.), Promep (to L.C.); the Jeanne Timmins Costello Fellowship of the Montreal Neurological Institute (to Y.H.); MRI infrastructure from the Canada Foundation for Innovation; Alberta Science and Research Authority; Alberta Heritage Foundation for Medical Research; the University of Alberta Hospital Foundation. Fiber-tracking software (DTI studio) was kindly provided by Drs Hangyi Jiang and Susumu Mori (NIH grant P41 RR15241).
Acknowledgments
Conflict of Interest: None declared.
References
- Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 2007;3:e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Amaral LAN, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci USA. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Burgel U, Mohlberg H, Uylings HB, Zilles K. Broca's region revisited: cytoarchitecture and intersubject variability. J Comp Neurol. 1999;412:319–341. doi: 10.1002/(sici)1096-9861(19990920)412:2<319::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Barrick TR, Lawes IN, Mackay CE, Clark CA. White matter pathway asymmetry underlies functional lateralization. Cereb Cortex. 2007;17:591–598. doi: 10.1093/cercor/bhk004. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Batagelj V, Mrvar A. Pajek—program for large network analysis. Connections. 1998;21:47–57. [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system—a technical review. NMR Biomed. 2002;15:435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage. 2007;34:144–55. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50:1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- Catani M, Jones DK, Donato R, Ffytche DH. Occipito-temporal connections in the human brain. Brain. 2003;126:2093–2107. doi: 10.1093/brain/awg203. [DOI] [PubMed] [Google Scholar]
- Cavanna AE, Trimble MR. The precuneus: a review of its functional anatomy and behavioural correlates. Brain. 2006;129:564–83. doi: 10.1093/brain/awl004. [DOI] [PubMed] [Google Scholar]
- Concha L, Beaulieu C, Gross DW. Bilateral limbic diffusion abnormalities in unilateral temporal lobe epilepsy. Ann Neurol. 2005;57:188–196. doi: 10.1002/ana.20334. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci USA. 1999;96:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LD, Rodrigues FA, Travieso G, Boas PRV. Characterization of complex networks: a survey of measurements. Adv Phys. 2007;56:167–242. [Google Scholar]
- Crick F, Jones E. Backwardness of human neuroanatomy. Nature. 1993;361:109–110. doi: 10.1038/361109a0. [DOI] [PubMed] [Google Scholar]
- Crosby EC, Humphrey T, Lauer EW. Correlative anatomy of the nervous system. New York: Macmillian; 1962. [Google Scholar]
- Eguiluz VM, Chialvo DR, Cecchi GA, Baliki M, Apkarian AV. Scale-free brain functional networks. Phys Rev Lett. 2005;94:018102. doi: 10.1103/PhysRevLett.94.018102. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci USA. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman LC. Set of measures of centrality based on betweenness. Sociometry. 1977;40:35–41. [Google Scholar]
- Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Kahng B, Kim D. Universal behavior of load distribution in scale-free networks. Phys Rev Lett. 2001;87:278701. doi: 10.1103/PhysRevLett.87.278701. [DOI] [PubMed] [Google Scholar]
- Gong G, Jiang T, Zhu C, Zang Y, Wang F, Xie S, Xiao J, Guo X. Asymmetry analysis of cingulum based on scale-invariant parameterization by diffusion tensor imaging. Hum Brain Mapp. 2005;24:92–98. doi: 10.1002/hbm.20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, Meuli R, Thiran JP. Mapping human whole-brain structural networks with diffusion MRI. PLoS ONE. 2007;2:e597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci. 2008;28(18):4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highley JR, Walker MA, Esiri MM, Crow TJ, Harrison PJ. Asymmetry of the uncinate fasciculus: a post-mortem study of normal subjects and patients with schizophrenia. Cereb Cortex. 2002;12:1218–1224. doi: 10.1093/cercor/12.11.1218. [DOI] [PubMed] [Google Scholar]
- Hilgetag CC, Burns GA, O'Neill MA, Scannell JW, Young MP. Anatomical connectivity defines the organization of clusters of cortical areas in the macaque monkey and the cat. Philos Trans R Soc Lond B Biol Sci. 2000;355:91–110. doi: 10.1098/rstb.2000.0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag CC, O'Neill MA, Young MP. Hierarchical organization of macaque and cat cortical sensory systems explored with a novel network processor. Philos Trans R Soc Lond B Biol Sci. 2000;355:71–89. doi: 10.1098/rstb.2000.0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag CC, ONeill MA, Young MP. Indeterminate organization of the visual system. Science. 1996;271:776–777. doi: 10.1126/science.271.5250.776. [DOI] [PubMed] [Google Scholar]
- Honey CJ, Kotter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci USA. 2007;104:10240–10245. doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodriguez EJ, Aleman-Gomez Y, Melie-Garcia L. Studying the human brain anatomical network via diffusion-weighted MRI and Graph Theory. Neuroimage. 2008;40:1064–1076. doi: 10.1016/j.neuroimage.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Jones DK, Simmons A, Williams SCR, Horsfield MA. Non-invasive assessment of axonal fiber connectivity in the human brain via diffusion tensor MRI. Magn Reson Med. 1999;42:37–41. doi: 10.1002/(sici)1522-2594(199907)42:1<37::aid-mrm7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. Modelling the development of cortical networks. Neurocomputing. 2004;58–60:297–302. [Google Scholar]
- Kaiser M, Martin R, Andras P, Young MP. Simulation of robustness against lesions of cortical networks. Eur J Neurosci. 2007;25:3185–3192. doi: 10.1111/j.1460-9568.2007.05574.x. [DOI] [PubMed] [Google Scholar]
- Kobbert C, Apps R, Bechmann I, Lanciego JL, Mey J, Thanos S. Current concepts in neuroanatomical tracing. Prog Neurobiol. 2000;62:327–351. doi: 10.1016/s0301-0082(00)00019-8. [DOI] [PubMed] [Google Scholar]
- Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- Lebel C, Walker L, Leemans A, Phillips L, Beaulieu C. Microstructural maturation of the human brain from childhood to adulthood. Neuroimage. 2008;40:1044–55. doi: 10.1016/j.neuroimage.2007.12.053. [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Mesulam MM. Principles of behavioural and cognitive neurology. New York: Oxford University Press; 2000. [Google Scholar]
- Micheloyannis S, Pachou E, Stam CJ, Vourkas M, Erimaki S, Tsirka V. Using graph theoretical analysis of multi channel EEG to evaluate the neural efficiency hypothesis. Neurosci Lett. 2006;402:273–277. doi: 10.1016/j.neulet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. Fiber tracking: principles and strategies—a technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- Nolte J. The human brain: an introduction to its functional anatomy. St Louis (MO): Mosby-Year Book; 1993. [Google Scholar]
- Nucifora PG, Verma R, Melhem ER, Gur RE, Gur RC. Leftward asymmetry in relative fiber density of the arcuate fasciculus. Neuroreport. 2005;16:791–794. doi: 10.1097/00001756-200505310-00002. [DOI] [PubMed] [Google Scholar]
- Parker GJ, Alexander DC. Probabilistic anatomical connectivity derived from the microscopic persistent angular structure of cerebral tissue. Philos Trans R Soc Lond B Biol Sci. 2005;360:893–902. doi: 10.1098/rstb.2005.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrides M. Lateral prefrontal cortex: architectonic and functional organization. Philos Trans R Soc Lond B Biol Sci. 2005;360:781–795. doi: 10.1098/rstb.2005.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005a;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Schwarzbauer C, Bullmore E. Undirected graphs of frequency-dependent functional connectivity in whole brain networks. Philos Trans R Soc Lond B Biol Sci. 2005b;360:937–946. doi: 10.1098/rstb.2005.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Young MP. The connectional organization of neural systems in the cat cerebral cortex. Curr Biol. 1993;3:191–200. doi: 10.1016/0960-9822(93)90331-h. [DOI] [PubMed] [Google Scholar]
- Sporns O, Chialvo DR, Kaiser M, Hilgetag CC. Organization, development and function of complex brain networks. Trends Cogn Sci. 2004;8:418–425. doi: 10.1016/j.tics.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kotter R. Identification and classification of hubs in brain networks. PLoS ONE. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman GM. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. The small world of the cerebral cortex. Neuroinformatics. 2004;2:145–162. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- Stam CJ. Functional connectivity patterns of human magnetoencephalographic recordings: a ‘small-world’ network? Neurosci Lett. 2004;355:25–28. doi: 10.1016/j.neulet.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. Small-world networks and functional connectivity in Alzheimer's disease. Cereb Cortex. 2007;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Schwartz C, Lin RT, Khan AA, Toga AW. Three-dimensional statistical analysis of sulcal variability in the human brain. J Neurosci. 1996;16:4261–4274. doi: 10.1523/JNEUROSCI.16-13-04261.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Mapping brain asymmetry. Nat Rev Neurosci. 2003;4:37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proc Natl Acad Sci USA. 1994;91:5033–5037. doi: 10.1073/pnas.91.11.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vernooij MW, Smits M, Wielopolski PA, Houston GC, Krestin GP, van der Lugt A. Fiber density asymmetry of the arcuate fasciculus in relation to functional hemispheric language lateralization in both right- and left-handed healthy subjects: a combined fMRI and DTI study. Neuroimage. 2007;35:1064–1076. doi: 10.1016/j.neuroimage.2006.12.041. [DOI] [PubMed] [Google Scholar]
- Wakana S, Jiang HY, Nagae-Poetscher LM, van Zijl PCM, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Westbury CF, Zatorre RJ, Evans AC. Quantifying variability in the planum temporale: a probability map. Cereb Cortex. 1999;9:392–405. doi: 10.1093/cercor/9.4.392. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Xu DR, Mori S, Solaiyappan M, van Zijl PCM, Davatzikos C. A framework for callosal fiber distribution analysis. Neuroimage. 2002;17:1131–1143. doi: 10.1006/nimg.2002.1285. [DOI] [PubMed] [Google Scholar]
- Young MP. The organization of neural systems in the primate cerebral cortex. Proc R Soc Lond B Biol Sci. 1993;252:13–18. doi: 10.1098/rspb.1993.0040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]