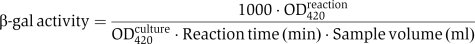Increased RNA polymerase availability directs resources towards growth at the expense of maintenance (original) (raw)
Abstract
Nutritionally induced changes in RNA polymerase availability have been hypothesized to be an evolutionary primeval mechanism for regulation of gene expression and several contrasting models have been proposed to explain how such ‘passive' regulation might occur. We demonstrate here that ectopically elevating Escherichia coli RNA polymerase (Eσ70) levels causes an increased expression and promoter occupancy of ribosomal genes at the expense of stress-defense genes and amino acid biosynthetic operons. Phenotypically, cells overproducing Eσ70 favours growth and reproduction at the expense of motility and damage protection; a response reminiscent of cells with no or diminished levels of the alarmone guanosine tetraphosphate (ppGpp). Consistently, we show that cells lacking ppGpp displayed markedly elevated levels of free Eσ70 compared with wild-type cells and that the repression of ribosomal RNA expression and reduced growth rate of mutants with constitutively elevated levels of ppGpp can be suppressed by overproducing Eσ70. We conclude that ppGpp modulates the levels of free Eσ70 and that this is an integral part of the alarmone's means of regulating a trade-off between growth and maintenance.
Keywords: passive regulation, ppGpp, RNA polymerase, stringent control, transcriptional trade-off
Introduction
The most pervasive means of altering the relative abundance of transcripts in the cell is to control the efficiency at which the RNA polymerase initiates transcription at different promoters. In Escherichia coli, RNA polymerase consists of the core enzyme (E) containing the α2ββ′ω subunits, and one of the seven possible sigma (σ) factors conferring promoter specificity to E by making sequence-specific contacts to DNA (Gross et al, 1992). The majority of genes (housekeeping genes) expressed during exponential growth of E. coli require σ70 (encoded by rpoD) for transcription initiation. This sigma factor directs E to genes encoding proliferation-related activities, including the genes of the protein synthesizing system (PSS; i.e. those encoding the ribosomes, tRNAs and factors required for translation). In addition, a large number of maintenance and stress-defense genes induced upon growth limitations are also dependent on the Eσ70 holoenzyme. Directing Eσ70 to appropriate promoters (or inhibiting promoter contacts) under changing conditions is to a large part accomplished by specific activators and repressors. However, Eσ70-dependent genes have been argued to be subjected to differential control also by changes in Eσ70 availability, a so-called passive regulation of gene expression.
Ole Maaløe was perhaps first in presenting a model for passive control of gene expression inspired by the conviction that the massive alterations of gene expression needed during drastically changing growth conditions, such as nutrient limitations, required a more robust control than those afforded by specific repression, de-repression, and activator circuits (Maaløe, 1979). This type of gene regulation might have been operational before the evolution of specific regulatory factors and maintained some of its importance in representing a regulatory baseline control on top of which more specific and pronounced effects can be achieved with activators and repressors. Maaløe suggested, for example, that the synthesis of the PSS is regulated by Eσ70 availability, which changes with changing growth conditions (Bremer and Dennis, 1996). Although details of Maaløe's original model has been found wanting, the assumption that individual promoters compete with each other for a limiting amount of free Eσ70 has been confirmed (Shepherd et al, 2001; Jishage et al, 2002; Bremer et al, 2003; Magnusson et al, 2003; Grigorova et al, 2006).
Several authors have presented updated models for passive regulation and endorsed passive control as an integral part of stringent control by the nucleotide guanosine tetraphosphate (ppGpp). Cells of E. coli elicit a swift down-regulation of the PSS during amino acid starvation—a response called the stringent response—and ppGpp, acting together with the protein DksA, is the effector molecule of this response (reviewed in Potrykus and Cashel, 2008). The nucleotide ppGpp is produced by RelA and/or SpoT. This occurs not only in response to amino acid limitation but also upon starvation for many different kinds of nutrients and in circumstances restricting growth (Cashel et al, 1996). A key feature in one passive model of gene regulation, referred to here as the ‘saturation model', is that altered availability of free Eσ70 affects different promoters differently based on the fact that promoters display diverse saturation properties (Jensen and Pedersen, 1990). For example, a reduction in Eσ70 availability is argued to specifically reduce expression from rRNA promoters and other promoters of the PSS as it is reasonable to assume that genes whose products (ribosomes in this case) are in high demand will possess promoters exhibiting high maximal velocity. Indeed, no other promoters seem to initiate transcription with the same high frequency as rrn promoters (estimated to have up to 90 initiations per minute per rrn gene at fast growth (Dennis et al, 2004)) and thus, despite an excess of about 2000 mRNA promoters, the small number of rRNA promoters are responsible for >50% of all transcripts made during exponential growth (Wagner, 2000). As rrn promoters are not saturated with Eσ70 under most growth conditions, their activities are argued to increase with increasing growth rates because of increasing concentrations of free Eσ70 (Jensen and Pedersen, 1990; Dennis et al, 2004). In contrast to rRNA genes (and other PSS genes), σ70-dependent promoters that are easily saturated, such as promoters driving stress-defense genes, would, in the saturation model, be repressed in relative terms, upon an increased pool size of Eσ70.
In contrast to the ‘saturation model' described above, an alternative model points to promoter affinity as the key feature behind differential control by alterations in the concentration of Eσ70 (Zhou and Jin, 1998; Barker et al, 2001a, 2001b), referred to here as the ‘Affinity Model'. This model argues for an increased, rather than decreased, availability of free Eσ70 during slow growth and a stringent response. Such an increase in Eσ70 concentration is suggested to be a result of Eσ70 falling off rRNA promoters because of decreased open complex stability elicited by ppGpp (Gaal et al, 1997; Barker et al, 2001b), whose concentration is inversely dependent on growth rate and is drastically elevated during nutrient exhaustion (Cashel, 1969; Ryals et al, 1982; Zacharias et al, 1989; Teich et al, 1999). As a consequence, promoters with relatively poor ability to recruit Eσ70—in the affinity model argued to be those requiring ppGpp for their induction (e.g. stress-defense genes and amino acid biosynthetic operons)—are suggested to be induced upon growth limitation and elevated ppGpp levels because of the increased availability of Eσ70. Although it has been argued that the data supporting this model place considerable constraints on models invoking hypothetical factors that might increase amino acid promoter activity in a ppGpp-dependent manner (Barker et al, 2001a), it was later shown that DksA is indeed such a factor (Brown et al, 2002; Paul et al, 2004a, 2005). Hence, the passive models for gene regulation need to be considered as working in concert with, or in parallel to, direct and active control by ppGpp/DksA.
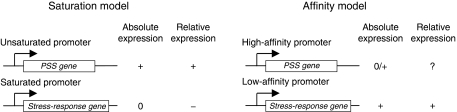
Both models of passive regulation suggest that alterations in the availability of free Eσ70 may re-direct transcriptional resources between growth-related activities (PSS) and maintenance/stress defenses but the predictions of the models are conflicting in almost every instance possible (Figure 1). For example, whereas the affinity model predicts that increased Eσ70 concentration would lead to an increased expression of maintenance and amino acid biosynthetic genes at the expense of PSS genes (i.e. a stringent-like pattern of gene expression) the saturation model predicts the exact opposite (i.e. a relaxed response, a phenotype linked to ppGpp deficiency). The affinity model argues for a direct correlation between ppGpp concentration and the pool size of free Eσ70, whereas the saturation model predicts an inverse correlation. In addition, overproduction of functional Eσ70 is in the saturation model envisaged to suppress the stringent phenotype of mutant cells with constitutively elevated levels of ppGpp, whereas the affinity model expects this to be ineffective or counterproductive. In this work, we report on a set of experiments designed to test these contrasting predictions experimentally.
Figure 1.
The predictions from the two models of passive regulation upon increase in Eσ70 availability. In the saturation model, an increase in Eσ70 availability would increase (+) absolute expression from unsaturated promoters, whereas saturated promoters will not be affected (0). A saturated promoter would in this case be relatively repressed (−). In contrast, the affinity model argues that an increase in Eσ70 availability would not benefit a high-affinity promoter (0/+), only promoters with low affinity would have an increase in absolute expression (+). Thus, in relative terms, a low-affinity promoter is predicted to have an increase in relative expression (+), whereas no prediction can be made for high-affinity promoters (?). The two models then make opposite predictions for relative expression from stress response genes (saturated, low affinity) upon an increase in Eσ70.
Results
Overproduction of E_σ_70 elevates expression of PSS genes
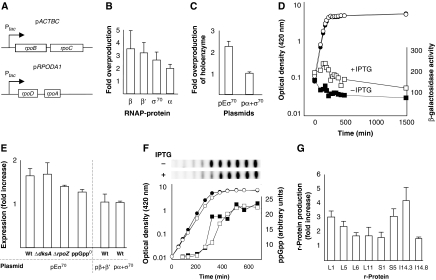
To test the effect of increasing Eσ70 holoenzyme availability, we used two plasmids encoding the subunits of core RNA polymerase (E) (i.e. the α, β, β′ subunits) and the housekeeping sigma factor σ70 under the control of P_tac_ promoters (Figure 2A) (Dykxhoorn et al, 1996). Western blot analysis confirmed that all Eσ70 subunit proteins were overproduced ∼2–3-fold during exponential growth by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) (Figure 2B). Further analysis showed that the two-fold increase was kept in stationary phase (data not shown). To asses whether this increase in subunits also resulted in an increased level of Eσ70 complexes, we measured the amount of σ70 that co-purified with the core enzyme using an affinity column with antibodies against β′. The over-expression of all subunits resulted in a more than two-fold increase of Eσ70 complexes, whereas overproduction of α and σ70 did not (Figure 2C). It should also be noted that this overproduction has but a marginal effect on growth (Figure 2D).
Figure 2.
Overproduction of Eσ70 elevates expression from PSS genes. (A) Expression vectors; p_ACTBC_ and p_RPODA1_, expressing β+β′ and σ70+α, respectively, under the control of a P_tac_ promoter (Dykxhoorn et al, 1996). (B) Quantification of Eσ70 subunit overproduction. Samples were taken in exponential growth phase. (C) Quantification of σ70 subunits co-purified with β′ (core enzyme) in a strain containing expression vectors p_ACTBC_ and p_RPODA1_ (pEσ70) and in a strain containing only p_RPODA1_ (pσ70+α). (D) Growth (circles) and expression from the rrnB_P1−41−+50 promoter (squares) with (open symbols; 1 mM IPTG) and without (closed symbols; no IPTG) overproduction of Eσ70. (E) Fold increase in expression from the rrnB_P1−41−+50 promoter upon Eσ70, β+β′, or σ70+α overproduction. The first four bars from the left show the effect of overproducing Eσ70 in wild-type (Wt), Δ_dksA, Δ_rpoZ, and ppGpp0 cells as indicated. The last two bars shows overproduction of only β and β′ subunits (pβ+β′) and of only σ70 and α subunits (pσ70+α) in wild-type (Wt) cells. (F) The effects of Eσ70 overproduction on the levels of ppGpp. Growth (circles) and ppGpp levels (squares) with (open symbols; 1 mM IPTG) and without (closed symbols; no IPTG) overproduction of Eσ70. The autoradiograms of the thin-layer chromatography (TLC) plates are shown in the upper panel. (G) The effects of Eσ70 overproduction on ribosomal protein production as detected on 2D gels. The bars indicate the difference in relative production of the particular protein with (1 mM IPTG) compared with without (no IPTG) Eσ70 overproduction.
We first examined the effect of Eσ70 overproduction on _rrnB_P1 promoter activity, which is under negative control by ppGpp (Cashel et al, 1996; Paul et al, 2004b). Expression from both promoter constructs, rrnB_P1−41−+50_-lacZ (Figure 2D) and rrnB_P1−51−+50_-lacZ (not shown), was increased about two-fold as a result of Eσ70 overproduction. This increase was observed both during growth and stationary phase (Figure 2D). Overproduction of only β and β′, or of only α and σ70 did not result in increased expression from _rrnB_P1 (Figure 2E), demonstrating that the effect on expression is a specific effect of increased Eσ70 concentration rather than of overproduction per se.
Earlier in vitro experiments suggested that the E subunit ω is required for Eσ70 sensitivity to ppGpp, but that this requirement is relieved in the presence of DksA (whose activity is independent of ω) (Vrentas et al, 2005). In line with the fact that ω seems of minor importance for Eσ70 function when DksA is present in the cell, we found an increased expression from rrnB_P1 upon overproduction of Eσ70 in mutants lacking ω (Δ_rpoZ) (Figure 2E).
As the rrn genes are negatively regulated by ppGpp, we tested whether the elevated _rrnB_P1 activity observed upon Eσ70 overproduction was a result of reduced levels of ppGpp. This was not the case as the concentration of ppGpp was not significantly altered during either growth or starvation by overproducing Eσ70 (Figure 2F). We also found that overproduction of Eσ70 effectively increased _rrnB_P1 expression in cells lacking DksA (Figure 2E), a protein influencing the regulatory activities of ppGpp (Paul et al, 2004a; Magnusson et al, 2007). As an additional control, Eσ70 was also overproduced in a ppGpp0 background and despite the fact that the lack of ppGpp itself elevates expression from _rrnB_P1, Eσ70 overproduction resulted in a further 1.3-fold increased expression (Figure 2E). Thus, we conclude that overproduction of Eσ70 does not change the ppGpp levels and that it elevates expression from _rrnB_P1 independent of both DksA and ppGpp.
Non-equilibrium pH 2D polyacrylamide gel electrophoresis (NEPHGE) was used to examine the pattern of ribosomal protein (r-protein) production upon Eσ70 overproduction. The r-proteins analysed are shown on a standard NEPHGE gel (Supplementary Figure S1). Pulse labelling demonstrated that the rates of synthesis of all r-proteins analysed were markedly higher in the cells overproducing Eσ70 (Figure 2G) ranging from 1.5- to 4-fold induction. The data suggest that production of the PSS is elevated upon increased availability of Eσ70, in line with predictions of the saturation model.
Overproduction of E_σ_70 attenuates expression of stress-defense genes and amino acid biosynthetic operons
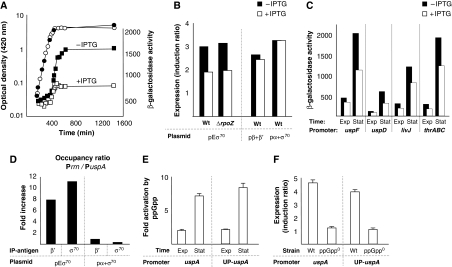
We next tested the effect of Eσ70 overproduction on the expression of the Eσ70-dependent usp genes (Nystrom and Neidhardt, 1996; Gustavsson et al, 2002; Nachin et al, 2005), which, in contrast to genes of the PSS, are positively regulated by ppGpp (Kvint et al, 2000; Gustavsson et al, 2002). The expression from all the _usp_-promoters tested, uspA/uspD/uspF (Figure 3A and C), and uspC/uspE (not shown), were markedly attenuated upon Eσ70 overproduction. The expressions from promoters of amino acid biosynthetic genes, like promoters of the usp genes, also require ppGpp. Similar to the usp promoters, the expression from both livJ and thrABC promoters (Figure 3C) were negatively affected by Eσ70 overproduction. As a control, we overproduced only β and β′ or only α and σ70, which confirmed that the decreased induction of the uspA promoter is specific to Eσ70 overproduction (Figure 3B).
Figure 3.
Expression of stress-defense genes and amino acid biosynthetic operons is attenuated upon Eσ70 overproduction. (A) Growth (circles) and expression from the P_uspA_ promoter (squares) with (open symbols; 1 mM IPTG) and without (closed symbols; no IPTG) overproduction of Eσ70. (B) Induction ratios (stationary phase promoter activity/exponential growth promoter activity) for the uspA promoter with (open bars; +1 mM IPTG) and without (closed bars; -IPTG) overproduction of Eσ70, β+β′, or σ70+α. The first four bars from the left show the effect of overproducing Eσ70 (pEσ70) in wild-type (Wt) and Δ_rpoZ_ cells as indicated. The last four bars shows overproduction of only β and β′ subunits (pβ+β′) and of only σ70 and α subunits (pσ70+α) in wild-type (Wt) cells. (C) Expression from P_uspF_, P_uspD_, P_livJ_, and P_thrABC_ promoters during exponential growth (Exp) and in stationary phase (Stat) with (open bars; +1 mM IPTG) and without (closed bars; -IPTG) overproduction of Eσ70 (D) DNA isolated from formaldehyde-fixed cells with (1 mM IPTG) and without (no IPTG) overproduction of Eσ70 (pEσ70) or σ70 and α (pσ70+α) alone was subjected to immunoprecipitation using antibodies against either the β′ or the σ70 subunit of Eσ70. PCR-based amplification of co-precipitated DNA was done using specific primers to the uspA promoter or to the rrn promoters. The Eσ70 promoter occupancy ratio (P_rrn_/P_uspA_) of cells not subjected to IPTG was set to 1 and all other values are expressed relative to that. Representative data are shown. (E, F) Effect on ppGpp-dependent expression from a uspA promoter upon addition of an UP element. (E) Fold induction of the P_uspA_ and UP-P_uspA_ promoters by ppGpp (wild-type promoter activity/ppGpp0 promoter activity) during exponential growth (Exp) and in stationary phase (Stat). (F) Induction ratios (stationary phase promoter activity/exponential growth promoter activity) for the P_uspA_ and UP-P_uspA_ promoters in wild-type (Wt) and ppGpp0 strains.
In addition, we confirmed that Eσ70 overproduction decreased the induction also in mutants lacking ω (Δ_rpoZ_) (Figure 3B) and that a Δ_rpoZ_ strain, in the absence of Eσ70 overproduction plasmids, do not exhibit altered uspA promoter activity (Supplementary Figure S2). Further, overproduction of ω cannot restore uspA expression in cells lacking either ppGpp or DksA (Supplementary Figure S2). Thus, we conclude that the effects of overproducing Eσ70 are not because of the absence of simultaneous overproduction of the ω subunit.
Repression of σ70-dependent stress-defense genes and amino acid biosynthetic operons upon ectopic elevation of Eσ70 levels is in accord with the saturation model (Jensen and Pedersen, 1990; Dennis et al, 2004), which argues that such genes are driven by promoters that are much more easily saturated than rrn promoters and would not, relative to rrn, benefit from increased concentrations of free Eσ70. In other words, the apparent repression of promoters like P_uspA_ upon Eσ70 overproduction is predicted to be a relative rather than absolute phenomenon and because of the fact that rrn promoters, in comparison, have a superior capacity to accommodate high levels of Eσ70. We tested this notion by chromatin immunoprecipitation (ChIP) using antibodies against β′ and σ70. This analysis demonstrated that Eσ70 overproduction in vivo resulted in an 8- and 11-fold increase in the ratio of promoter occupancy at rrn and uspA promoters, for β′ and σ70, respectively (Figure 3D). In contrast, overproduction of only α and σ70 did not significantly alter the occupancy ratio, in line with the lack of effect on expression from these promoters (Figures 2E and 3B).
In contrast to the saturation model, the affinity model predicts that Eσ70 overproduction would benefit promoters positively regulated by ppGpp, such as the P_uspA_ and P_thrABC_ tested here, as the defining characteristics of such promoters, according to this model, is their poor ability to recruit Eσ70. It has been reported that P_argI_, a promoter that is positively regulated by ppGpp, becomes less dependent on ppGpp by inserting an UP element just upstream the −35 hexamer and thereby increasing the promoter-Eσ70 association rate, supporting the affinity model (Barker et al, 2001a). We constructed a derivative of the uspA promoter fused to the UP element (see Materials and methods) and tested to what extent this diminished the promoters' requirement for ppGpp. The fold activation by ppGpp in both exponential and stationary phase was similar for the wt P_uspA_ and the UP-P_uspA_ promoters (Figure 3E) and the induction of the two promoters upon glucose starvation was the same in both the wt and ppGpp0 (Δ_relA_ Δ_spoT_) genetic backgrounds (Figure 3F). Thus, we conclude that the ppGpp-dependent expression from a uspA promoter is insensitive to addition of an UP-element to the promoter, suggesting that it is insensitive to increased Eσ70 affinity.
Overproduction of E_σ_70 creates a phenocopy of relaxed cells
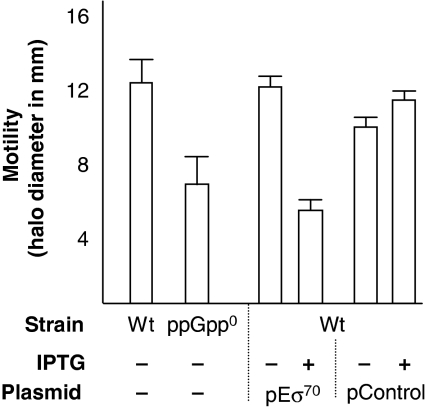
The results presented suggest that cells with increased levels of Eσ70 holoenzyme exhibit a ‘relaxed' phenotype, that of cells lacking ppGpp, with respect to gene expression of stringently regulated promoters. We, therefore, tested whether strains with elevated Eσ70 levels exhibited any other of the phenotypes associated with a relaxed response. Indeed, we observed that cells overproducing Eσ70 were, similar to a ppGpp0 strain (Magnusson et al, 2007), defective in motility on 0.3% agar plates (Figure 4).
Figure 4.
Overproduction of Eσ70 decreases motility, a phenocopy of a relaxed cell. Motility measured as the diameter of the halo formed after 24 h incubation on soft agar plates of wild-type (Wt), ppGpp0, and a Eσ70 overproduction strain and vector control strain with and without addition of 1 mM IPTG.
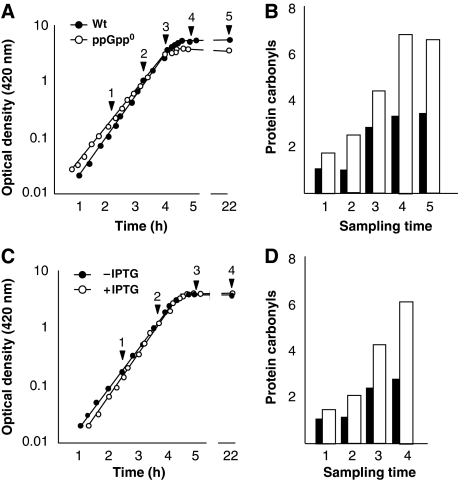
As a ppGpp0 mutant and an Eσ70 overproducing strain seem to direct its transcriptional resources predominantly to growth and the production of the PSS at the expense of stress-defense systems, we hypothesized that such cells may show signs of diminished maintenance activities and, as a consequence, elevated damage to its cellular constituents. Protein carbonylation serves as a general diagnostic marker of a diminished performance of maintenance systems and increases as a consequence of, for example, diminished oxidative stress defenses, decreased translational proof-reading, reduced proteolytic activities, or mutations in chaperone genes (Dukan et al, 2000; Ballesteros et al, 2001; Fredriksson et al, 2006). As shown in Figure 5A and B, protein carbonyl levels were markedly higher in the relaxed ppGpp0 strain during both exponential growth and upon growth arrest. Similarly, Eσ70 overproduction elevated carbonylation during both growth and growth arrest (Figure 5C and D), suggesting, again, that Eσ70 overproduction mimics a relaxed phenotype. However, Eσ70 overproduction did not result in amino acid auxotrophy (not shown), a phenotype seen in ppGpp0 strains (Xiao et al, 1991).
Figure 5.
Overproduction of Eσ70 increases protein carbonylation, a phenocopy of a relaxed cell. (A) Growth of wild-type (closed symbols) and ppGpp0 (open symbols) cells. The arrows indicate where samples for protein carbonylation measurements were taken. (B) Quantified carbonylation levels of wild-type (closed bars) and ppGpp0 (open bars) cells. (C) Growth of a wild-type strain containing the Eσ70 overproduction plasmids with (open symbols) or without 1 mM IPTG (closed symbols). The arrows indicate where samples for protein carbonylation measurements were taken. (D) Quantified carbonylation levels, with (open bars) and without (closed bars), overproduction of Eσ70.
The pool size of free E_σ_70 is elevated in cells lacking ppGpp
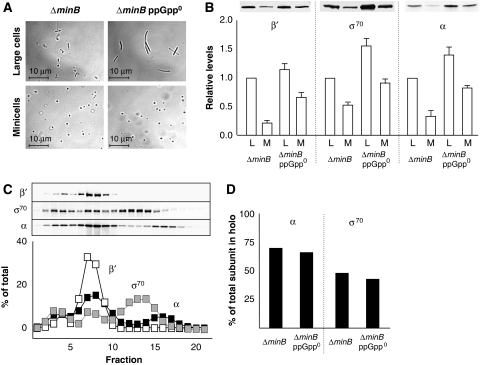
As overproduction of Eσ70 mimics a relaxed response, we asked whether cells lacking ppGpp display an elevated concentration of free Eσ70 and whether such elevation is, in fact, an integral part of the relaxed response. To measure the levels of cytoplasmic free Eσ70, a minicell approach was used (Rogerson and Stone, 1974; Shepherd et al, 2001). A strain with an inactivated minB locus allows septum formation at the cell poles resulting in the possibility to form either two normal (large) cells or one chromosome-free minicell and a filamentous-like cell (de Boer et al, 1988). The Eσ70 content in the minicells will represent free Eσ70 as they are devoid of DNA. Large cells and chromosome-free minicells from wild-type (Δ_minB_) and ppGpp0 (Δ_minB_ Δ_relA_ Δ_spoT_) were isolated (Figure 6A) and assayed for their content of Eσ70. The concentration of free Eσ70, as estimated from measurements of α, β′, and σ70 levels, were clearly higher in the ppGpp0 cell (∼60% of total Eσ70 was free) compared with the wild-type cell (∼30% of total Eσ70 was free), demonstrating that relaxed cells indeed have elevated concentrations of free Eσ70 (Figure 6B).
Figure 6.
A ppGpp0 strain displays elevated levels of free Eσ70. (A) A typical image of a 1000 × magnification of isolated large cell and minicell fractions from Δ_minB_ and Δ_minB_ ppGpp0 strains. (B) Relative levels of Eσ70 subunits β′ (left panel), σ70 (centre panel), and α (right panel) in large cells (L) and minicells (M) from Δ_minB_ and Δ_minB_ ppGpp0 strains. Upper panel shows representative blots of β′, σ70, and α, respectively. Equal protein concentration was loaded. (C) Western blot and the corresponding quantifications of gel-filtrated fractions from the Δ_minB_ ppGpp0 minicell lysate. (D) The relative amount of total subunit that co-elutes with β′ (holo) in Δ_minB_ and Δ_minB_ ppGpp0 cells.
To ascertain that the increased concentration of Eσ70 subunits in ppGpp0 cells represented increased levels of free holoenzyme, rather than free subunits, we fractionated crude cell extracts by gel filtration and the content of α, β′, and σ70 subunits in each fraction were quantified (one example, Δ_minB_ ppGpp0 minicells, is shown in Figure 6C). This analysis demonstrated that there was only a minor difference between wild-type and ppGpp0 cells in the relative amount of α and σ70 bound to E (Figure 6D). However, the somewhat lower fractions of bound α and σ70 in the ppGpp0 strain corresponds well with the fact that the increase in total levels of these subunits, compared with wild type, is higher than the increase of β′ (Figure 6B). Thus, the increased concentrations of α, β′, and σ70 subunits in the absence of ppGpp reflect a higher level of free Eσ70 holoenzyme, which seems to be limited by the concentration of β′ (and probably β).
Overproduction of E_σ_70 counteracts the effect of elevating ppGpp
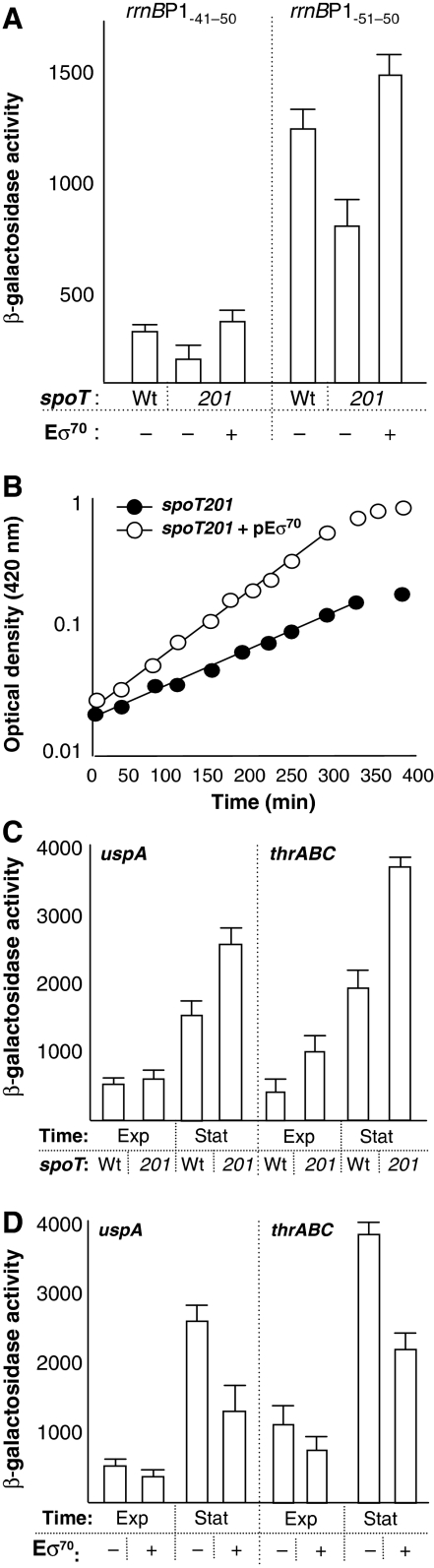
If, as predicted by the saturation model, stringency is partly accomplished by a reduction in free Eσ70 levels, an overproduction of Eσ70 should to some extent repress the stringent response. To test this possibility, we analysed expression from rrn, uspA, and thrAB promoters in a spoT201 mutant strain exhibiting constitutively elevated levels of ppGpp because of a diminished ppGpp hydrolysis activity of the mutated SpoT protein. The spoT201 allele results in 2.5-fold increase in ppGpp concentration and mutants carrying this allele display the mildest growth rate reduction of all mutant spoT alleles that have been examined (Sarubbi et al, 1988). In line with earlier studies of this strain (Sarubbi et al, 1988), the spoT201 allele represses expression from the rrnB_P1 promoter (Figure 7A). Interestingly, this repression could be suppressed by overproducing Eσ70 in the spoT201 strain (Figure 7A), indicating that the reduced expression of rrn genes in cells with elevated ppGpp levels is, indeed, at least partly because of limitation in Eσ70 availability. In fact, such a limitation in Eσ70 seems to be partly responsible for the reduced growth rate of the s_poT201 mutant, as Eσ70 overproduction greatly shortened the generation time of the cells (from 105 to 70 min; Figure 7B). As seen in Figure 7C, introducing the s_poT201_ allele elevates both uspA and thrABC promoter activity and overproducing Eσ70 totally suppressed this up-regulation of uspA and thrABC (Figure 7D). Taken together, the data demonstrate that elevating Eσ70 counteracts stringency and argues for limitations in free Eσ70 being an integral regulatory component of a stringent response of E. coli.
Figure 7.
Overproduction of Eσ70 suppresses the effect of elevated ppGpp levels. (A) Gene expression from P_rrnB_ P1−41−50 and P_rrnB_ P1−51−50 during exponential growth in wild type (wt) and a strain with spoT201 (201) with or without overproduction of Eσ70. (B) Growth rate of spoT201 cells without (closed circles; no IPTG) and with (open circles; 1 mM IPTG) overproduction of Eσ70. (C) Expression from P_uspA_ and P_thrABC_ during exponential growth (Exp) and stationary phase (Stat) in wild-type (Wt) and spoT201 (201) cells. (D) Expression from P_uspA_ and P_thrABC_ during exponential growth (Exp) and stationary phase (Stat) in the spoT201 strain with (1 mM IPTG) and without (no IPTG) overproduction of Eσ70.
Discussion
We demonstrate here that a two-fold overproduction of Eσ70 elevates the production of PSS at the expense of stress-defense genes (_usp_s) and amino acid biosynthetic operons—a response typical of cells with a diminished ability to make ppGpp. These results are in line with the saturation model (Jensen and Pedersen, 1990; Dennis et al, 2004), which argues that PSS genes possess unsaturated promoters exhibiting high maximal initiation velocity, whereas promoters of, for example, stress-defense genes (_usp_s) and amino acid biosynthetic operons are often saturated (or close to saturated) with Eσ70 and exhibit lower maximal initiation velocity (Figure 8). Thus, the PSS promoters are limited by the rate of Eσ70 recruitment and, in contrast to saturated promoters, become sensitive to the concentration of free Eσ70. Elevated levels of Eσ70 would, therefore, be expected to specifically boost the expression from _rrn-_promoters. The fact that Eσ70 overproduction indeed caused a marked elevation of promoter occupancy and transcription from rrn, demonstrates that _rrn_-promoters do not work at their maximal capacity even during exponential growth in rich medium. This is in line also with data from the Squires laboratory, which has demonstrated that the expression from individual rrn operons can increase to compensate for the deletion of other rrn operons (Condon et al, 1992, 1993, 1995). It is likely that the rrnB_P1_-lacZ reporter constructs underestimate the initiation capacity of a native _rrnB_P1, because these constructs lack the exceptionally fast elongation rate of natural rrn genes (Dennis et al, 2009). Thus, the clearing rate of the rrnB_P1_-lacZ might be limited by the transcription rate and queuing rather than intrinsic promoter properties.
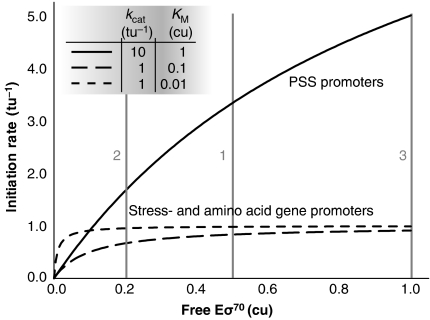
Figure 8.
Illustration of the mechanism of passive regulation by the saturation model. The transcription initiation rate in arbitrary time units−1 (tu−1) is plotted as a function of Eσ70 in arbitrary concentration units (cu), using the classic Michaelis–Menten expression, but with the Eσ70 as the ‘substrate' and the promoter as the ‘enzyme': 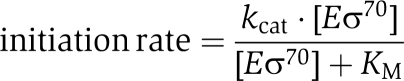 The figure illustrates how promoter-Eσ70 interactions, which results in different maximal initiation rates (_k_cat) or half promoter saturation concentrations (_K_M), leads to different responses to changes in the concentration of free Eσ70. Assume, for example, that the concentration of free Eσ70 is 0.5 cu in wild-type cells at exponential growth (grey line 1), and that it decreases to 0.2 cu in stationary phase (grey line 2) or increases to 1.0 cu by over-expression of Eσ70 from the plasmids (grey line 3). The initiation rate for PSS promoters (with high _k_cat and high _K_M) depends strongly on the concentration of free Eσ70. In contrast, the initiation rates from stress and amino acid biosynthesis genes (with low _k_cat and low _K_M) show only minor changes or remain unaltered at higher concentration of Eσ70, but this results in a decreased expression relative to the total transcription in the cell.
The figure illustrates how promoter-Eσ70 interactions, which results in different maximal initiation rates (_k_cat) or half promoter saturation concentrations (_K_M), leads to different responses to changes in the concentration of free Eσ70. Assume, for example, that the concentration of free Eσ70 is 0.5 cu in wild-type cells at exponential growth (grey line 1), and that it decreases to 0.2 cu in stationary phase (grey line 2) or increases to 1.0 cu by over-expression of Eσ70 from the plasmids (grey line 3). The initiation rate for PSS promoters (with high _k_cat and high _K_M) depends strongly on the concentration of free Eσ70. In contrast, the initiation rates from stress and amino acid biosynthesis genes (with low _k_cat and low _K_M) show only minor changes or remain unaltered at higher concentration of Eσ70, but this results in a decreased expression relative to the total transcription in the cell.
In view of the fact that elevating the pool size of Eσ70 favours functions related to growth at the expense of maintenance and that reducing Eσ70 levels has the opposite effect (Magnusson et al, 2003), one might ask whether alterations in the pool size of free Eσ70 comprise a normal, physiologically relevant, control mechanism for allocating resources between growth and maintenance activities in E. coli. If so, it is expected that the levels of free, and transcription available, Eσ70 should rise with the quality of the growth medium and this has been argued to be the case (Jensen and Pedersen, 1990; Bremer and Dennis, 1996). Specifically, Bremer et al (2003) derived a theory to calculate free Eσ70 concentrations from the concentrations of total RNA polymerase and promoters in a model system with estimated Michaelis–Menten constants for the transcription initiation on different promoter (compare with Figure 8). According to such calculations, the concentration of free Eσ70 is about 0.4 and 1.2 μM at growth rates corresponding to 1.0 and 2.5 doublings/h, respectively, that is, the free Eσ70 concentration increases with increasing growth rates. Using a minicell approach, we report here that cells lacking ppGpp displayed a two-fold higher concentration of free Eσ70 compared with wild-type cells. Thus, we believe that growth medium regulation of free Eσ70 levels requires ppGpp and that this is an integral part of this alarmone's means of regulating gene expression. In line with this, we found that in cells with constitutively elevated ppGpp levels, the expression levels, from both positively and negatively regulated promoters, could be restored, and the slow growth rate could be improved by providing them with more Eσ70. The data strongly suggest that the nutritional quality of the environment is regulating the growth rate of cells by altering the availability of free Eσ70 and that this control is influenced by the alarmone ppGpp.
How, then, does the nutritional quality affect Eσ70 availability mechanistically through alterations in ppGpp levels? As ppGpp is known to increase transcriptional pausing and reduce the rate of transcription elongation (Kingston et al, 1981; Sorensen et al, 1994; Krohn and Wagner, 1996), it has been suggested that the alarmone sequesters Eσ70 in the elongating stage—more so the poorer the media (Jensen and Pedersen, 1990). However, it should be noted that the reduction in transcription elongation has been argued to be too small to effectively reduce the concentration of free Eσ70 (Bremer and Ehrenberg, 1995). Another mechanism for reducing Eσ70 availability may be linked to the role of ppGpp as a master regulator of sigma factor competition as seen by data demonstrating that alternative sigma factors; σS, σ32, and σ54, compete significantly better against σ70 in the presence of ppGpp both in vivo and in vitro (Jishage et al, 2002; Laurie et al, 2003). In addition, the fraction of σ70 bound to E is increased at the expense of both σS and σ32 in cells lacking ppGpp (Jishage et al, 2002) and elevated ppGpp levels have been shown to reduce the fraction of σ70 bound to E in vivo (Hernandez and Cashel, 1995). Thus, ppGpp seems to prime the Eσ70 in accordance with environmental signals such that the transcriptional apparatus will be primarily occupied with transcription of σ70-dependent housekeeping genes as long as the ppGpp levels are low, which signals that the nutritional status of the environment is favourable for growth (reviewed in Nystrom, 2004). In addition, it has been demonstrated that σ70 occasionally remains attached to E during transcriptional elongation (Bar-Nahum and Nudler, 2001; Mukhopadhyay et al, 2001) and that the fraction of σ70 that remains attached increases during nutritional stress (Bar-Nahum and Nudler, 2001). Although it is unknown whether this increase in elongating Eσ70 complexes is a consequence of increased ppGpp levels, it has been suggested that it promotes pausing at promoter-like sequences in the coding region (Mooney and Landick, 2003), and it is reasonable to assume that this reduces the availability of free Eσ70. Surprisingly, a recent theoretical study (Klumpp and Hwa, 2008) suggests that neither of the suggested effects of ppGpp can result in a substantial decrease of the concentration of free Eσ70. However, the reason for this result is that their model propose that a large majority of all Eσ70 is non-specifically bound to DNA at all growth rates (Klumpp and Hwa, 2008), which thereby creates a large pool of non-transcribing Eσ70 that masks any change in the concentration of free Eσ70. To have such a large fraction of the Eσ70 inactivated by non-specific binding is remarkable from an evolutionary perspective and, as demonstrated experimentally here using the minicell approach, the pool size of free Eσ70 is at least two-fold higher in cells lacking ppGpp.
The regulatory design of higher organisms is proposed to comprise a trade-off between activities dedicated to growth and those devoted to homeostasis. The trade-off is, in the disposable soma hypothesis, argued to be a consequence of limited resources in any one organism and is regulated by the nutritional quality of the environment (Kirkwood, 1977). Trade-offs between reproduction and survival may not be restricted to organisms with a soma distinct from the germ line and has been suggested to be part of the intrinsic regulatory design also of unicellular prokaryotes (Nystrom, 2004). We propose that nutritionally induced alterations in Eσ70 availability elicit such a trade-off between proliferation and maintenance and that this trade-off is regulated by the alarmone ppGpp. However, it should be noted that a stringent down-regulation of rRNA genes can occur in the absence of ppGpp accumulation (and transcription factors) by any condition effectively sequestering and/or reducing the availability of free Eσ70. Indeed, pyrimidine limitation, reducing transcription elongation rates, was shown to cause a phenocopy of a stringent response in the absence of ppGpp accumulation (Vogel et al, 1991) and a genetic approach to reduce cellular Eσ70 availability is likewise effective in repressing rRNA expression both in the presence and absence of ppGpp (Magnusson et al, 2003). This passive regulation in response to limitations in building blocks might represent an ancient regulatory baseline control in the cell on top of which more specific and pronounced effects could be achieved with the evolution of specific effector molecules, activators, and repressors.
Materials and methods
Bacterial strains and growth conditions
The E. coli strains used in this work are listed in Supplementary Table S1 (Supplementary data). Transformation of the Eσ70 overproduction plasmids (Dykxhoorn et al, 1996) was performed by standard methods. The spoT201 (Sarubbi et al, 1988) allele and Δ_minB_∷kan cassette (de Boer et al, 1989) were transduced into the different strains by standard P1 transduction. The presence of the spoT201 allele was confirmed by sequencing. The UP-P_uspA_ promoter was constructed such that the −60 to −39 part of the _rrnB_P1 promoter UP element (Ross et al, 1993) was fused into the same position upstream of the uspA promoter. All promoter-lacZ constructs in this work were incorporated into bacteriophage λRS45 and integrated in the E. coli chromosomal λ att site as described earlier (Farewell et al, 1996). Cultures were grown in Erlenmeyer flasks in M9-defined medium supplemented with a limiting concentration of glucose (0.08%) and all the amino acids in excess (Wanner et al, 1977) and thiamine (10 μM) at 37°C. Carbenicillin (50 μg/ml) or/and chloramphenicol (30 μg/ml) was added in all experiments with strains containing plasmids. For the ppGpp measurement, the cells were grown in MOPS buffered media with low phosphate (0.33 mM) supplemented with a limiting concentration of glucose (0.08%) and all the amino acids in excess and thiamine (10 μM). Minicell strains were grown at 37°C in MOPS buffered Luria-Bertani (LB) medium supplemented with 0.1% glucose and 50 μg/ml kanamycin as described in Shepherd et al (2001). Large cells and minicells were isolated using sucrose gradient centrifugation according to Shepherd et al (2001).
Measurement of cellular components
Relative β-galactosidase levels were determined as has been described (Miller, 1972) with modifications (Albertson and Nystrom, 1994). The β-galactosidase activity is expressed as:
Western blot analysis of Eσ70 subunit levels was performed with primary antibodies from NeoClone as described (Jishage and Ishihama, 1995). IRDye 800CW-labelled goat anti-mouse immunoglobulin G antibodies (LI-COR Biosciences) were used for detection and blots were analysed with the Odyssey infrared imaging system and software (LI-COR Biosciences).
Estimating holoenzyme content using affinity chromatography
Cultures were grown as described above in the presence or absence of 1 mM IPTG. The cells were harvested at OD 1 by centrifugation and freezing the pellet in −80°C. Pellets were dissolved in lysis buffer (50 mM Tris–HCl [pH 8.1 at 4°C], 0.1 mM EDTA, 0.5 M NaCl, 1 mM DTT, 5 μg/ml DNaseI, 0.1 mM PMSF and 10% glycerol) and cells were disrupted using a Freezer/Mill 6870 (Spex Sampleprep). The cell lysate was cleared by centrifugation, aliquoted, and freezed in −80°C. Cell lysate containing 100 μg total proteins were diluted in 10 times 50 mM Tris–HCl [pH 8.1 at 4°C], 0.1 mM EDTA, before adding to a column packed with 100 μl Softag 1 resin (Neoclone). Affinity purification was performed according to the manufacturer and the content of the eluate was quantified by western blot analysis as described above.
Determination of ppGpp levels
Cultures were continuously labelled for at least two generations before sampling with 75 μCi/ml of [32P]orthophosphate (Amersham Biosciences, Uppsala, Sweden) in low-phosphate (0.33 mM) MOPS medium. Measurements were essentially done according to Gentry et al (1993). In short, samples (100 μl) were mixed with 20 μl cold 13 M formic acid and immediately frozen in dry ice. Nucleotides were extracted by three freeze-thaw cycles and cell debris was removed by centrifugation. Subsequently, the supernatant was added onto thin-layer chromatography (TLC) plates (Polygram CEL 300 PEI, Machery-Nagel, Düren, Germany). Levels of ppGpp were quantified using a phosphoimager (Personal FX, Bio-Rad) and the images were analysed using the QuantityOne software (Bio-Rad).
Resolution of proteins by 2D polyacrylamide gel electrophoresis
Samples were taken in stationary phase and 1 ml of the culture was mixed with 10 μl of [35S]-methionine (10 mCi/ml, 1000 Ci/mmol, Amersham Biosciences) for pulse labelling. Incorporation was allowed to proceed for 10 m at 37°C followed by a chase (3 min) with excess of non-radioactive methionine (50 μl, 0.2 M). The samples were processed for resolution on 2D polyacrylamide gels according to O'Farrell (1978) with modifications (O'Farrell, 1978). Electrophoresis in the first dimension was carried out between pH 3.5 and pH 10 and the gels were run according to the NEPHGE protocol (non-equilibrium pH gel electrophoresis). The second dimension was run on 11.5% polyacrylamide gels. Radio-labelled proteins were detected in a phosphoimager (Personal FX, Bio-Rad) and the images were analysed using the PDQuest 6.2 software (Bio-Rad). Alphanumeric (A-N) designations and/or protein names were assigned to protein spots after matching them to the reference 2D images of the gene–protein database of E. coli (VanBogelen et al, 1992) or after identification using mass spectrometry.
ChIP assays and real-time PCR analysis
ChIP assays were performed essentially as described earlier (Lin and Grossman, 1998). Cells were grown in M9-defined media described above, and formaldehyde was added to a final concentration of 1% at an optical density of 0.7 (420 nm). After 20 min of incubation, glycine was added to a final concentration of 0.5 M, and cells were harvested by centrifugation and washed twice with Tris-buffered saline (pH 7.5). Cells were resuspended in 1000 μl of lysis buffer (10 mM Tris at pH 8.0, 20% sucrose, 50 mM NaCl, 10 mM EDTA, 10 mg/ml lysozyme) and incubated at 37°C for 30 min. A measure of 500 μl of IP buffer (50 mM HEPES-KOH at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) and 1 mM PMSF were added to the cell extract. DNA was sheared by sonication to an average size of ∼500–1000 bp. Insoluble cellular material was removed by centrifugation for 10 min, and the supernatant was transferred to a fresh tube. An aliquot of this supernatant were kept for use as the ‘input' sample. Proteins were immunoprecipitated by diluting a fraction of the cross-linked cell extract with IP buffer to a final volume of 500 μl. This was then incubated with 20 μl of Protein A-Sepharose beads (Amersham-Pharmacia) and either RNAP β′ subunit mouse monoclonal (NeoClone, W0001) or RNAP σ70 subunit mouse monoclonal (NeoClone, W0004) for at least 4 h at 4°C with gentle mixing. Samples were then washed twice with IP buffer, twice with IP buffer+500 mM NaCl, and once with wash buffer (10 mM Tris–HCl at pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet-P40, 0.5% sodium deoxycholate). Immunoprecipitated complexes were eluted by incubation of beads with 100 μl 2 × elution buffer (100 mM Tris–HCl at pH 7.8, 10 mM EDTA, 1% SDS) at RT for 30 min. Cross-links, of the eluates and the corresponding ‘input' samples, were reversed by incubation for 6 h at 65°C in elution buffer+100 μl NaCl solution containing 0.2 mg/ml Proteinase K. DNA was purified using a PCR purification kit (Qiagen). Co-precipitated DNA was analysed in triplicate by quantitative PCR using the Bio-Rad iQ5 detection system (Bio-Rad). Primer sequences are listed in Supplementary Table S2 (Supplementary data).
Motility assays
Bacterial cells were picked from colonies grown on LB agar plates and inoculated onto low agar LB plates (0.3%). The plates were incubated at room temperature for 24 h and motility was assessed quantitatively by measuring the diameter of the circular halo formed by the growing motile bacteria cells.
Measurement of protein carbonylation
Analysis of carbonylated proteins was performed using the chemical and immunological reagents of the OxyBlot Oxidized Protein Detection Kit. The carbonyl groups in the protein side chains were derivatized to 2,4-dinitrophenylhydrazone by reaction with 2,4-dinitrophenylhydrazine. The derivatized proteins were analysed immunochemically on 1D western blots onto polyvinylidene difluoride membranes as described earlier (Fredriksson et al, 2006).
Determination of α, _β_′, and _σ_70 as free subunits or in the holoenzyme
Isolated large cells and minicells were resuspended in reconstitution buffer (R-buffer) (10 mM Tris–HCl [pH 7.6 at 4°C], 0.1 mM DTT, 0.1 mM EDTA, 200 mM NaCl, and 5% glycerol (Jishage et al, 2002). The cells were disrupted using French press (Spectronic Instruments) and the cell debris was removed by centrifugation. The clear supernatant was precipitated with 50% ammonium sulphate and the precipitate was again resuspended in R-buffer and applied to a Superdex 200 PC 3.2/30 column (GE Healthcare) connected to a Smart chromatography system (Pharmacia LKB). Fractions of 50 μl were collected and their content was quantified by western blot analysis as described above.
Supplementary Material
Supplementary Material
Review Process Article
Acknowledgments
We sincerely thank Thomas Linn, Mike Cashel, Rick Gourse, and Piet AJ de Boer for sharing strains, Alexandra Grönvall and Linda Rosenquist for their contributions to the early stage of this work, and colleagues of the Nyström laboratory for discussion and suggestions on the paper. This work was sponsored by a grant from the Swedish Natural Science Research Council, and an award from the Göran Gustafsson foundation for scientific research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albertson NH, Nystrom T (1994) Effects of starvation for exogenous carbon on functional mRNA stability and rate of peptide chain elongation in Escherichia coli. FEMS Microbiol Lett 117: 181–187 [DOI] [PubMed] [Google Scholar]
- Ballesteros M, Fredriksson A, Henriksson J, Nystrom T (2001) Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J 20: 5280–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nahum G, Nudler E (2001) Isolation and characterization of sigma(70)-retaining transcription elongation complexes from Escherichia coli. Cell 106: 443–451 [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL (2001a) Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol 305: 689–702 [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL (2001b) Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol 305: 673–688 [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis P, Ehrenberg M (2003) Free RNA polymerase and modeling global transcription in Escherichia coli. Biochimie 85: 597–609 [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP (1996) Modulation of chemical composition and other parameters of the cell by growth rate. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC (ed). Vol. 2, 2nd edn, pp 1553–1569. Washington: ASM Press [Google Scholar]
- Bremer H, Ehrenberg M (1995) Guanosine tetraphosphate as a global regulator of bacterial RNA synthesis: a model involving RNA polymerase pausing and queuing. Biochim Biophys Acta 1262: 15–36 [DOI] [PubMed] [Google Scholar]
- Brown L, Gentry D, Elliott T, Cashel M (2002) DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol 184: 4455–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M (1969) The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem 244: 3133–3141 [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D (1996) The stringent response In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC (ed). Vol. 1, 2nd edn, pp 1458–1496. Washington: ASM Press [Google Scholar]
- Condon C, French S, Squires C, Squires CL (1993) Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J 12: 4305–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Liveris D, Squires C, Schwartz I, Squires CL (1995) rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol 177: 4152–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Philips J, Fu ZY, Squires C, Squires CL (1992) Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J 11: 4175–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI (1988) Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol 170: 2106–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56: 641–649 [DOI] [PubMed] [Google Scholar]
- Dennis P, Ehrenberg M, Fange D, Bremer H (2009) Varying rate of RNA chain elongation during rrn transcription in E. coli. J Bacteriol 191: 3740–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Ehrenberg M, Bremer H (2004) Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol Rev 68: 639–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, Nystrom T (2000) Protein oxidation in response to increased transcriptional or translational errors. PNAS 97: 5746–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, St Pierre R, Linn T (1996) Synthesis of the beta and beta' subunits of Escherichia coli RNA polymerase is autogenously regulated in vivo by both transcriptional and translational mechanisms. Mol Microbiol 19: 483–493 [DOI] [PubMed] [Google Scholar]
- Farewell A, Diez A, DiRusso C, Nystrom T (1996) Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J Bacteriol 178: 6443–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson A, Ballesteros M, Dukan S, Nystrom T (2006) Induction of the heat shock regulon in response to increased mistranslation requires oxidative modification of the malformed proteins. Mol Microbiol 59: 350–359 [DOI] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL Jr, Gourse RL (1997) Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278: 2092–2097 [DOI] [PubMed] [Google Scholar]
- Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M (1993) Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J Bacteriol 175: 7982–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova IL, Phleger NJ, Mutalik VK, Gross CA (2006) Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci USA 103: 5332–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Lonetto M, Losick R (1992) Bacterial sigma factors. In Transcriptional Regulation, SL M (ed)., pp 129–176. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Gustavsson N, Diez A, Nystrom T (2002) The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol Microbiol 43: 107–117 [DOI] [PubMed] [Google Scholar]
- Hernandez VJ, Cashel M (1995) Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J Mol Biol 252: 536–549 [DOI] [PubMed] [Google Scholar]
- Jensen KF, Pedersen S (1990) Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol Rev 54: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Ishihama A (1995) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of sigma 70 and sigma 38. J Bacteriol 177: 6832–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Kvint K, Shingler V, Nystrom T (2002) Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev 16: 1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Nierman WC, Chamberlin MJ (1981) A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem 256: 2787–2797 [PubMed] [Google Scholar]
- Kirkwood TB (1977) Evolution of ageing. Nature 270: 301–304 [DOI] [PubMed] [Google Scholar]
- Klumpp S, Hwa T (2008) Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc Natl Acad Sci USA 105: 20245–20250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn M, Wagner R (1996) Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J Biol Chem 271: 23884–23894 [DOI] [PubMed] [Google Scholar]
- Kvint K, Hosbond C, Farewell A, Nybroe O, Nystrom T (2000) Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol Microbiol 35: 435–443 [DOI] [PubMed] [Google Scholar]
- Laurie AD, Bernardo LM, Sze CC, Skarfstad E, Szalewska-Palasz A, Nystrom T, Shingler V (2003) The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J Biol Chem 278: 1494–1503 [DOI] [PubMed] [Google Scholar]
- Lin DC, Grossman AD (1998) Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685 [DOI] [PubMed] [Google Scholar]
- Maaløe O (1979) Regulation of the protein synthesizing machinery—ribosomes, tRNA, factors and so on In Biological Regulation and Development, Goldberger RF (ed)., Vol. 1, pp 487–542. New York: Plenum Publishing Corp [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T (2007) Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol 189: 5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Nystrom T, Farewell A (2003) Underproduction of sigma 70 mimics a stringent response. A proteome approach. J Biol Chem 278: 968–973 [DOI] [PubMed] [Google Scholar]
- Miller J (1972) Experiments in Molecular Genetics. Cold Spring Harbour, N.Y.: Cold Spring Harbour Laboratory [Google Scholar]
- Mooney RA, Landick R (2003) Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev 17: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH (2001) Translocation of sigma(70) with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106: 453–463 [DOI] [PubMed] [Google Scholar]
- Nachin L, Nannmark U, Nystrom T (2005) Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol 187: 6265–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T (2004) Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54: 855–862 [DOI] [PubMed] [Google Scholar]
- Nystrom T, Neidhardt FC (1996) Effects of overproducing the universal stress protein, UspA, in Escherichia coli K-12. J Bacteriol 178: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH (1978) The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14: 545–557 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL (2004a) DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118: 311–322 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL (2005) DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA 102: 7823–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL (2004b) rRNA transcription in Escherichia coli. Annu Rev Genet 38: 749–770 [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62: 35–51 [DOI] [PubMed] [Google Scholar]
- Rogerson AC, Stone JE (1974) Beta-beta' subunits of ribonucleic acid polymerase in episome-free minicells of Escherichia coli. J Bacteriol 119: 332–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL (1993) A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262: 1407–1413 [DOI] [PubMed] [Google Scholar]
- Ryals J, Little R, Bremer H (1982) Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol 151: 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarubbi E, Rudd KE, Cashel M (1988) Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet 213: 214–222 [DOI] [PubMed] [Google Scholar]
- Shepherd N, Dennis P, Bremer H (2001) Cytoplasmic RNA polymerase in Escherichia coli. J Bacteriol 183: 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MA, Jensen KF, Pedersen S (1994) High concentrations of ppGpp decrease the RNA chain growth rate. Implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J Mol Biol 236: 441–454 [DOI] [PubMed] [Google Scholar]
- Teich A, Meyer S, Lin HY, Andersson L, Enfors S, Neubauer P (1999) Growth rate related concentration changes of the starvation response regulators sigmaS and ppGpp in glucose-limited fed-batch and continuous cultures of Escherichia coli. Biotechnol Prog 15: 123–129 [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Sankar P, Clark RL, Bogan JA, Neidhardt FC (1992) The gene-protein database of Escherichia coli: edition 5. Electrophoresis 13: 1014–1054 [DOI] [PubMed] [Google Scholar]
- Vogel U, Pedersen S, Jensen KF (1991) An unusual correlation between ppGpp pool size and rate of ribosome synthesis during partial pyrimidine starvation of Escherichia coli. J Bacteriol 173: 1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL (2005) Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev 19: 2378–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R (2000) Transcription Regulation in Prokaryotes. Oxford: Oxford University Press [Google Scholar]
- Wanner BL, Kodaira R, Neidhardt FC (1977) Physiological regulation of a decontrolled lac operon. J Bacteriol 130: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M (1991) Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266: 5980–5990 [PubMed] [Google Scholar]
- Zacharias M, Goringer HU, Wagner R (1989) Influence of the GCGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth-rate control and stringent sensitivity. EMBO J 8: 3357–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YN, Jin DJ (1998) The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like ‘stringent' RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA 95: 2908–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Review Process Article