CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium (original) (raw)
Abstract
Joubert syndrome (JS) is a developmental brain disorder characterized by cerebellar vermis hypoplasia, abnormal eye movement, ataxia and mental retardation. Mutations in CEP290 mutations are responsible for the cerebello–oculo–renal subtype of JS that includes kidney cysts and retinal degeneration, two phenotypes commonly linked to ciliopathies. CEP290 mutations are also associated with Meckel–Gruber syndrome and Bardet–Biedl syndrome (BBS). Here we demonstrate that CEP290 interacts with a centriolar satellite protein PCM-1, which is implicated in BBS4 function. CEP290 binds to PCM-1 and localizes to centriolar satellites in a PCM-1- and microtubule-dependent manner. The depletion of CEP290 disrupts subcellular distribution and protein complex formation of PCM-1. In accord with PCM-1’s role in microtubule organization, CEP290 knockdown causes the disorganization of the cytoplasmic microtubule network. Moreover, we show that both CEP290 and PCM-1 are required for ciliogenesis and are involved in the ciliary targeting of Rab8, a small GTPase shown to collaborate with BBS protein complex to promote ciliogenesis. Our results suggest that PCM-1 is a potential mediator that may link CEP290 with BBS proteins in common molecular pathways.
INTRODUCTION
Joubert syndrome (JS) often presents phenotypes which significantly overlap with cilium-related disorders (ciliopathies) such as Senior–Lőken syndrome, Meckel–Gruber syndrome (MKS) and Bardet–Biedl syndrome (BBS), implicating ciliary dysfunction in JS (1,2). CEP290 is one of several genes mutated in JS. Nonsense or frame-shift mutations in CEP290 are responsible for a majority of the oculorenal form of JS (3,4), whereas an intronic mutation in CEP290 that creates a strong splice-donor site is the single most common identified cause of Leber congenital amaurosis, a severe retinal dystrophy, without associated cerebellar abnormalities (5). CEP290 is known to localize to the centrosome and basal body of ciliated kidney cells and to the connecting cilium of retinal photoreceptors (3,6). Previously, it has been suggested that CEP290 may be present in complex with several microtubule-based transport proteins including RPGR-interacting protein 1, p150Glued, p50dynamitin, Kif3A and kinesin-associated protein 3 (6,7). Consistent with a potential involvement of CEP290 in protein transport, a mutant mouse line (rd16) harboring an in-frame CEP290 deletion exhibits the mislocalization of phototransduction proteins in photoreceptors and odorant-signaling proteins in the cilia of olfactory receptor neurons (6,7).
Genetic heterogeneity is a characteristic feature of most ciliopathies: multiple genes have been identified for each of these distinct disorders (1,8). Although ciliopathy genes encode proteins with diverse functional domains, several biochemical studies have demonstrated physical interactions between the proteins involved in each type of ciliopathy, suggesting that they may act collaboratively in some common molecular pathways (9–12). For example, it was shown that seven BBS proteins (BBS1, 2, 4, 5, 7, 8 and 9) form a stable complex, which was termed BBSome, providing an explanation for the same pleiotropic phenotype of the mutations in the BBS genes (12). Given the substantial overlap in clinical manifestations of several ciliopathies, it is expected that various proteins that are associated with distinct ciliopathies may also interact physically and/or functionally. Moreover, recent genetic studies have demonstrated that JS, MKS and BBS are allelic disorders with causative mutations in some of the same genes. Mutations in CEP290 have been identified in fetuses with MKS and in families showing several features of MKS (13). In addition, mutations in three BBS genes (BBS2, BBS4 and BBS6) were shown to cause MKS-like phenotypes (14). Further evidence of the allelism between these disorders comes from the identification of mutations in CEP290 and MKS3 genes in BBS patients (15). Although these findings prove interlinked genetic interactions between JS, MKS and BBS, it remains to be shown whether JS proteins are directly implicated in the molecular pathways that involve MKS or BBS proteins.
Centriolar satellites are non-membranous 70–100 nm granules scattered around the centrosome of many types of animal cells, and similar structures also exist around the basal bodies (equivalent to the centrosome) of ciliated cells (16,17). PCM-1 was identified as the major component of centriolar satellites (17,18). PCM-1 plays a role in the recruitment of centrosomal proteins including centrin, pericentrin and ninein, and is required for the organization of the cytoplasmic microtubule network (19). The interaction between BBS4 and PCM-1 was first identified by a yeast two-hybrid screen using BBS4 cDNA as bait (20), and affinity purification confirmed that PCM-1 is associated with the stable BBS complex (12). BBS4 appears to act as an adaptor that connects p150glued subunit of dynein transport machinery with PCM-1 and thus assists the centrosomal recruitment of PCM-1 and its associated centrosomal proteins required for the organization of microtubule radiations from the centrosome (20). This model was supported by the finding that both BBS1 and BBS4 knockout mice exhibit mislocalization of PCM-1 and disorganization of the dendritic microtubule network in the olfactory epithelium (21).
The Rab family of Ras-like small GTPases is a key regulator of vesicle trafficking (22). Recently, it has been demonstrated that GTP-bound Rab8 enters the primary cilium and is required for the biogenesis of ciliary membrane (12,23). The depletion of either Rab8 or Rabin8, a GDP/GTP exchange factor specific for Rab8, causes ciliogenesis defect (12). BBS1, a subunit of the BBSome, was shown to directly bind to Rabin8, and centrosomal localization of BBS4, another subunit of the complex, is disrupted in the absence of Rabin8. A current model incorporating these findings is that the BBSome may potentiate Rabin8 function through direct physical association at the base of the cilium and thus mediate a localized Rab8 activation to promote the docking and fusion of vesicles carrying ciliary membrane and transmembrane proteins. However, it is not clear whether PCM-1 is also involved in the functional interaction between BBS complex and Rab8 for ciliogenesis.
Here, we report physical and functional interaction between CEP290 and PCM-1 and show that both CEP290 and PCM-1 are required for the ciliary localization of Rab8. We demonstrate that CEP290 is implicated in the molecular pathways that have been shown to involve BBS4, and thus provide a clue to understand the underlying mechanisms of the genetic link between JS and BBS.
RESULTS
Recruitment of CEP290 to centriolar satellites requires PCM-1 and microtubules
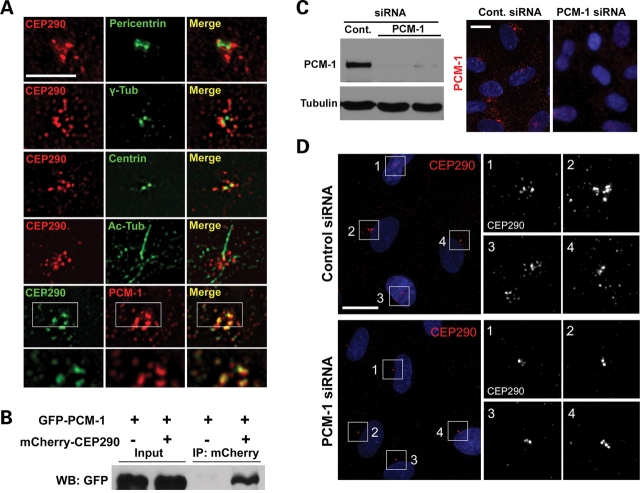
The current model suggests that CEP290 exerts its major effects at the centrosome and the basal body because preliminary studies have identified localization at the vicinity (3,4,6) and CEP290 co-immunoprecipitates with three centrosomal proteins, pericentrin, γ-tubulin and centrin as well as with a centriolar satellite protein PCM-1 (6). To gain insight into its function, we examined the localization of CEP290 in relation to these centrosomal proteins at a higher magnification. Both CEP290 and pericentriolar components, pericentrin and γ-tubulin, show distribution in the vicinity of the centrosome as has been reported in hTERT-RPE cells (Fig. 1A). However, unexpectedly, exact co-localization between CEP290 and these two proteins was not observed. It was also apparent that centriolar marker centrin does not perfectly co-localize with CEP290 (Fig. 1A). Remarkably, anti-CEP290 staining was not limited to the proximal area of the centrosome, but instead was detected in granular structures scattered beyond the other centrosomal proteins. Similar CEP290 distribution was observed in ciliated hTERT-RPE cells after serum starvation to induce ciliogenesis (Fig. 1A). In contrast to other centrosomal proteins, PCM-1 showed extensive overlap with CEP290, suggesting that CEP290 localizes to centriolar satellites (Fig. 1A).
Figure 1.
CEP290 localizes to centriolar satellites and is associated with PCM-1. (A) Double-immunofluorescent localization of CEP290 in hTERT-RPE cells. Anti-CEP290 antibody labels granular structures scattered around the centrosome. CEP290 is adjacent to, but not overlapping with, pericentrin and γ-tubulin. CEP290 only partially overlaps with centrin. CEP290;Ac-tub double-labeling shows that ciliated cells exhibit similar CEP290 immunostaining patterns. GFP-tagged CEP290, which recapitulated endogenous CEP290 localization at lower expression levels, extensively co-localizes with centriolar satellite marker PCM-1. Bottom panels are higher magnification images of an individual optical section captured by a ×100 objective. (B) HEK293T cells were co-transfected with GFP–PCM-1 and either mCherry–CEP290 or mCherry plasmid. Cell lysates were immunoprecipitated with anti-mCherry antibody and then immunoblotted with anti-GFP antibody. GFP–PCM-1 was co-immunoprecipitated with mCherry–CEP290. (C) PCM-1 knockdown was assessed by immunoblotting and immunofluorescence staining 3 days after PCM-1 siRNA transfection. (D) hTERT-RPE cells were transfected with siRNAs for 3 days and stained with anti-CEP290 antibody. Depletion of PCM-1 leads to the reduction of the number of CEP290 granules scattered around the centrosome. Right panels are higher magnification view of the boxed areas in the left panels. Cell nuclei were stained with Hoechst. Scale bars: (A) 5 µm; (C) 15 µm; (D) 20 µm.
To explore functional interaction between CEP290 and PCM-1, we first confirmed that CEP290 associates with PCM-1 using co-immunoprecipitation (Fig. 1B). Next, we tested whether PCM-1 is required for the centriolar satellite localization of CEP290, by examining CEP290 localization after the depletion of PCM-1 from hTERT-RPE cells using small interfering RNA (siRNA). Efficient PCM-1 depletion was confirmed by both immunoblotting and immunofluorescence staining (Fig. 1C). As shown in Fig 1D, PCM-1 knockdown resulted in a substantial decrease in the number of anti-CEP290-labeled granules scattered around the centrosome although CEP290 was not depleted from peri-centriolar regions. These results suggest that CEP290 localizes to centriolar satellites in a PCM-1-dependent manner, whereas peri-centriolar CEP290 localization is mediated by a distinct mechanism.
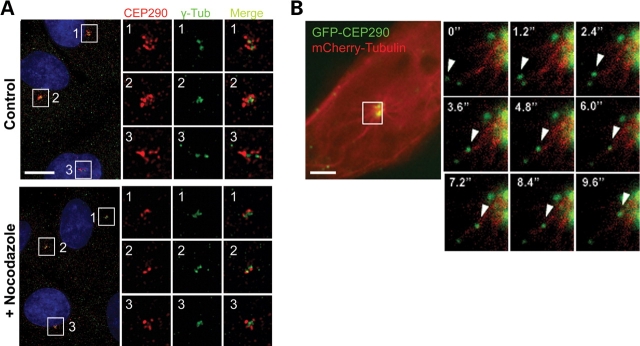
It was shown that centriolar satellites are released from the centrosome and are scattered into the cytoplasm when microtubule networks are disrupted by nocodazole treatment (17). On the other hand, polymerized microtubules are known to be dispensable for maintaining CEP290 at the centrosome (4,6). Given the centriolar satellite localization of CEP290, these two findings seem to be incompatible. To resolve this issue, we examined CEP290 localization after 20 μm nocodazole treatment for 2 h, which completely disassembled the microtubule architecture. In accord with previous reports (4,6), the disruption of microtubule networks did not completely inhibit the association of CEP290 with the centrosome, suggesting that polymerized microtubules are dispensable for the maintenance of, at least, a fraction of CEP290 that is tightly attached to the centrosome (Fig. 2A). However, there was a substantial decrease in the amount of the granular CEP290 staining located around the centrosome after nocodazole treatment (Fig. 2A). This observation supports the idea that the major fraction of CEP290 localizes to centriolar satellites.
Figure 2.
CEP290 moves along microtubules to the centrosome. (A) Cultured hTERT-RPE cells were treated with 20 µm nocodazole for 2 h and double-stained with anti-CEP290 and anti-γ-tubulin antibodies. Nocodazole treatment causes substantial decrease in granular CEP290 staining detected around the centrosome, suggesting that the recruitment of CEP290 to centriolar satellites requires intact microtubules. (B) Time-lapse observation of the movement of GFP-CEP290 along mCherry-labeled microtubules in live IMCD-3 cells. The boxed area in the left panel is magnified in the small panels. Numbers indicate the time lapse in seconds. Scale bars: (A) 10 µm; (B) 5 µm.
A previous report showed that PCM-1 granules move along microtubules toward the centrosome (17). To test whether CEP290 is also recruited to centriolar satellites along microtubules, we examined the behavior of GFP-tagged CEP290 in live cells. Both GFP-CEP290 and mCherry-tubulin were co-expressed transiently, and live-cell image was captured by high-speed fluorescence microscopy (Fig. 2B and Supplementary Material, Video S1). We frequently observed linear movements of GFP-CEP290 granules toward the centrosome along mCherry-labeled microtubules at maximum rates of ∼0.6–0.7 µm/s, which is similar to the maximum rate of cytoplasimc dynein (Fig. 2B). These observations indicate that CEP290 recruitment to centriolar satellites involves association with polymerized microtubules, and likely minus-end directed motors.
CEP290 modulates PCM-1 subcellular distribution and formation of complexes containing PCM-1
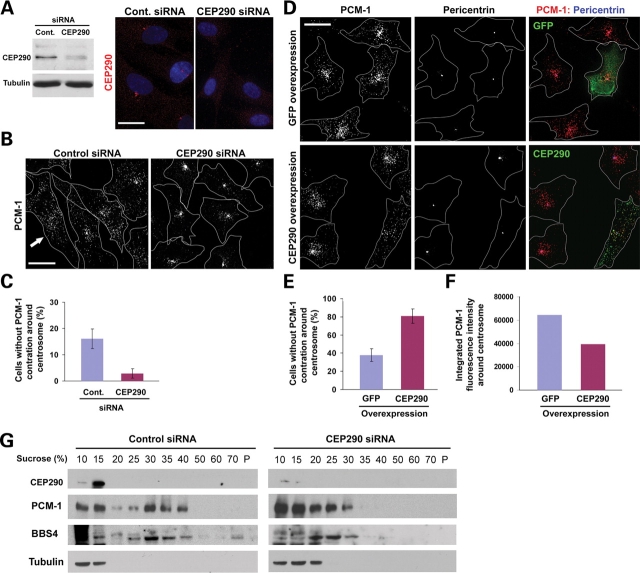
To test whether CEP290 is required for PCM-1 function, we depleted CEP290 from hTERT-RPE cells using siRNAs and assessed the localization of PCM-1 granules. Both immunoblotting and immunofluorescence staining showed a substantial reduction of CEP290 expression (Fig. 3A). As reported previously (18), PCM-1 granules were not only localized to peri-centrosomal area but also scattered throughout the cytoplasm (Fig. 3B). Furthermore, ∼16% of interphase control cells showed widely dispersed PCM-1 granules without clear enrichment around the centrosome, suggesting a dynamic shuttling of PCM-1 between the centrosome and the cytosol (Fig. 3B and C). Remarkably, CEP290 depletion greatly facilitates the concentric aggregation of PCM-1 granules at the centrosome, depleting scattered cytosolic pool of PCM-1 (Fig. 3B). Only 3% of CEP290-depleted cells showed widely dispersed PCM-1 staining without centrosomal concentration (Fig. 3C). These results show that CEP290 is not required for the centrosomal recruitment of PCM-1, but plays a role in the regulation of the dispersion of PCM-1 particles. To support this idea, we examined the effect of CEP290 overexpression on PCM-1 localization. As shown in Fig 3D, CEP290 overexpression facilitates the dispersion of PCM-1 granules from the centrosome. PCM-1 dispersion was observed in 81% of cells overexpressing CEP290 (Fig. 3E). Although the overexpression of GFP alone also resulted in a small increase in the number of cells showing dispersed PCM-1, the increase was incomparable with that of CEP290 overexpression (Fig. 3E). A decrease in integrated anti-PCM-1 immunofluorescence intensities around the centrosome confirms PCM-1 re-distribution in CEP290-overexpressing cells (Fig. 3F). Together these results demonstrate that CEP290 is involved in the regulation of dynamic PCM-1 distribution, and that the positioning of CEP290 and PCM-1 to pericentriolar granules is bi-dependent.
Figure 3.
CEP290 affects the localization and complex formation of PCM-1. (A) CEP290 knockdown was assessed by immunoblotting and immunofluorescence staining 3 days after CEP290 siRNA transfection. (B) hTERT-RPE cells were stained with anti-PCM-1 antibody 3 days after siRNA transfection. CEP290 knockdown causes the concentric accumulation of PCM-1, depleting widely dispersed PCM-1 pool. Arrow indicates a cell showing widely dispersed PCM-1 granules without apparent centrosomal concentration. Line drawings mark cell outline. (C) Histogram quantifying cells without noticeable concentration of PCM-1 granules around the centrosome as shown in (B) (arrow). More than 700 cells from three independent experiments were counted for each bar by two investigators (one of them was blind to the experiment). Error bars represent SD. Mitotic cells were excluded from the counting. (D) hTERT-RPE cells were stained with anti-PCM-1;anti-pericentrin antibodies 15 h after transfection with either GFP or GFP-CEP290 plasmid DNA. CEP290 overexpression facilitates the dispersion of centrosomal pool of PCM-1 granules. (E) Histogram quantifying the result shown in (D). More than 170 cells expressing transgenes from two independent experiments were counted for each bar by two investigators (one of them was blind to the experiment). (F) Histogram showing integrated pixel density of anti-PCM-1 immunofluorescence around the centrosome (7 µm2). Each bar is mean value (arbitrary unit) of 18 transgene-positive cells. (G) Cells were treated with nocodazole and cytochalasin to disassemble cytoskeleton 3 days after siRNA transfection and lysed in 0.5% NP-40 and 150 mm NaCl. Low-speed (1500 g) supernatants were fractionated on discontinuous sucrose gradients. PCM-1 and BBS4 proteins from CEP290-depleted lysates fail to migrate into higher density fractions, indicating that they form smaller protein complexes in the absence of CEP290. Scale bar: 20 µm.
It is likely that PCM-1 granules moving along polymerized microtubules are protein complexes that are shuttling between the cytosol and the centrosome to transport molecules into the centrosome (19,20). To test whether CEP290 depletion affects properties of the protein complexes that constitute PCM-1 granules, we examined the sedimentation property of PCM-1 in sucrose density gradient centrifugation. As reported previously (12), solubilized α-tubulin stayed near the top of the gradient (Fig. 3G). In the control sample, PCM-1 sedimented as two peaks: one near the top of the gradient and the other between the 30 and 40% steps (Fig. 3G). Importantly, CEP290 depletion dramatically altered the behavior of PCM-1 in the gradient. PCM-1 from cells transfected with CEP290 siRNA mainly stayed near the top of the gradient and only small fractions of PCM-1 sedimented beyond the 30% step (Fig. 3G). In addition, we found that sedimentation properties of BBS4 are also affected by CEP290 knockdown. In control sample, BBS4 co-sedimented with PCM-1 at 25–40% gradients, and small portions of BBS4 were detected even at the bottom of the gradient, suggesting that BBS4 is a part of a large protein complex (Fig. 3G). In contrast, BBS4 from CEP290-depleted cells did not move beyond 30% gradient and was mainly detected in fractions between 20 and 30% gradients. These results suggest that the assembly of the BBS–PCM-1 protein complex requires normal levels of CEP290.
It is noteworthy that even though CEP290 depletion affected the higher molecular size fractions of both PCM-1 and BBS4, CEP290 from control lysates was not co-sedimented with the higher molecular size fractions (Fig. 3G). Instead, CEP290 overlaps only with a lower molecular size fraction (15% sucrose) of PCM-1. CEP290 immunoreactivity is restricted to the centriolar satellites and pericentriolar space, whereas PCM-1 granules are detected throughout the cytoplasmic space although they are concentrated around the centrosome (Fig. 3A and B). Thus, the presence of PCM-1 which does not co-sediment with CEP290 is not unexpected. One possible hypothesis is that CEP290 might interact with smaller size intermediate PCM-1 complexes, supporting the assembly of large multisubunit PCM-1;BBS complexes, and CEP290 might then be released from the complexes and recycled. However, we cannot exclude the possibility that CEP290 is dissociated from higher molecular size BBS–PCM-1 complex during the sample preparation and centrifugation. The observation that CEP290 depletion causes an abnormal accumulation of PCM-1 at the centrosome (Fig. 3B) implies that smaller size PCM-1 complexes might be unable to efficiently shuttle between the centrosome and the cytosol.
Depletion of CEP290 interferes with microtubule organization and ciliogenesis
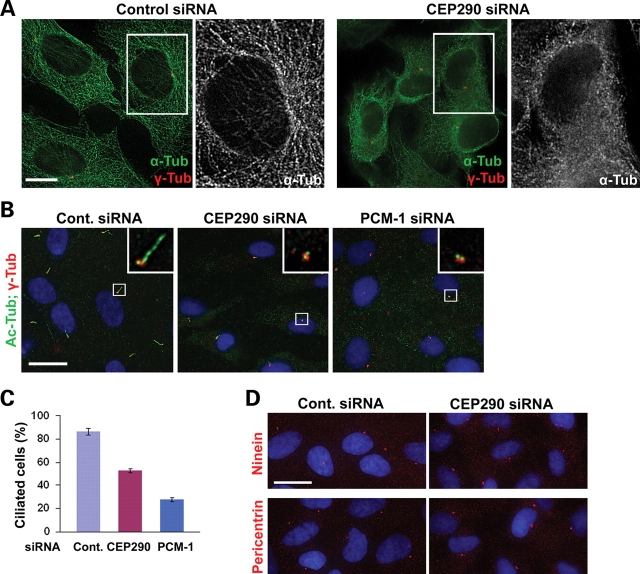
We speculated that if the changes in subcellular localization and sedimentation properties caused by CEP290 depletion reflect dysfunction of PCM-1, CEP290 knockdown might recapitulate part or all of PCM-1 loss-of-function phenotypes. It was shown that the organization of the microtubule network is disrupted in response to either PCM-1 or BBS4 knockdown (19,20). We therefore depleted CEP290 in hTERT-RPE cells and visualized the microtubule network by staining anti-α-tubulin antibody (Fig. 4A). Cells transfected with control siRNA displayed distinct microtubule networks. In contrast, CEP290-depleted cells showed disorganized diffuse tubulin staining around the nucleus, and polymerized microtubules were only apparent in cell periphery (Fig. 4A). In addition, unlike control cells, CEP290-depleted cells did not show prominent microtubule arrays radiating from the centrosome although centrosomal γ-tubulin staining was still evident. These results indicate that CEP290 is required for microtubule network organization and also supports a functional link between CEP290 and PCM-1;BBS4 complex. It is unlikely that the abnormal enhancement of PCM-1 accumulation around the centrosome in CEP90 depleted cells (Fig. 3B and C) is a consequence of microtubule collapse because the disruption of microtubule by nocodazole treatment causes a wide dispersion of PCM-1 granules (Supplementary Material, Fig. S1).
Figure 4.
Microtubule organization and ciliogenesis require CEP290 function. (A) The depletion of CEP290 from hTERT-RPE cells causes a collapse of microtubule networks, which is accompanied by an apparent decrease in cell size. (B–C) hTERT-RPE cells were shifted from 10% serum to serum-free medium 24 h after transfection with the indicated siRNAs and cultured for additional 60 h before fixation. Ciliated cells were identified by anti-Ac-tub antibody staining. Basal body was labeled by anti-γ-tub antibody. The depletion of either CEP290 or PCM-1 inhibits ciliogenesis. (C) Histogram quantifying the data shown in (B). More than 200 cells from two independent experiments were counted for each bar. Error bars represent SD. (D) Representative images of hTERT-RPE cells stained with the indicated antibodies after CEP290 siRNA transfection and serum starvation. Both ninein and pericentrin staining of CEP290-depleted cells were indistinguishable from those of control cells. Scale bars: (A) 10 µm; (B and D) 20 µm.
To further explore functional links between CEP290 and PCM-1, we tested the involvement of CEP290 and PCM-1 in serum starvation-induced ciliogenesis in hTERT-RPE cells. In agreement with recent reports (12,24), PCM-1 depletion caused a reduction in the number of cells displaying a primary cilium (Fig. 4B and C). As reported previously (24), we also found a decrease in ciliated cell number after CEP290 knockdown, supporting the involvement of CEP290 in the assembly of the primary cilium (Fig. 4B and C).
Pericentrin is known to interact with intraflagellar transport proteins and polycystin-2, and is required for ciliogenesis (25). Given that PCM-1 is involved in the recruitment of centrosomal proteins including pericentin (19), it is likely that a decrease in the levels of pericentrin at the basal body may contribute to the ciliogenesis defect observed in PCM-1-depleted cells. We therefore examined whether the mislocalization of centrosomal proteins underlies the ciliogenesis defect caused by CEP290 depletion. Unexpectedly, immunostaining for both pericentrin and ninein did not detect an obvious decrease in the levels of these proteins at the centrosome/basal body in the absence of CEP290 (Fig. 4D). This result suggests that CEP290 may exert its ciliogenic effect through a mechanism that is independent of ninein and pericentrin recruitment to the centrosome. This result also implies that CEP290 depletion does not interfere with PCM-1’s function in centrosomal protein recruitment although it disrupted subcellular localization and the complex formation of PCM-1.
A recent paper showed that primary cilia assembly is inhibited by the RNAi-mediated depletion of several centrosomal proteins including γ-, δ- and ε-tubulin, centrin2, centriolin, GCP2 and pericentrin (26). The depletion of all those proteins also results in the loss of centrosome integrity (loss, separation or fragmentation of centrioles) which causes the activation of a checkpoint that inhibits G1-S progression (26). It is likely that ciliogenesis defect observed in cells depleted of key centrosomal proteins is a secondary consequence of the loss of centrosome integrity. In contrast, we found no evidence that CEP290 knockdown can affect centriole structure (Supplementary Material, Fig. S2A). Furthermore, CEP290 depletion did not interfere with G1-S progression (Supplementary Material, Fig. S2B). Together with normal staining patterns of pericentriolar material makers (Fig. 4B and D), these observations suggest that CEP290’s role may be ciliogenesis pathway-specific.
CEP290 and PCM-1 are required for efficient recruitment of Rab8 to the primary cilium
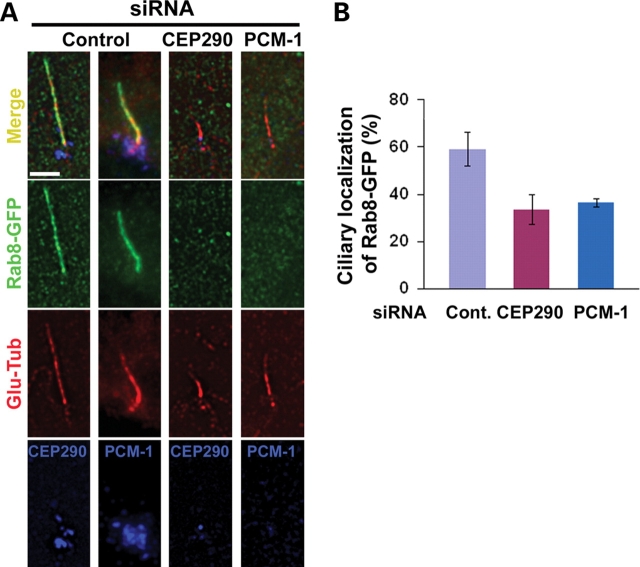
The Rab small GTPases regulate multiple aspects of vesicle transport, budding and fusion, and are also critical for defining membrane subdomain identity (22). Recent studies have shown that Rab8 enters the primary cilium and plays a role in the elongation of ciliary membrane (12,23). Importantly, the BBSome was shown to interact with Rabin8, a Rab8 GDP/GTP exchange factor, suggesting a functional interaction between the BBSome and Rab8 at the primary cilium (12). Since CEP290 depletion did not affect ninein and pericentirin localization to the centrosome, we next examined whether ciliogenic function of CEP290 and PCM-1 is also linked to Rab8 function at the primary cilium. Although the siRNAs used in this study efficiently depleted PCM-1 (Fig. 1C) or CEP290 (Fig. 3A), a subset of transfected cells still developed the primary cilium in response to serum starvation (Fig. 4C). Thus, we were able to examine CEP290/PCM-1 loss-of-function effect on ciliary targeting of Rab8. Remarkably, CEP290 knockdown cells showed substantially lower levels of ciliary localization of Rab8-GFP, suggesting that CEP290 is required for the efficient recruitment of Rab8 to the primary cilium (Fig. 5). This result supports the idea that CEP290 plays a specific role in ciliogenesis pathway. PCM-1 depletion also interferes with Rab8 translocation into the cilium (Fig. 5). Thus, PCM-1’s role in ciliogenesis is not limited to the recruitment of centrosomal proteins. These results provide a common proximal cause of the defective ciliogenesis observed in cells depleted with either CEP290 or PCM-1 and provide a potential link between JS and BBS.
Figure 5.
Both CEP290 and PCM-1 are involved in the regulation of Rab8 translocation into the primary cilium. (A) hTERT-RPE cells were shifted from 10% serum to serum-free medium 24 h after siRNA transfection, cultured for 24 h, and then re-transfected with Rab8-GFP for 36 h. Cells were triple-labeled with Rab8-GFP, anti-Glu-tub antibody (a ciliary marker) and anti-CEP290/PCM-1. (B) Histogram quantifying the data shown in (A). Ciliated Rab8-GFP+ cells that show substantial reduction of CEP290 or PCM-1 were compared with ciliated Rab8-GFP+ control cells. One hundred and fifteen control cells, 50 CEP290-depleted cells and 48 PCM-1-depleted cells from two independent experiments were counted for each bar. Error bars represent SD. The depletion of either CEP290 or PCM-1 interferes with Rab8-GFP translocation into the primary cilium. Scale bar: (A) 5 µm.
DISCUSSION
Genetic heterogeneity of ciliopathies may be ascribed to the inherent complexity of the molecular mechanisms that underlie the formation and function of the primary cilium. It is conceivable that the perturbation of various cellular processes, such as the coordination of intraflagellar transport motors (27), biogenesis of ciliary membrane (12) or centriole migration to the apical membrane (11), can converge on defective ciliogenesis or ciliary dysfunction, resulting in a range of overlapping clinical outcomes. An alternative, but not mutually exclusive, explanation for the heterogeneity is that a group of diverse proteins associated with each ciliopathy can act collaboratively in a common molecular pathway possibly as a multisubunit protein complex. Mutations in each subunit may result in the dysfunction of the entire complexes. Recently, this hypothesis has been gaining acceptance due to several lines of evidence. In addition to the identification of the BBSome (12), the biochemical interaction between two MKS proteins, MKS1 and meckelin, was demonstrated (11). Moreover, physical interactions between nephronopthisis proteins, NPHP1, inversin, NPHP3 and NPHP4, have been reported (9,10,28). Our data presented here further develop the idea of the interlinked actions of ciliopathy proteins. We provide evidence that proteins implicated in different types of ciliopathies may also interact in a common molecular pathway.
BBS4 binds to PCM-1 and acts together at centriolar satellites for protein recruitment and microtubule organization (20). Similar to BBS4, CEP290 localizes to centriolar satellites and binds to PCM-1. Remarkably, either the depletion or the overexpression of CEP290 caused redistribution of PCM-1. Furthermore, CEP290 knockdown resulted in the disorganization of cytoplasimc microtubule network in hTERT-RPE cells. These results suggest that CEP290 may be involved in a specific molecular pathway that requires functions of both PCM-1 and BBS4. However, it is unlikely that CEP290 and BBS4 play a redundant role on PCM-1 function because knockdown phenotypes are distinct: the depletion of CEP290 resulted in an abnormal concentric accumulation of PCM-1, whereas BBS4 knockdown caused the dispersion of PCM-1 (20). This observation suggests that, unlike BBS4, CEP290 might act to either destabilize PCM-1–dynein motor interaction or promote the interaction between PCM-1 and kinesin motors, facilitating the movement of PCM-1 back to the cytosol. Indeed, CEP290 can interact with components of both dynein and kinesin motor machineries (6,7). It was also shown that PCM-1 granules move along the microtubule not only toward the centrosome but also toward the cell periphery, suggesting that PCM-1 distribution is regulated by coordinated actions between plus end- and minus end-directed microtubule motors (17). It is likely that CEP290 and BBS4 together may mediate this coordination.
It is noteworthy that the abnormal concentric accumulation of PCM-1 granules in CEP290-depleted cells was accompanied by a change in biochemical properties of both PCM-1 and BBS4 proteins. The apparent decrease in molecule size in sucrose density gradient indicates that PCM-1 and BBS4 form smaller complexes in the absence of CEP290. This result suggests that CEP290 may be required for the formation of the BBSome, which is known to contain at least seven BBS proteins as well as PCM-1. However, the observation that CEP290 from control cell lysate was mainly detected in smaller molecular size fractions excludes the possibility that CEP290 is incorporated into the large BBS–PCM-1 complex as a scaffold. In agreement with this, BBS4 was not detected in CEP290 immunoprecipitates which contain PCM-1 and several other centrosomal proteins (7). Interestingly, CEP290 has the consensus sequence of ATP/GTP-binding motif, GxxxxGKS/T, which is typically present in ATPase/GTPase (P-loop NTPase) (29), and CEP290 from all vertebrates whose sequence data are available contains this exact motif. Thus, it is conceivable that CEP290 facilitates BBSome assembly in a catalytic manner, utilizing ATP binding. PCM-1 granules are not only concentrated at the centrosomal area but also scattered throughout the cytoplasm, whereas CEP290 is mainly detected around the centrosome. However, CEP290 knockdown mainly affects non-centrosomal pool of PCM-1. This result also suggests that CEP290 may modulate the dynamic shuttling of PCM-1 in a non-stoichiometric or catalytic manner.
The exact role of PCM-1 in the assembly of the primary cilium remains to be elucidated. In particular, in vivo functions of PCM-1 are totally unknown: PCM-1 has not been reported to be associated with any ciliopathy, and the animal model of PCM-1 loss-of-function is not available. Besides, a recent report suggested that PCM-1 may be dispensable for centriole formation and ciliogenesis in multi-ciliated tracheal epithelial cells (30). However, previous results (12,24) and the data presented here clearly demonstrate that PCM-1 function is required for the formation of the non-motile primary cilium. Given its known role in the recruitment of major centrosomal proteins (19), it is not surprising that the depletion of PCM-1 interferes with ciliogenesis. In addition, the interdependence of PCM-1 with ciliopathy proteins, BBS4 (12,20) and CEP290, for proper subcellular localization further supports the connection between PCM-1 and the primary cilium. Moreover, our data suggest that PCM-1 is involved in the regulation of a membrane-trafficking pathway mediated by Rab8, which may be coordinated with the assembly of the ciliary axoneme. The dispensability of PCM-1 for tracheal epithelial cell ciliogenesis suggests that molecular pathways for the assembly of motile multi-cilia are distinct from those of the primary cilium. This may explain why dysfunction of motile multi-cilia is not common in JS and BBS.
Previously, it was shown that ciliogenesis is inhibited by the depletion of two BBS complex subunits (BBS1 or BBS5), whereas the localization of PCM-1, BBS4 and centrosomal proteins are unaffected by the depletion (12). Importantly, CEP290 knockdown also interferes with ciliogenesis and Rab8 targeting to the primary cilium without apparent defect in the recruitment of centrosomal proteins. These results raise the possibility that Rab8 transport to the cilium and centrosomal protein recruitment mediated by PCM-1-BBS4 function are unlinked molecular processes. One possible scenario would be that PCM-1-BBS4 is a bi-functional module that can either form transient complexes with dynein transport machinery for recruiting centrosomal proteins or fit into the BBSome, which serves as a docking site for Rab8 activation machinery at the basal body. In this scenario, CEP290 might function to coordinate the assembly and disassembly of the complexes. The elucidation of detailed mechanisms of the link between CEP290 and PCM-1/BBS requires additional biochemical study. In summary, our results identify PCM-1 as a molecule that can interact with both CEP290 and BBS proteins.
MATERIALS AND METHODS
Cell culture and transfection
hTERT-RPE, IMCD-3 and HEK293T cells were grown as suggested by American Type Culture Collection. Cells were transfected with 20–40 nm siRNAs using Lipofectamine RNAi-Max (Invitrogen). Sequences of pooled Cep290 siRNA duplex oligonucleotides are 5′-GGAUUCGGAUGAAAUGAAAUU-3′, 5′-GGAAUUGACUUACCUGAUGUU-3′, 5′-GAAAGUUAAUGAGCAAUUGUU-3′ and 5′-GAAGUAGAGUCCCUCAGAAUU-3′ (Dharmacon). The sequence of PCM-1 siRNA duplex oligonucleotides is 5′-GGCUUUAACUAAUUAUGGATT-3′ (Dharmacon). Non-target siRNA was used for control transfection (Dharmacon). Plasmid vector for expressing GFP-tagged mouse CEP290 was described previously (3). GFP–PCM-1 plasmid was a gift from Andreas Merdes (University of Edinburgh). Rab8-GFP plasmid was a gift from Maxence Nachury (Genentech). mCherry–tubulin plasmid was obtained from Clontech, Inc. To generate mCherry–CEP290 vector, GFP coding region from GFP-CEP290 plasmid was replaced with mCherry sequence. Cells were transfected with plasmid DNAs using Lipofectamine2000 (Invitrogen).
Immunofluorescence and imaging
For indirect immunofluorescence, cells were fixed in either methanol or 4% PFA depending on antigen. Primary antibodies used for immunofluorescence are rabbit anti-CEP290 antibody (a gift from Dr Hemant Khanna, University of Michigan); rabbit anti-PCM-1 antibody (a gift from Andreas Merdes, University of Edinburgh); mouse anti-centrin (a gift from Jeffrey Salisbury, Mayo Clinic), mouse-anti-Ac-tub (Zymed); rabbit-anti-Glu-tub (Chemicon); mouse-anti-pericentrin (Covance); mouse-anti-α-tub (Sigma); rabbit-anti-γ-tub (Sigma); goat-anti-γ-tub (Santa Cruz); mouse-anti-GFP (Genetex) and anti-BrdU (ABcam). Alexa 488-, Alexa 594- and Alexa-647-conjugated secondary antibodies (Molecular Probes) were applied for 1 h at room temperature. DNA was stained with Hoechst 33342.
Images were obtained using a DeltaVision Spectris Deconvolution System (Applied Precision) equipped with an Olympus IX70 microscope and a cooled charge-coupled device system. Image stacks were collected with a Z-step size of 0.2 µm and then deconvolved with DeltaVision software (Applied Precision). Settings were enhanced ratio with noise filtering set to medium and 10 deconvolution cycles. The number of Z stacks collected was variable (between 0.6 and 1.2 µm), depending on magnification. Deconvoluted images were projected into one picture using DeltaVision software's quick projection tool with maximum intensity setting. Exposure times and settings for image processing were constant for all samples to be compared within any given experiment. Images were sized and placed in figures using Adobe Photoshop 7.0. Alexa-647 fluorescence was pseudo-colored to blue in Figure 5A bottom panels. For the quantification of immunofluorescence intensity, integrated pixel density was measured using Image J software. Rollin ball radius for background subtraction was set to 25.
Immunoblotting and immunoprecipitation
For immunoblotting, cells were extracted with RIPA lysis buffer and boiled with SDS sample buffer. Same volumes of lysates were loaded for paired experiments. Primary antibodies used for western blotting are rabbit anti-mouse CEP290 antibody raised against a fragment spanning amino acid 2286–2304; anti-PCM-1; anti-GFP (Genetex); anti-BBS4 (Santa Crutz). Bound antibodies were detected using HRP-conjugated secondary antibodies (PIERCE). For co-immunoprecipitation, cells were extracted with EBC lysis buffer, and cell lysates were incubated with anti-DsRed/mCherry antibody (Clontech), which does not cross-react with GFP, and Protein A-agarose beads (PIERCE).
Sucrose density gradient centrifugation
hTERT-RPE cells were treated with 20 µg/ml nocodazole and 10 µg/ml cytochalasin B for 15 min and lysed in 50 mm Tris (pH 8.0), 150 mm NaCl, 1 mm EGTA, 1 mm MgCl2, 10% glycerol, 0.5% NP-40, 1 mm DTT and protease inhibitor cocktail (Roche). Lysates were centrifuged at 1500 g for 5 min at 4°C, and DNase I (2 µg/ml; Sigma) was added to the supernatants. Supernatants were filtered through 35 µm cell-strainer cap (BD Falcon) and fractionated on discontinuous sucrose gradients at 100 000 g for 16 h at 4°C in SW 41 Ti rotor. The discontinuous sucrose gradients comprise 10 steps (1 ml each) of 10–70% sucrose prepared in 50 mm Tris (pH 8.0), 150 mm NaCl, 1 mm EGTA, 1 mm MgCl2, 10% glycerol, 0.05% NP-40 and 1 mm DTT.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
[Supplementary Data]
FUNDING
This work was supported by NIH grants R01 NS052455 and the Burroughs Wellcome Fund to J.G.G. J.K. is supported in part by 2007 NARSAD Young Investigator Award. Imaging was supported by the UCSD Neurosciences Microscopy Shared Facility Award P30 NS047101. J.G.G. is an investigator of the Howard Hughes Medical Institute.
ACKNOWLEDGEMENTS
We thank Dr H. Khanna for CEP290 antibody, Dr A. Merdes for PCM-1 antibody and plasmid, Dr J.L. Salisbury for Centrin antibody, and Dr M.V. Nachury for Rab8-GFP plasmid.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hildebrandt F., Otto E. Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 2005;6:928–940. doi: 10.1038/nrg1727. [DOI] [PubMed] [Google Scholar]
- 2.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 3.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 4.Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 5.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G., et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwen D.P., Koenekoop R.K., Khanna H., Jenkins P.M., Lopez I., Swaroop A., Martens J.R. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl Acad. Sci. USA. 2007;104:15917–15922. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams M., Smith U.M., Logan C.V., Johnson C.A. Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J. Med. Genet. 2008;45:257–267. doi: 10.1136/jmg.2007.054999. [DOI] [PubMed] [Google Scholar]
- 9.Mollet G., Silbermann F., Delous M., Salomon R., Antignac C., Saunier S. Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum. Mol. Genet. 2005;14:645–656. doi: 10.1093/hmg/ddi061. [DOI] [PubMed] [Google Scholar]
- 10.Arts H.H., Doherty D., van Beersum S.E., Parisi M.A., Letteboer S.J., Gorden N.T., Peters T.A., Marker T., Voesenek K., Kartono A., et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 11.Dawe H.R., Smith U.M., Cullinane A.R., Gerrelli D., Cox P., Badano J.L., Blair-Reid S., Sriram N., Katsanis N., Attie-Bitach T., et al. The Meckel–Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum. Mol. Genet. 2007;16:173–186. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- 12.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 13.Baala L., Audollent S., Martinovic J., Ozilou C., Babron M.C., Sivanandamoorthy S., Saunier S., Salomon R., Gonzales M., Rattenberry E., et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am. J. Hum. Genet. 2007;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karmous-Benailly H., Martinovic J., Gubler M.C., Sirot Y., Clech L., Ozilou C., Auge J., Brahimi N., Etchevers H., Detrait E., et al. Antenatal presentation of Bardet–Biedl syndrome may mimic Meckel syndrome. Am. J. Hum. Genet. 2005;76:493–504. doi: 10.1086/428679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Al-Fadhel M., Lewis R.A., Eyaid W., Banin E., et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet–Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 16.Berns M.W., Rattner J.B., Brenner S., Meredith S. The role of the centriolar region in animal cell mitosis. A laser microbeam study. J. Cell Biol. 1977;72:351–367. doi: 10.1083/jcb.72.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo A., Tsukita S. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 2003;116:919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- 19.Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J.C., Badano J.L., Sibold S., Esmail M.A., Hill J., Hoskins B.E., Leitch C.C., Venner K., Ansley S.J., Ross A.J., et al. The Bardet–Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 21.Kulaga H.M., Leitch C.C., Eichers E.R., Badano J.L., Lesemann A., Hoskins B.E., Lupski J.R., Beales P.L., Reed R.R., Katsanis N. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat. Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 22.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurczyk A., Gromley A., Redick S., San Agustin J., Witman G., Pazour G.J., Peters D.J., Doxsey S. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 2004;166:637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 27.Ou G., Blacque O.E., Snow J.J., Leroux M.R., Scholey J.M. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 28.Olbrich H., Fliegauf M., Hoefele J., Kispert A., Otto E., Volz A., Wolf M.T., Sasmaz G., Trauer U., Reinhardt R., et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat. Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 29.Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 30.Vladar E.K., Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]