Histone Methylation Regulator PTIP Is Required for PPARγ and C/EBPα Expression and Adipogenesis (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 1.
Summary
PPARγ and C/EBPα cooperate to control preadipocyte differentiation (adipogenesis). However, the factors that regulate PPARγ and C/EBPα expression during adipogenesis remain largely unclear. Here we show PTIP, a protein that associates with histone H3K4 methyltransferases, regulates PPARγ and C/EBPα expression in mouse embryonic fibroblasts (MEFs) and during preadipocyte differentiation. PTIP deletion in MEFs leads to marked decreases of PPARγ expression and PPARγ-stimulated C/EBPα expression. Further, PTIP is essential for induction of PPARγ and C/EBPα expression during preadipocyte differentiation. Deletion of PTIP impairs the enrichment of H3K4 trimethylation and RNA polymerase II on PPARγ and C/EBPα promoters. Accordingly, _PTIP_−/− MEFs and preadipocytes all show striking defects in adipogenesis. Furthermore, rescue of the adipogenesis defect in _PTIP_−/− MEFs requires co-expression of PPARγ and C/EBPα. Finally, deletion of PTIP in brown adipose tissue significantly reduces tissue weight in mice. Thus, by regulating PPARγ and C/EBPα expression, PTIP plays a critical role in adipogenesis.
Introduction
Peroxisome Proliferator-Activated Receptor-γ (PPARγ) is considered the master regulator of adipogenesis. It is a member of the nuclear receptor superfamily of ligand-activated transcription factors and is both necessary and sufficient for adipogenesis (Farmer, 2006; Rosen et al., 2002). PPARγ has two isoforms, PPARγ1 and PPARγ2, generated by usage of two distinct promoters and alternative splicing (Zhu et al., 1995). PPARγ1 is ubiquitously expressed while PPARγ2 expression is restricted to adipose tissues. However, both isoforms are strongly induced during preadipocyte differentiation in vitro and both are highly expressed in adipose tissues in animals. PPARγ1 is induced earlier than PPARγ2 and is maintained at a level higher than PPARγ2 during preadipocyte differentiation (Morrison and Farmer, 1999). Data from PPARγ2 isoform-specific knockout mice indicate that PPARγ2 is not absolutely required for adipogenesis in vivo (Rosen and MacDougald, 2006), suggesting that PPARγ1 may be critical for adipogenesis. Although the functional difference between endogenous PPARγ1 and PPARγ2 remains unclear, ectopic expression of either PPARγ1 or PPARγ2 is sufficient to stimulate immortalized, non-adipogenic MEFs to differentiate into adipocytes (Mueller et al., 2002).
C/EBPα (CCAAT/enhancer binding protein α) is another principal adipogenic transcription factor and is strongly induced in the early phase of preadipocyte differentiation. Ectopic expression of C/EBPα stimulates non-adipogenic MEFs to undergo adipogenesis while deletion of C/EBPα in mice results in almost complete absence of white adipose tissue (WAT) (Farmer, 2006). PPARγ and C/EBPα positively regulate each other's expression and cooperate to control preadipocyte differentiation (adipogenesis) (Farmer, 2006; Rosen and MacDougald, 2006). However, the factors and the underlying mechanisms that regulate the induction of PPARγ and C/EBPα expression during adipogenesis remain unclear.
PTIP (Pax transactivation domain-interacting protein) is a ubiquitously expressed nuclear protein that associates with active chromatin. Knockout of PTIP in mice leads to lethality by embryonic day 9.5 (Patel et al., 2007). PTIP carries six tandem BRCT domains that are predominantly found in proteins involved in DNA damage response. Indeed, ectopically expressed PTIP has been implicated in cellular response to DNA damage (Manke et al., 2003). However, the physiological function of endogenous PTIP has remained largely unclear.
Histone lysine methylation has been implicated in both gene activation and repression, depending on the specific lysine residue that gets methylated. For example, methylation on histone H3 lysine 4 (H3K4) associates with gene activation while methylation on histone H3 lysine 27 (H3K27) associates with gene repression (Li et al., 2007). We and others recently show that endogenous PTIP is a component of a histone methyltransferase (HMT) complex that contains histone H3K4 methyltransferases MLL3 and MLL4 (also known as ALR), and the JmjC domain-containing protein UTX (Cho et al., 2007; Issaeva et al., 2007; Patel et al., 2007). Further, we and others demonstrate UTX is a histone H3K27-specific demethylase (Hong et al., 2007; Swigut and Wysocka, 2007). Thus, endogenous PTIP associates with two H3K4 methyltransferases and one H3K27 demethylase. Since both methylation of H3K4 and demethylation of H3K27 presumably associate with gene activation, these data strongly suggest a role of PTIP in gene activation.
In this report, we identify PTIP as a factor that regulates PPARγ and C/EBPα expression in MEFs as well as during preadipocyte differentiation. In MEFs, PTIP deletion leads to over 10 fold decrease of PPARγ expression and over 5 fold decrease of PPARγ-stimulated C/EBPα expression. In preadipocytes, while PTIP is dispensable for the basal level expression of PPARγ and C/EBPα before differentiation, it is essential for the robust induction of PPARγ and C/EBPα but not C/EBPβ during differentiation. Further, we show PTIP is required for the enrichment of methyltransferase MLL4, H3K4 trimethylation (H3K4me3) and RNA polymerase II (Pol II) on PPARγ and C/EBPα promoters. Accordingly, PTIP-deficient MEFs and white and brown preadipocytes all show striking defects in adipogenesis. Finally, tissue-specific deletion of PTIP significantly reduces brown adipose tissue weight and leads to cold intolerance in mice.
Results
PTIP Is Required for PPARγ Expression in MEFs
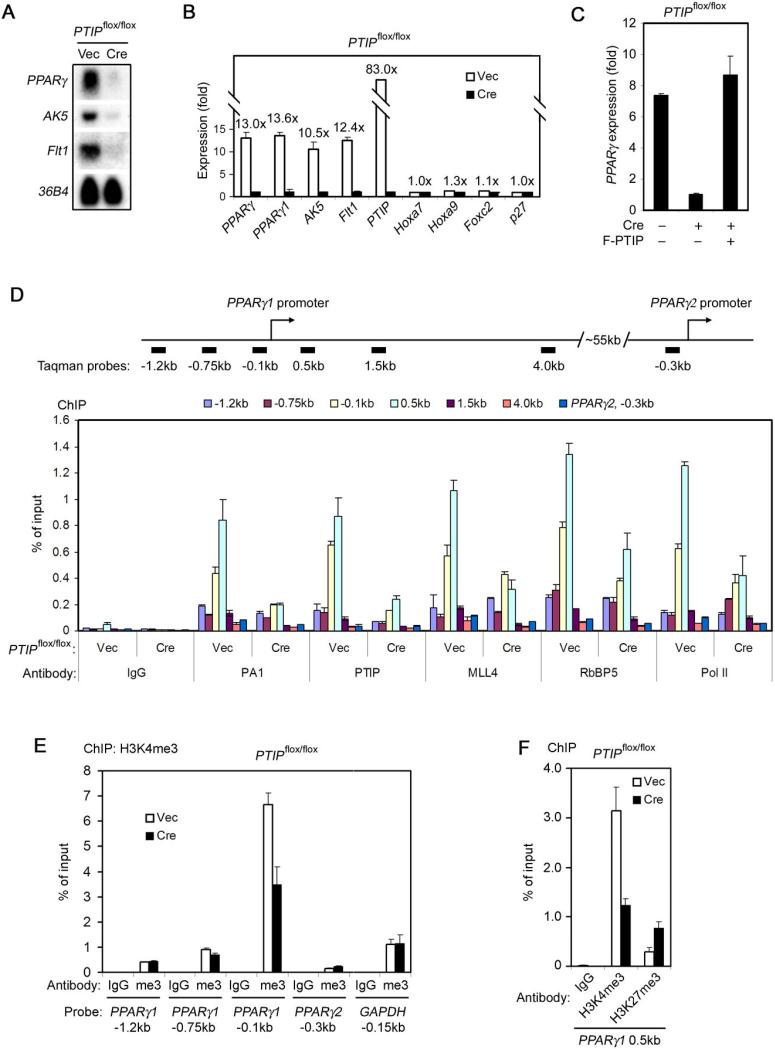
To identify PTIP-regulated genes, immortalized PTIP conditional knockout _PTIP_flox/flox MEFs were infected with retroviruses expressing Cre recombinase. By microarray analysis using the whole mouse genome expression arrays, we found that deletion of PTIP by Cre led to over 2.5 fold decrease of expression of ∼ 60 genes. Among these, PPARγ was the most significant one (Supplemental Table S1). Northern blot and quantitative reverse-transcriptase PCR (qRT-PCR) confirmed that PTIP deletion led to over ten fold decrease of PPARγ expression in _PTIP_flox/flox cells (Figure 1A-B). Isoform-specific qRT-PCR revealed that PPARγ1 was the only isoform expressed in MEFs where PPARγ2 expression was undetectable. Accordingly, we observed over ten fold decrease of PPARγ1 expression in PTIP-deficient MEFs (Figure 1B). Expression of ectopic PTIP could fully rescue PPARγ level in PTIP-deficient cells, confirming that PTIP is required for PPARγ expression in MEFs (Figure 1C).
Figure 1. PTIP Is Required for PPARγ Expression in MEFs.
(A) – (B) Immortalized _PTIP_flox/flox MEFs were infected with retrovirus MSCVpuro expressing Cre or vector (Vec) alone. Total RNA was extracted for confirmation of putative PTIP-regulated genes identified in microarray analysis (Supplemental Table S1). (A) Northern blot; (B) qRT-PCR. PPARγ2 mRNA level is below detection limit in qRT-PCR. AK5 and Flt1 stand for Adenylate Kinase 5 and FMS-like tyrosine kinase 1, respectively. 36B4 is the loading control.
(C) Ectopic PTIP can rescue PPARγ expression in PTIP-deficient cells. _PTIP_flox/flox MEFs were sequentially infected with retroviruses MSCVhygro expressing Cre and MSCVpuro expressing FLAG-tagged PTIP (F-PTIP). PPARγ expression was analyzed by qRT-PCR.
(D) ChIP assay of binding of the PTIP-associated HMT complex subunits to PPARγ1 and PPARγ2 promoters in _PTIP_flox/flox MEFs. Top panel, schematic of mouse PPARγ1 and PPARγ2 promoters. The black bars indicate locations of Taqman probes used for PCR quantitation of ChIP assays. Pol II, RNA polymerase II.
(E) ChIP assay of H3K4me3 on PPARγ1 and PPARγ2 promoters in _PTIP_flox/flox MEFs. me3, H3K4me3.
(F) ChIP assay of H3K4me3 and H3K27me3 on PPARγ1 promoter in _PTIP_flox/flox MEFs.
All results are representative of two – four independent experiments.
To investigate how PTIP regulates PPARγ expression in MEFs, we performed chromatin immunoprecipitation (ChIP) assays. Endogenous PTIP associated with PPARγ1 promoter with a signal peaked around the transcription start site. In contrast, no significant enrichment of PTIP was found on PPARγ2 proximal promoter in MEFs (Figure 1D). Representative subunits of PTIP-associated histone H3K4 methyltransferase complex, such as PA1, RbBP5 and the enzymatic subunit MLL4 (Cho et al., 2007), were also enriched on the PPARγ1 but not the PPARγ2 proximal promoter in MEFs (Figure 1D). H3K4me3, an epigenetic mark for active genes, was highly enriched on the PPARγ1 but not the PPARγ2 proximal promoter, consistent with the lack of PPARγ2 expression (Figure 1E). PTIP deletion not only reduced enrichment of PTIP, PA1, RbBP5 and MLL4 on the PPARγ1 promoter, but also decreased H3K4me3 signal and the recruitment of Pol II to the same promoter (Figure 1D-E). Interestingly, PTIP deletion also led to significant increase of histone H3K27 trimethylation (H3K27me3), an epigenetic mark for repressed genes, on PPARγ1 promoter (Figure 1F). These data indicate that PTIP and the associated HMT complex directly regulate PPARγ1 expression in MEFs, and suggest that PTIP regulates PPARγ1 expression by modulating the recruitment of histone H3K4 methyltransferase and Pol II to the target gene promoters.
PTIP Is Required for Adipogenesis in Primary MEFs and White and Brown Preadipocytes
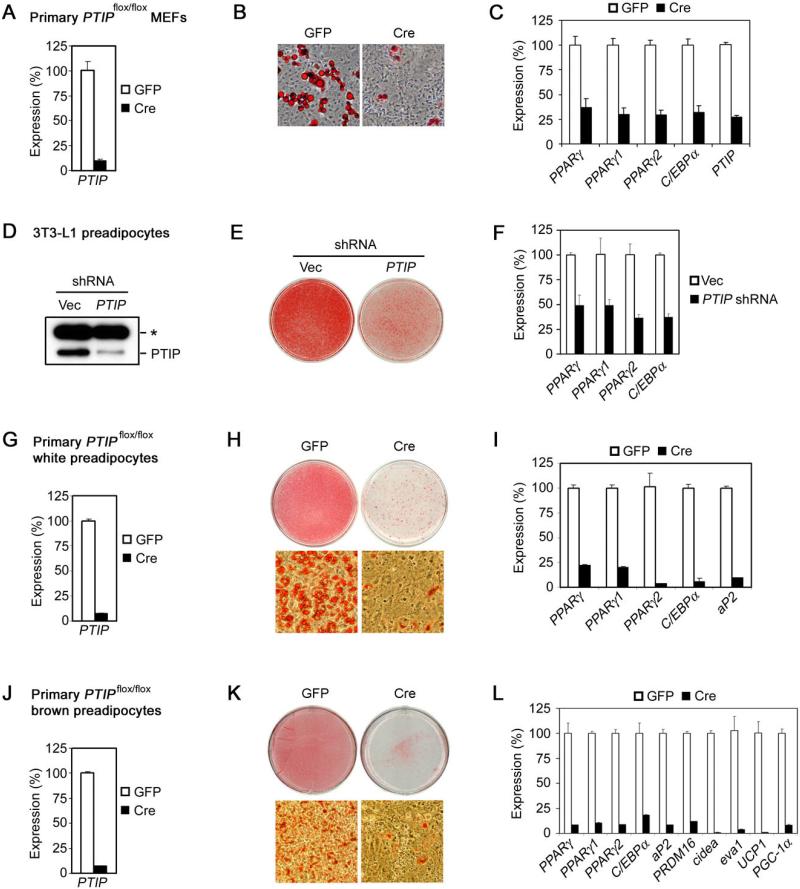
Since PTIP was required for PPARγ expression in MEFs, we tested whether PTIP was required for adipogenesis. Primary _PTIP_flox/flox MEFs were infected with adenovirus expressing Cre (Ad-Cre) to acutely delete PTIP gene (Figure 2A). In adipogenesis assays, approximately 20−30% of primary MEFs infected with the control virus differentiated into adipocytes as determined by Oil Red O staining. Deletion of PTIP led to a severe adipogenesis defect and significantly reduced expression of adipogenesis markers including PPARγ and C/EBPα (Figure 2B-C).
Figure 2. PTIP Is Required for Adipogenesis of Primary MEFs and Preadipocytes.
(A) – (C) PTIP is required for adipogenesis of primary MEFs. Primary _PTIP_flox/flox MEFs were infected with adenovirus expressing Cre (Ad-Cre) or control virus expressing GFP alone, followed by adipogenesis assay.
(A) To determine Ad-Cre-mediated gene deletion efficiency, PTIP expression was analyzed by qRT-PCR before induction of adipogenesis.
(B) Morphological differentiation under the phase-contrast microscope (Oil Red O staining).
(C) qRT-PCR analysis of gene expression after adipogenesis.
(D) – (F) PTIP is required for differentiation of 3T3-L1 white preadipocyte cell line. 3T3-L1 cells were infected with pSUPER.retro-based retroviral shRNA construct targeting PTIP or vector alone. After puromycin selection, cells were induced to undergo adipogenesis for 8 days.
(D) Western blot of PTIP expression before induction of differentiation. The asterisk indicates a non-specific band.
(E) Morphological differentiation.
(F) qRT-PCR analysis of gene expression 8 days after induction of differentiation.
(G) – (L) PTIP is required for differentiation of primary white (G – I) and brown (J – L) preadipocytes. Primary white and brown preadipocytes isolated from _PTIP_flox/flox mice were infected with Ad-Cre or control virus expressing GFP, followed by adipogenesis assay.
(G) and (J) qRT-PCR analysis of Cre-mediated PTIP gene deletion before differentiation.
(H) and (K) Morphological differentiation. Top panels, stained dishes. Lower panels, representative fields under the microscope.
(I) and (L) After differentiation, total RNA was isolated and subjected to qRT-PCR analysis of expression of genes common to white and brown adipocytes (PPARγ, PPARγ1, PPARγ2, C/EBPα, aP2) and genes specific for brown adipocytes (PRDM16, cidea, eva1, UCP1, PGC-1α) (Seale et al., 2007). All results are representative of two – four independent experiments.
The white preadipocyte cell line 3T3-L1 has been extensively used to study adipogenesis (Farmer, 2006). Stable knockdown of PTIP expression in 3T3-L1 led to severe defects in adipogenesis and the associated expression of adipogenesis markers, indicating that PTIP was also important for differentiation of 3T3-L1 (Figure 2D-F).
Further, we investigated whether PTIP was required for differentiation of primary preadipocytes. Primary white and brown preadipocytes isolated from _PTIP_flox/flox mice were infected with Ad-Cre (Figure 2G and 2J). PTIP deletion by Cre led to a nearly complete block of adipogenesis in both white (Figure 2H) and brown preadipocytes (Figure 2K). Consistent with the morphology, PTIP deletion blocked expression of markers for white (Figure 2I) and brown adipocytes (Figure 2L). These results are consistent with the requirement of PTIP for PPARγ expression in MEFs, and indicate that PTIP is required for adipogenesis in primary MEFs and preadipocytes.
Both PTIP-Deficient and C/EBPα-Deficient MEFs Show Defects in PPARγ-Stimulated Adipogenesis
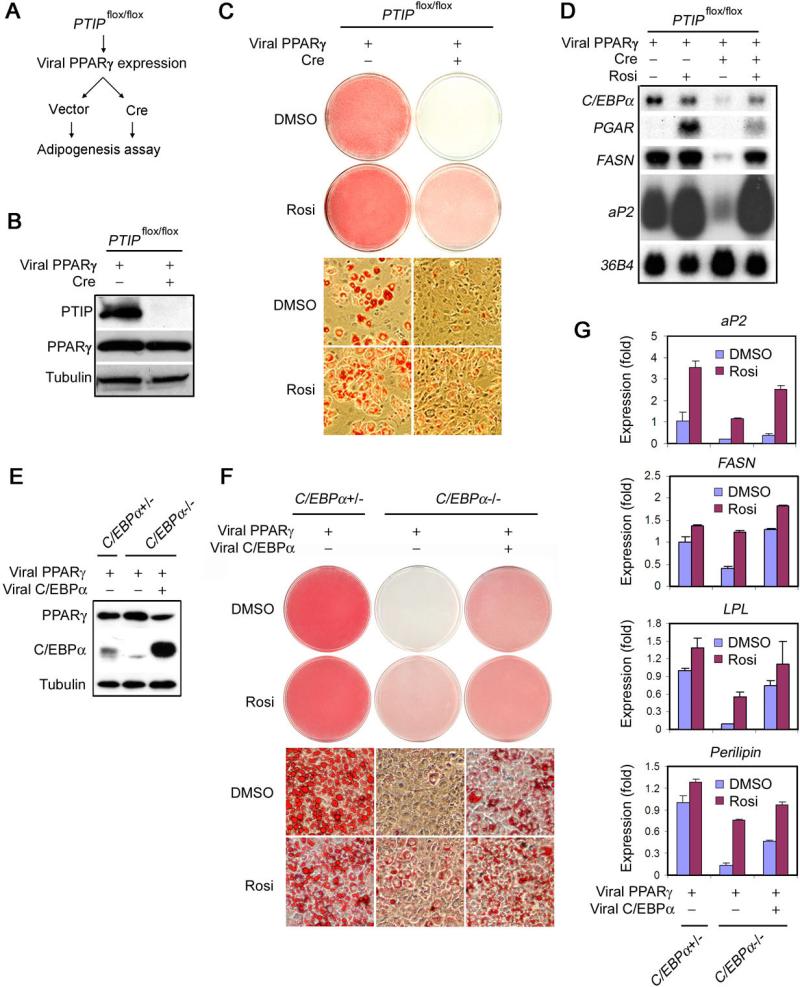
Ectopic PPARγ is sufficient to stimulate immortalized, non-adipogenic wild type MEFs to differentiate into adipocytes even in the absence of synthetic PPARγ ligand (Ge et al., 2002; Rosen et al., 2002). To investigate whether the severe adipogenesis defects in PTIP-deficient cells was solely due to the defective expression of endogenous PPARγ, we tested whether ectopic PPARγ could stimulate adipogenesis in PTIP-deficient MEFs.
The immortalized, non-adipogenic _PTIP_flox/flox MEFs were sequentially infected with retroviruses expressing PPARγ and Cre (Figure 3A-B). Adipogenesis was induced in the presence of synthetic PPARγ ligand rosiglitazone (Rosi) or vehicle DMSO alone. Consistent with the previous report (Ge et al., 2002), ectopic PPARγ induced about 60−70% of cells to undergo adipogenesis in the presence of DMSO. Rosi treatment further stimulated differentiation (Figure 3C left panels). Surprisingly, ectopic PPARγ failed to induce adipogenesis in PTIP-deficient MEFs treated with DMSO. In the presence of Rosi, ectopic PPARγ induced about 60−70% of PTIP-deficient MEFs to differentiate into immature adipocytes that mainly contained small lipid droplets (Figure 3C right panels). However, the expression of several adipocyte markers including C/EBPα was impaired (Figure 3D). Thus, PTIP-deficient MEFs show a severe defect in PPARγ-stimulated adipogenesis in the absence of Rosi, and a partial defect in the presence of Rosi.
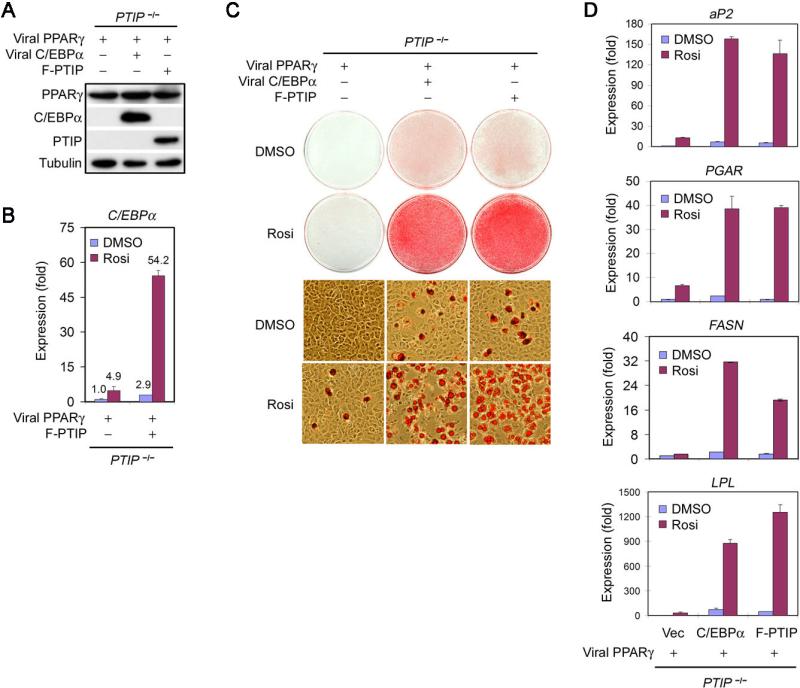
Figure 3. Both PTIP-Deficient and C/EBPα-Deficient MEFs Show Defects in PPARγ-Stimulated Adipogenesis.
(A) – (D) PTIP-deficient MEFs show significant defects in PPARγ-stimulated adipogenesis. Immortalized _PTIP_flox/flox MEFs expressing ectopic PPARγ were infected with MSCVpuro expressing Cre or vector alone, followed by adipogenesis assay in the presence or absence of synthetic PPARγ ligand Rosi.
(A) Schematic of the PPARγ-stimulated adipogenesis assay.
(B) Western blot of endogenous PTIP and ectopic PPARγ expression before adipogenesis.
(C) Morphological differentiation (Oil Red O staining). Top panels, stained dishes. Lower panels, representative fields under the microscope.
(D) Northern blot analysis of gene expression after adipogenesis.
(E) – (G) C/EBPα-deficient MEFs show significant defects in PPARγ-stimulated adipogenesis. Immortalized C/EBPα+/− and _C/EBPα_−/− MEFs infected with MSCVpuro-PPARγ2 were then infected with MSCVhygro expressing C/EBPα, followed by adipogenesis assay for 8 days in the presence or absence of Rosi.
(E) Western blot of PPARγ and C/EBPα expression before adipogenesis.
(F) Morphological differentiation.
(G) qRT-PCR analysis of gene expression after adipogenesis. PGAR is also known as Angiopoietin-like 4 (Angptl4). FASN stands for Fatty acid synthase. LPL stands for Lipoprotein lipase. All results are representative of two – four independent experiments.
MEFs have the capacity to differentiate into a variety of cell types besides adipocytes, as exemplified by MyoD-stimulated myogenesis (Ge et al., 2002). We found that PTIP was dispensable for MyoD-stimulated myogenesis in MEFs (Figure S1). Thus, PTIP is selectively required for PPARγ-stimulated adipogenesis, relative to MyoD-stimulated myogenesis, and indicate the differential involvement of PTIP in different fibroblast differentiation pathways.
C/EBPα is required for adipogenesis in vivo (Farmer, 2006). Ectopic PPARγ can induce adipogenesis in _C/EBPα_−/− MEFs in the presence of synthetic PPARγ ligand. However, _C/EBPα_−/− cells show relatively immature phenotype and consistently accumulate less lipid in smaller droplets than the C/EBPα+/+ cells (Wu et al., 1999), suggesting that _C/EBPα_−/− cells have a partial defect in PPARγ-stimulated adipogenesis in the presence of synthetic PPARγ ligand. Expression of ectopic PPARγ from MSCVpuro vector induces adipogenesis of MEFs even in the absence of synthetic PPARγ ligand (Ge et al., 2002; Rosen et al., 2002). Using MSCVpuro to express PPARγ in C/EBPα+/− and _C/EBPα_−/− MEFs, we found that _C/EBPα_−/− cells showed a severe defect in PPARγ-stimulated adipogenesis in the absence of synthetic PPARγ ligand Rosi, and a partial defect in the presence of Rosi. Such defects could be rescued by ectopic C/EBPα (Figure 3E-G). These results were remarkably similar to what we observed in PTIP-deficient MEFs, and suggest that the defective PPARγ-stimulated adipogenesis in PTIP-deficient MEFs may be due to the defective C/EBPα expression.
PTIP Directly Regulates C/EBPα Expression in MEFs
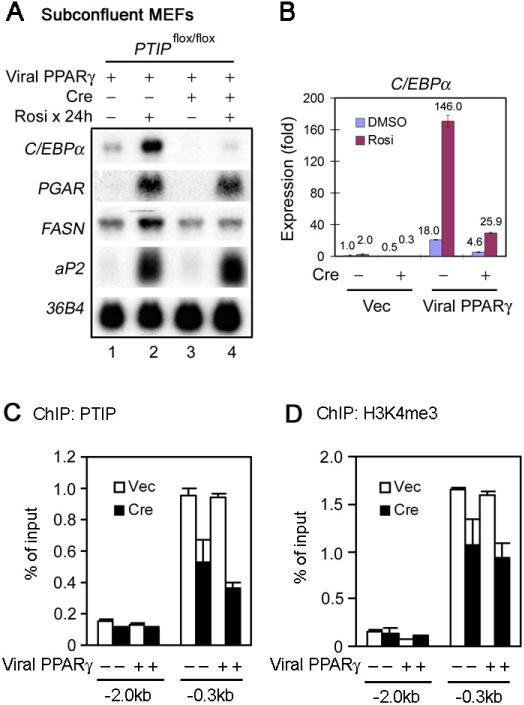
Next we investigated how PTIP regulates C/EBPα gene expression. The expression of PPARγ-regulated genes including C/EBPα during adipogenesis results from combined action of multiple adipogenic transcription factors including PPARγ (Farmer, 2006). In addition, there exists a possibility that the defective C/EBPα expression in PTIP-deficient, immature adipocytes may be secondary to the adipogenesis defect. Therefore, it was unclear from Figure 3D whether the defective PPARγ-stimulated adipogenesis in PTIP-deficient MEFs was directly due to the failure in C/EBPα expression. We have shown previously that in subconfluent, undifferentiated MEFs, ectopic PPARγ activates expression of endogenous target genes including C/EBPα and aP2 in a receptor- and ligand-dependent manner (Ge et al., 2008). Using this approach, we found that PTIP-deficient MEFs show a severe defect in PPARγ- and ligand-stimulated expression of endogenous C/EBPα but not aP2 (Figure 4A-B). By ChIP assay, we found that endogenous PTIP associated with the C/EBPα promoter with a signal peaked around the transcription start site (Figure 4C). H3K4me3 signal was highly enriched on the C/EBPα proximal promoter but was reduced upon PTIP deletion (Figure 4D). These results indicate that PTIP directly regulates C/EBPα expression in MEFs.
Figure 4. PTIP Directly Regulates C/EBPα Expression in MEFs.
(A) – (B) PTIP-deficient MEFs show over 5 fold decrease of PPARγ- and ligand-stimulated expression of endogenous C/EBPα. MEFs described in Figure 3A-D were treated with 0.5μM Rosi or DMSO for 24 h, followed by Northern blot (A) and qRT-PCR analysis (B). When RNA samples were collected, cell density was less than 80−90%.
(C) ChIP assay of PTIP recruitment to the C/EBPα promoter in _PTIP_flox/flox MEFs.
(D) ChIP assay of H3K4me3 enrichment on the C/EBPα promoter in _PTIP_flox/flox MEFs. All results are representative of three independent experiments.
Rescue of the Adipogenesis Defect in _PTIP_−/− MEFs Requires Co-expression of PPARγ and C/EBPα
Ectopic C/EBPα is unable to stimulate adipogenesis in PPARγ-deficient MEFs (Rosen et al., 2002). Consistent with the requirement of PTIP for PPARγ expression in MEFs, we found that PTIP was required for C/EBPα-stimulated adipogenesis and the associated expression of adipogenesis markers including endogenous C/EBPα and PPARγ in MEFs (Figure S2). Thus, PTIP-deficient MEFs showed significant defects in both PPARγ- and C/EBPα-stimulated adipogenesis.
Next, we tested whether co-expression of ectopic PPARγ and C/EBPα could rescue the adipogenesis defects in PTIP-deficient MEFs. For this purpose, _PTIP_−/− cell lines were established by infecting immortalized _PTIP_flox/flox MEFs with Ad-Cre to permanently delete PTIP gene. Cells were serial diluted and single colonies were isolated. After confirming complete deletion of PTIP gene, _PTIP_−/− MEFs were infected with retroviruses expressing PPARγ, followed by infection with retroviruses expressing C/EBPα or PTIP (Figure 5A). Ectopic PTIP could rescue the PPARγ- and ligand-stimulated expression of endogenous C/EBPα in subconfluent, undifferentiated _PTIP_−/− MEFs (Figure 5B). As expected, _PTIP_−/− MEFs were refractory to PPARγ-induced adipogenesis. However, such defects could be rescued by ectopic expression of either C/EBPα or PTIP (Figure 5C-D). These results indicate that rescue of the adipogenesis defect in _PTIP_−/− MEFs requires co-expression of PPARγ and C/EBPα, and suggests that the adipogenesis defect in PTIP-deficient cells is mainly due to the defective expression of both PPARγ and C/EBPα.
Figure 5. Rescue of the Adipogenesis Defect in _PTIP_−/− MEFs Requires Co-expression of PPARγ and C/EBPα.
_PTIP_−/− MEFs were established by infecting immortalized _PTIP_flox/flox MEFs with Ad-Cre, followed by isolation of single colonies. _PTIP_−/− MEFs were infected with MSCVpuro expressing PPARγ, followed by infection with MSCVhygro expressing C/EBPα or F-PTIP.
(A) Western blot analysis of ectopic PPARγ, C/EBPα and PTIP expression before induction of adipogenesis. Cells were cultured in the absence of Rosi.
(B) Ectopic PTIP rescues PPARγ- and ligand-stimulated expression of endogenous C/EBPα in subconfluent, undifferentiated _PTIP_−/− MEFs. Cells were treated with 0.5 μM Rosi or DMSO for 24 h. C/EBPα expression was analyzed by qRT-PCR.
(C) Cells were induced to undergo adipogenesis for 8 days and were stained with Oil Red O.
(D) qRT-PCR analysis of gene expression after adipogenesis.
All results are representative of two – four independent experiments.
PTIP Is Required for Induction of PPARγ and C/EBPα during Adipogenesis
We next investigated how PTIP regulates PPARγ and C/EBPα expression during adipogenesis. Interestingly, in contrast to the requirement of PTIP for PPARγ expression in MEFs, PTIP deletion in undifferentiated preadipocytes had little effect on the basal level of PPARγ, which was ∼ 5.4 fold lower in undifferentiated preadipocytes than in MEFs (Figure 6A and data not shown). Expression of PPARγ and C/EBPα is markedly induced during 3T3-L1 white preadipocyte differentiation (Morrison and Farmer, 1999) (Figure S3A). Similarly, during brown preadipocyte differentiation, expression of PPARγ1 and C/EBPα was markedly induced as early as day 1, well before the appearance of immature adipocytes on day 3, and peaked at day 4. PPARγ2 was induced relatively late (day 3) and remained a minor isoform throughout differentiation. PTIP deletion by Cre in brown preadipocytes almost completely prevented the induction of PPARγ1, PPARγ2 and C/EBPα, but had little effect on the induction of C/EBPβ, which is another adipogenic transcription factor that works upstream of both PPARγ and C/EBPα (Farmer, 2006) (Figure 6B-C).
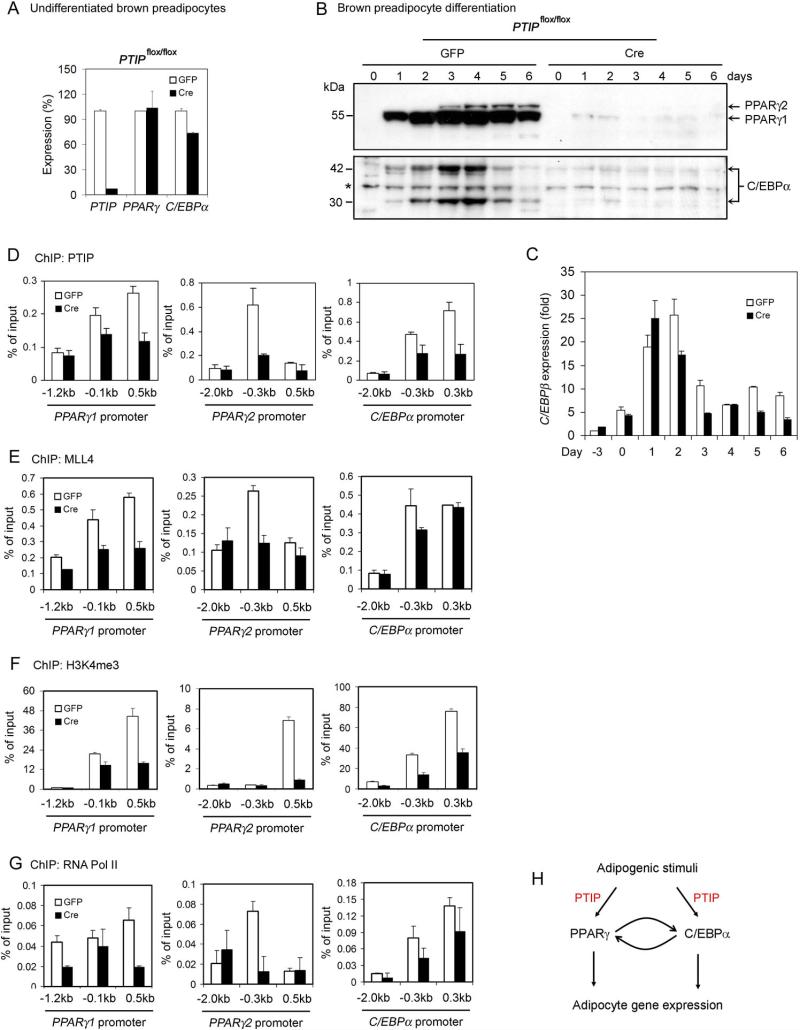
Figure 6. PTIP Is Required for Induction of PPARγ and C/EBPα during Adipogenesis.
(A) – (B) PTIP is essential for induction of PPARγ and C/EBPα expression during preadipocyte differentiation. Adipogenesis of primary _PTIP_flox/flox brown preadipocytes was performed as in Figure 2J-L. The basal level of PPARγ and C/EBPα expression in undifferentiated preadipocytes was determined by qRT-PCR (A). PPARγ and C/EBPα protein expression during preadipocyte differentiation was analyzed by Western blot (B). Whole cell extracts were collected every 24 h following induction of differentiation with IBMX, DEX and indomethacin at day 0. The 54.5 kDa PPARγ1 and 57.5 kDa PPARγ2 bands, as well as the 30 kDa and 42 kDa C/EBPα isoforms are indicated by arrows. The asterisk indicates a non-specific band.
(C) qRT-PCR analysis of C/EBPβ expression during differentiation of _PTIP_flox/flox brown preadipocytes. RNA samples were collected every 24 h starting from three days before induction of adipogenesis (d-3).
(D) – (G) ChIP assays of the recruitment of PTIP, MLL4 and Pol II, and H3K4me3 signal on PPARγ1, PPARγ2 and C/EBPα promoters in differentiating (day 4) _PTIP_flox/flox brown preadipocytes.
(H) Model depicting the role of PTIP in transcriptional regulation of preadipocyte differentiation (adipogenesis). PTIP regulates adipogenesis through direct modulation of PPARγ and C/EBPα expression. All results are representative of two – four independent experiments.
We performed ChIP assays to investigate how PTIP regulates PPARγ and C/EBPα genes during preadipocyte differentiation. In 3T3-L1 cells before differentiation (day −3), PTIP and H3K4me3 were enriched around the transcription start sites on the PPARγ1, PPARγ2 and C/EBPα proximal promoters. In differentiating 3T3-L1 cells (day 4) that expressed the peak levels of PPARγ1, PPARγ2 and C/EBPα, we observed increased enrichment of PTIP and H3K4me3 on the three promoters (Figure S3B-C). PTIP expression did not change significantly during preadipocyte differentiation (Figure S3A), suggesting that the increased binding of PTIP to the three promoters is likely due to gene-specific recruitment of PTIP. Similarly, in differentiating brown preadipocytes (day 4) that expressed the peak levels of PPARγ1, PPARγ2 and C/EBPα, we observed enrichment of PTIP, MLL4 and H3K4me3 around the transcription start sites on the three promoters. PTIP deletion not only reduced enrichment of PTIP and MLL4 on these promoters, but also decreased H3K4me3 signal and the recruitment of RNA Pol II to the same promoters (Figure 6D-G). Taken together, these results indicate that PTIP is required for the robust induction of PPARγ and C/EBPα during preadipocyte differentiation, and suggest the involvement of the PTIP-associated histone H3K4 methyltransferase in regulation of PPARγ and C/EBPα expression during adipogenesis.
Deletion of PTIP in Brown Adipose Tissue (BAT) Reduces Tissue Weight and Leads to Cold Intolerance in Mice
To investigate PTIP function in adipocytes in vivo, adipose-specific PTIP knockout mice (KO) were generated by crossing PTIP conditional knockout mice (Patel et al., 2007) with aP2-Cre mice expressing Cre under the control of adipose-specific aP2 promoter. The aP2-Cre mice used were from line 1, which expressed Cre predominantly in BAT (Abel et al., 2001). As PTIP is required for PPARγ expression and aP2 is a downstream target of PPARγ, this approach is expected to delete PTIP after formation of fat depots, allowing largely normal adipogenesis in mice (He et al., 2003).
Although KO mice had similar body weight as the control mice, they displayed over 50% decrease of BAT weight (Figure 7A-B and Figure S4). Expression of markers common to WAT and BAT (PPARγ, C/EBPα, aP2), BAT-selective genes (PRDM16, cidea, eva1, ntrk3) (Seale et al., 2007), brown fat thermogenic genes (UCP1 and PGC-1α) and mitochondrial components (cox5b and cox8b) also decreased significantly in the residual BAT of KO mice (Figure 7D). In contrast, no significant change of WAT weight was found in the KO mice, consistent with the ∼ 9.3 fold higher level of Cre expression in BAT than in WAT of KO mice, and with the lack of PTIP gene deletion in the WAT of KO mice (Figure 7A-C and Figures S4 and S5). These results are consistent with previous reports that deletion of PPARγ in BAT leads to marked decrease of tissue weight in mice (He et al., 2003).
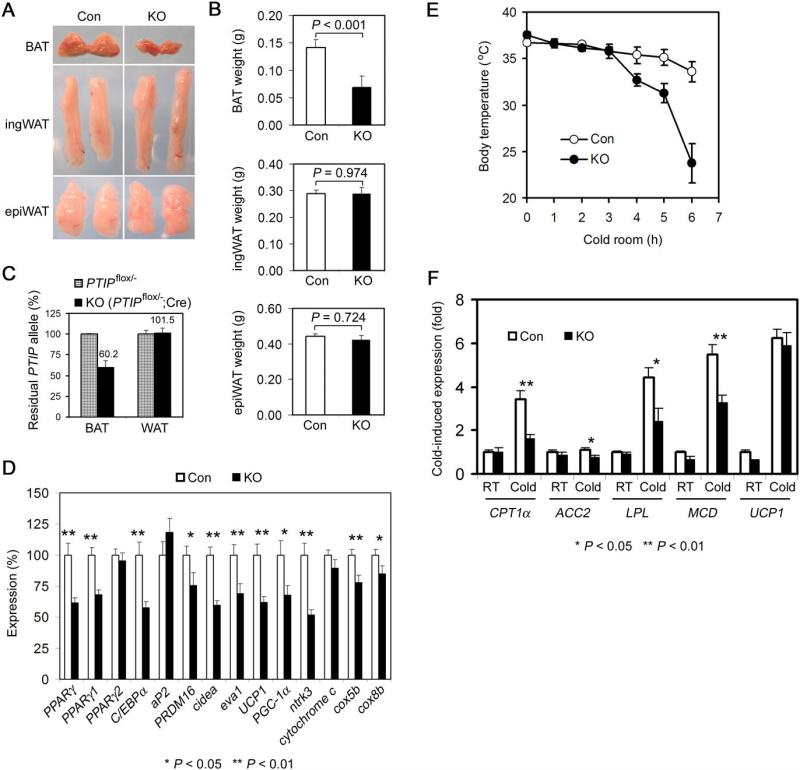
Figure 7. Deletion of PTIP in Brown Adipose Tissue Reduces Tissue Weight and Leads to Cold Intolerance in Mice.
(A) – (D) PTIP deletion in brown adipose tissue (BAT) decreases both tissue weight and expression of brown adipocyte-enriched genes. Adipose-specific PTIP knockout mice (KO) carried the genotype of _PTIP_flox/−;Cre. Other littermates were used as control (Con) (see Experimental Procedures). All data were from 12−16 week-old mice.
(A) Representative pictures of interscapular BAT, inguinal WAT (ingWAT) and epididymal WAT (epiWAT) isolated from Con and KO mice.
(B) Quantitation of BAT and WAT weights in Con (n = 22) and KO (n = 8) mice. No significant differences in body weight, BMI and adipose tissue weights were found among groups of control mice (Figure S4).
(C) Lack of aP2-Cre-mediated deletion of PTIP gene in WAT of KO mice as determined by quantitative PCR (n = 8).
(D) qRT-PCR analysis of gene expression in BAT of Con (n = 12) and KO (n = 8) mice.
(E) – (F) PTIP deletion in BAT leads to cold intolerance in mice.
(E) The body temperatures of mice housed in 4°C were measured every hour for 6h (n = 7−9).
(F) qRT-PCR analysis of cold-induced gene expression in BAT of mice after cold treatment for 6h (n = 6).
All values are the mean +/− SEM. *P < 0.05; **P < 0.01. Similar results were obtained from male and female mice but only the male mice data are shown here.
The main function of BAT is to burn fat for heat generation (thermogenesis). The decrease of BAT size in KO mice did not lead to significant change of serum perimeters such as blood glucose, free fatty acid, triglyceride and insulin levels, likely because BAT only accounts for a small percentage of the fat mass in mice (data not shown). However, when exposed to environmental cold, the KO mice were unable to maintain body temperatures and were cold intolerant (Figure 7E), consistent with previous report that ablation of BAT led to cold intolerance in mice (Lowell et al., 1993). In the residual BAT of KO mice, cold-induced expression of genes that are important for fatty acid catabolism, such as CPT1a, LPL and MCD (Yu et al., 2002), was impaired (Figure 7F), suggesting that the cold intolerance of the KO mice is due to, at least in part, the combined effects of the reduced BAT size and the impaired expression of cold-induced genes important for fatty acid catabolism.
Discussion
Elucidating the mechanisms that regulate PPARγ and C/EBPα expression is crucial to understanding the regulation of adipogenesis. In this paper, we report the histone methylation regulator PTIP is required for PPARγ and C/EBPα expression and adipogenesis. We show (i) that PTIP deletion leads to severe defects in PPARγ expression and PPARγ-stimulated C/EBPα expression in MEFs (Figures 1 and 4); (ii) that similar to C/EBPα-deficient MEFs, PTIP-deficient MEFs show significant defects in PPARγ-stimulated adipogenesis (Figure 3); (iii) that rescue of the adipogenesis defect in _PTIP_−/− MEFs requires co-expression of PPARγ and C/EBPα (Figure 5); (iv) that PTIP is required for induction of PPARγ and C/EBPα during preadipocyte differentiation. PTIP deletion impairs the enrichment of H3K4 methyltransferase MLL4, H3K4me3, and Pol II to the PPARγ and C/EBPα promoters (Figures 6 and S3); (v) that PTIP is required for adipogenesis of primary MEFs and white and brown preadipocytes (Figure 2); and (vi) that deletion of PTIP in BAT significantly reduces tissue weight and leads to cold intolerance in mice (Figures 7, S4 and S5). Thus, by regulating PPARγ and C/EBPα expression, PTIP plays a critical role in adipogenesis.
Regulation of PPARγ Expression by PTIP
Using the mouse whole genome expression arrays, we find that PTIP deletion in MEFs leads to over 2.5 fold decrease of expression of ∼ 60 genes, which account for less than 0.5% of total genes expressed in MEFs. Consistent with the specific association of PTIP with the MLL3- and MLL4-containing HMT complex, PTIP deletion has no effect on expression of Hoxa7, Hoxa9, Foxc2 and p27/Kip1, which are known targets of the homologous MLL1- and MLL2-containing HMT complexes (Figure 1B) (Milne et al., 2005). PTIP is required for PPARγ1 expression in MEFs where PPARγ2 expression is undetectable. These results strongly suggest a gene-specific regulatory role of PTIP in MEFs. In undifferentiated preadipocytes, PTIP is dispensable for the basal level expression of PPARγ and C/EBPα. However, PTIP is essential for the induction of PPARγ and C/EBPα but not C/EBPβ during preadipocyte differentiation (Figure 6), suggesting that PTIP plays a specific role in regulating PPARγ and C/EBPα expression during adipogenesis.
Several transcription factors, such as SREBP1c/ADD1 and E2F family members, have been shown to regulate PPARγ1 expression (Farmer, 2006). Multiple subunits of the PTIP-associated HMT complex are capable of interacting directly with sequence-specific transcription factors. For example, PTIP interacts with PAX2 (Patel et al., 2007). Ncoa6 interacts directly with multiple transcription factors and nuclear receptors including PPARγ in vitro or in cells (Lee et al., 2006; Mahajan and Samuels, 2005). It is possible that PTIP and the associated HMT complex may be recruited to the PPARγ1 promoter through their direct interactions with promoter-bound transcription factors.
C/EBPα directly binds to and activates PPARγ2 promoter and is required for the induction of PPARγ2 during adipogenesis (Farmer, 2006). In silico analysis predicts that PTIP may use its BRCT repeats to interact with phosphorylated C/EBPs (Miller, 2006). It remains to be determined whether phosphorylated C/EBPα and C/EBPβ recruit PTIP and the associated HMT complex to the PPARγ1 and PPARγ2 promoters during adipogenesis.
Regulation of C/EBPα Expression by PTIP
Activation of C/EBPα expression by PPARγ is considered a critical step in the transcriptional cascade that leads to adipogenesis (Farmer, 2006; Rosen and MacDougald, 2006). However, it is unclear how PPARγ activates C/EBPα expression. It has been suggested that PPARγ may directly activate C/EBPα gene expression (Hamm et al., 2001). The recent genome-wide analysis of PPARγ-binding sites in 3T3-L1 cells has identified several putative PPARγ target regions downstream of the C/EBPα gene but the functional PPARγ response elements remain to be identified (Nielsen et al., 2008).
Although PTIP deletion in MEFs only leads to 1.9 fold decrease of basal level of C/EBPα expression, the PPARγ- and ligand-stimulated 146 fold increase of C/EBPα expression is severely impaired (Figure 4A-B). Interestingly, the recruitment of PTIP to the C/EBPα promoter is independent of PPARγ (Figure 4C), suggesting that PTIP may not act through PPARγ to regulate C/EBPα expression. PPARγ may work with another transcription factor that works upstream of C/EBPα and requires PTIP to activate C/EBPα expression.
The induction of C/EBPα and PPARγ1 expression appears to be synchronized in the early phase of differentiation of both white and brown preadipocytes (Figure 6B and (Morrison and Farmer, 1999)). As PTIP directly regulates expression of both PPARγ1 and C/EBPα, our data suggest a possibility that PTIP may work with yet-to-be-identified upstream transcription factor(s) to coordinate and synchronize the induction of C/EBPα and PPARγ1 during preadipocyte differentiation. Future work will be needed to identify the upstream transcription factor(s) that may work with PTIP to regulate PPARγ1 and C/EBPα expression.
Regulation of Adipogenesis by PTIP
Preadipocyte differentiation (adipogenesis) is under the control of cooperative actions between PPARγ and C/EBPα, both of which are required for adipogenesis in vivo. Studies using preadipocyte differentiation in cell culture and gene knockout in mice have suggested a transcriptional cascade that controls adipogenesis. In the early phase of preadipocyte differentiation, adipogenic stimuli induce low level expression of PPARγ and C/EBPα, which then stimulate each other's expression in a positive feed back loop. The resulting high levels of PPARγ and C/EBPα activate adipocyte gene expression and lipid accumulation and ultimately lead to phenotypic conversion from fibroblasts to adipocytes (Rosen et al., 2002) (also see Figure 6H). PTIP is required for the induction of both PPARγ and C/EBPα during preadipocyte differentiation (Figures 6). In addition, PTIP-deficient MEFs show over 5 fold decrease of PPARγ-stimulated C/EBPα expression (Figure 4), which would impair the positive feedback loop between PPARγ and C/EBPα during adipogenesis. Thus, PTIP appears to regulate multiple steps in the transcriptional cascade that controls adipogenesis. Consistently, PTIP is required for adipogenesis in all in vitro differentiation systems that we have tested, including adipogenesis of primary MEFs, PPARγ- and C/EBPα-stimulated adipogenesis of immortalized MEFs, differentiation of 3T3-L1 preadipocytes as well as primary white and brown preadipocytes (Figures 2, 3, 5 and S2). Taken together, our data support a model that PTIP regulates adipogenesis through direct modulation of PPARγ and C/EBPα expression (Figure 6H).
Several transcriptional coactivators, such as Ncoa6 and TRAP220/Med1, have been shown to be required for PPARγ-stimulated adipogenesis in MEFs (Ge et al., 2002; Qi et al., 2003). Ncoa6 and PTIP are both subunits of the MLL3/MLL4 histone H3K4 methyltransferase complex. The adipogenesis defects seen in both PTIP- and Ncoa6-deficient MEFs suggest that the MLL3/MLL4 complex likely plays an important role in regulation of adipogenesis. TRAP220 is a subunit of the general transcription coactivator complex Mediator. _TRAP220_−/− MEFs show a severe defect in PPARγ-stimulated adipogenesis even in the presence of synthetic PPARγ ligand. However, unlike PTIP, TRAP220 is dispensable for PPARγ-stimulated C/EBPα expression in subconfluent MEFs (Ge et al., 2008), indicating that the adipogenesis defect in TRAP220-deficient MEFs likely involves a mechanism distinct from that in the PTIP-deficient cells.
Epigenetic Regulation of PPARγ Expression and Adipogenesis
Considerable evidence suggests that epigenetic mechanisms including histone methylation are critical for lineage specification of pluripotent stem cells. Recent mappings of histone methylation in pluripotent and lineage-committed cells have revealed an unexpected high frequency of colocalization of the “activating” H3K4me3 and the “repressive” H3K27me3 on promoters of developmental regulators. These bivalent modifications were proposed to poise key developmental genes for lineage-specific activation or repression (Bernstein et al., 2006). Interestingly, H3K4me3 and H3K27me3 also colocalize on the PPARγ1 promoter in MEFs (Mikkelsen et al., 2007). We previously show that endogenous PTIP associates with both histone H3K4 methyltransferases MLL3 and MLL4 and histone H3K27 demethylase UTX (Cho et al., 2007; Hong et al., 2007). The PTIP-associated HMT complex directly regulates PPARγ1 gene in MEFs. Deletion of PTIP in MEFs leads to significant decrease of H3K4me3 and increase of H3K27me3 on PPARγ1 proximal promoter (Figure 1D-F). Since both methylation of H3K4 and demethylation of H3K27 presumably associate with gene activation, it will be particularly interesting to investigate the roles of MLL3, MLL4 and UTX, and the potential synergy between the associated H3K4 methyltransferase and H3K27 demethylase activities, in regulation of PPARγ expression during adipogenesis.
Experimental Procedures
Plasmids and Antibodies
The following retroviral plasmids have been described: MSCVpuro-PPARγ2, MSCVhygro-PPARγ2, MSCVhygro-C/EBPα, MSCVpuro-MyoD (Ge et al., 2008; Ge et al., 2002). MSCVpuro-F-PTIP expresses FLAG-tagged PTIP (Cho et al., 2007). Cre cDNA was cloned into MSCVpuro and MSCVhygro (Clontech) to generated MSCVpuro-Cre and MSCVhygro-Cre. The PTIP knockdown construct targeting mouse PTIP sequence TGACAGAGATGACCTGAAG was constructed in retroviral vector pSUPER.retro.puro (OligoEngine). All plasmids were confirmed by DNA sequencing.
The following antibodies were generated by immunizing rabbits with His-tagged recombinant proteins and were affinity purified with antigens: anti-PTIP#3 was generated against 274−472aa of mouse PTIP; anti-PA1#2 was generated against full length human PA1; anti-MLL4#3 was generated against 3020−3192aa of human MLL4. Anti-MHC antibody MF20 was described (Ge et al., 2002). Mouse IgG (sc-2025), anti-C/EBPα (sc-61), anti-MyoD (sc-760), anti-myogenin (sc-52903), anti-PPARγ (sc-7273), and anti-α-tubulin (sc-5286) were from Santa Cruz. Rabbit IgG (I-5006) was from Sigma. Anti-RbBP5 (A300−109A) was from Bethyl. Anti-H3K4me3 (ab8580) and anti-RNA Pol II (ab5408) were from Abcam. Anti-H3K27me3 (07−449) was from Millipore.
Isolation of Primary Cells, Cell Culture, and Virus Infection
3T3-L1 preadipocyte cell line was cultured in DMEM + 10% bovine serum (Invitrogen). Primary brown preadipocytes were cultured in DMEM containing 25 mM glucose, 20 mM HEPES (pH7.5), and 20% fetal bovine serum (FBS). All other cells were routinely cultured in DMEM plus 10% FBS (Hyclone). All chemicals were from Sigma unless indicated.
Primary MEFs isolated from E13.5 _PTIP_flox/flox embryos (Patel et al., 2007) were immortalized by following the 3T3 protocol (Ge et al., 2002). To isolate primary white preadipocytes, inguinal WAT from 4−8 week _PTIP_flox/flox mice was excised, minced, and digested with 1 mg/ml collagenase I in the isolation buffer (123 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 5 mM glucose, 100 mM HEPES (pH7.5), 4% BSA and 100 units/ml penicillin and streptomycin) at 37°C for 45 min. After digestion, the slurry was passed through a 100 μm mesh cell strainer (Falcon) and cells were collected by centrifugation. Primary brown preadipocytes were isolated from interscapular BAT of 1−2 days old _PTIP_flox/flox pups following a published protocol (Klein et al., 1999).
Retrovirus infection of MEFs was done as described (Ge et al., 2002). Adenoviruses expressing Cre recombinase and GFP (Ad5CMV-Cre-GFP) or GFP alone (Ad5CMV-GFP) were obtained from Baylor Vector Development Laboratory. Adenovirus infection of primary MEFs and white and brown preadipocytes that had limited life span in culture were done at 100 moi as described (Ge et al., 1999).
Adipogenesis and Myogenesis Assays
PPARγ-stimulated adipogenesis and MyoD-stimulated myogenesis in immortalized MEFs were performed as described (Ge et al., 2002). Adipogenesis was induced in the presence of 0.5 μM rosiglitazone (Rosi) (Cayman Chemical) or vehicle DMSO alone for 8 days. After adipogenesis, cells were either stained for lipid droplets with Oil Red O or total RNA was extracted for gene expression analysis by Northern blot or qRT-PCR.
Adipogenesis of 3T3-L1 was induced as described (Nielsen et al., 2008). For adipogenesis of primary MEFs, two days after cells reached confluence, cells were treated with medium supplemented with 0.5 mM 3-isobutyl-1-methyl-xanthine (IBMX), 1 μM dexamethasone (DEX), 10 μg/ml insulin and 0.5 μM Rosi. Two days later, cells were changed to medium containing insulin and Rosi. The medium was replenished at 2-day intervals for 12 days.
For adipogenesis of primary white preadipocytes, cells were plated at a density of 1 × 106 per 10cm dish four days before induction of adipogenesis. At day 0, cells were treated with medium supplemented with 0.5 mM IBMX, 1 μM DEX, 5 μg/ml insulin, 0.2 nM T3 and 0.5 μM Rosi. Two days later, cells were changed to medium containing insulin, T3 and Rosi. The medium was replenished at 2-day intervals. At day 4, small lipid droplets appeared in differentiating cells. Adipogenesis of primary brown preadipocytes was carried out as described (Klein et al., 1999). At day 6 − 8 post induction, cells were either stained with Oil Red O or subjected to gene expression analysis by qRT-PCR.
Western Blot, Northern Blot, Microarray and qRT-PCR
Western blot and Northern blot were done as described (Ge et al., 2008). For microarray analysis, total RNA was sequentially purified with Trizol (Invitrogen) and RNeasy kit (Qiagen) and analyzed on Mouse Genome 430 2.0 Array (Affymetrix) at local microarray facility following standard protocols. Microarray data have been deposited in NCBI GEO data base (accession number GSE16263). For qRT-PCR, purified total RNA was reverse transcribed using random hexamers and SuperScript First-Strand Synthesis System (Invitrogen). The resulting first-strand cDNAs were quantified with Taqman or Sybr-Green assays on PRISM 7900HT System using standard curve and relative quantitation method with 18S rRNA as control (Applied Biosystems). The sequences of Sybr-Green primers are available upon request. _PPARγ1_- and _PPARγ2_-specific Taqman probes and primers were custom-synthesized: PPARγ1 forward primer 5'-AAAGAAGCGGTGAACCACTGATA-3', Taqman probe 5'-FAM-ACCCTTTACTGAAATTACC-3', and reverse primer 5'-AATGGCATCTCTGTGTCAACCA-3'; PPARγ2 forward primer 5'-CGCTGATGCACTGCCTATGA-3', Taqman probe 5’-FAM-CACTTCACAAGAAATTAC-3’; PPARγ2 reverse primer is the same as that of PPARγ1. Data are presented as means +/− SD.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (Nelson et al., 2006) with minor modifications. The cross-linked chromatin was sheared to ∼ 400 bp fragments with a Bioruptor UCD-200TM (Diagenode). Each immunoprecipitation was performed with 1∼2 μg antibody in the presence of secondary antibody conjugated to Dynabeads (Invitrogen). PCR quantitation of precipitated genomic DNA relative to inputs was performed in triplicate using Taqman probes. The sequences of Taqman probes and primers are available upon request.
Mouse Breeding, Genotyping and Cold Tolerance Test
Mice carrying two floxed PTIP alleles (_PTIP_flox/flox) or one floxed and one null alleles (_PTIP_flox/−) (Patel et al., 2007) were crossed with aP2-Cre transgenic mice line 1 (Abel et al., 2001). The progeny were intercrossed to generate _PTIP_flox/− mice carrying aP2-Cre transgene (_PTIP_flox/−;Cre), which were used as adipose-specific PTIP knockout mice (KO). Other littermates, such as wild type (_PTIP_flox/flox and _PTIP_flox/+), heterozygous (_PTIP_flox/− and PTIP+/−), and _PTIP_flox/+;Cre mice, were used as control (Con). No statistically significant differences in the body and tissue weights were found among the 3 groups of control mice (Figure S4). Genotyping of PTIP conditional knockout mice was done as described (Patel et al., 2007). Mice carrying aP2-Cre transgene were detected by PCR using primers 5'-CCTGTTTTGCACGTTCACCG-3' and 5'-ATGCTTCTGTCCGTTTGCCG-3' and the PCR product was 250 bp. The efficiency of aP2-Cre-mediated deletion of PTIP gene in adipose tissues of KO mice was measured by quantitative PCR of genomic DNA using a Taqman probe specific for the floxed exon I of PTIP gene: 5'-FAM-CTGTCAGGTGGCCGCACG-3', and a Taqman probe specific for GAPDH gene as internal control. After normalizing with GAPDH allele, the residual PTIP allele in KO mice (_PTIP_flox/−;Cre) was compared with that in the _PTIP_flox/− mice.
Mice were fed with standard rodent chow ad libitum. Twelve to sixteen week-old, age- and gender-matched mice with a mixed 129Sv and C57BL/6J background were used for this study. For cold tolerance test, mice that were housed singly and unrestrained and had free access to food and water were moved to the cold room with an ambient temperature of 4°C. Body temperature were measured every hour using mouse rectal probe (Thermalert TH-5). When the body temperature drops below 24°C, mice were moved out of the cold room. At the end of experiments, mice were euthanized and tissues were collected. Data are presented as means +/− SEM. Differences were analyzed with Student's t test. Similar results were obtained from male and female mice but only male mice data are shown. All mouse work is approved by the Animal Care and Use Committee of NIDDK, NIH.
Supplementary Material
01
Acknowledgement
We thank G. Dressler for PTIP conditional knockout mice, B. Kahn for aP2-Cre mice line one, G. Darlington for C/EBPα+/− and _C/EBPα_−/− cell lines, D. Lane for 3T3-L1 cell line, H. Yu, G. Poy, W. Jou and T. Chanturiya for technical assistance, and A. McPherron for critical reading of the manuscript. This work was supported by the Intramural Research Program of the NIDDK, NIH to K.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel ED, Peroni O, Kim JK, Kim Y-B, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Cho Y-W, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP Associates with MLL3- and MLL4-containing Histone H3 Lysine 4 Methyltransferase Complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Cho Y-W, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative Mechanisms by Which Mediator Subunit MED1/TRAP220 Regulates Peroxisome Proliferator-Activated Receptor {gamma}-Stimulated Adipogenesis and Target Gene Expression. Mol Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast GC. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci U S A. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Hamm JK, Park BH, Farmer SR. A Role for C/EBPbeta in Regulating Peroxisome Proliferator-activated Receptor gamma Activity during Adipogenesis in 3T3-L1 Preadipocytes. J Biol Chem. 2001;276:18464–18471. doi: 10.1074/jbc.M100797200. [DOI] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proceedings of the National Academy of Sciences. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Fasshauer M, Ito M, Lowell BB, Benito M, Kahn CR. beta 3-Adrenergic Stimulation Differentially Inhibits Insulin Signaling and Decreases Insulin-induced Glucose Uptake in Brown Adipocytes. J Biol Chem. 1999;274:34795–34802. doi: 10.1074/jbc.274.49.34795. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee D-K, Dou Y, Lee J, Lee B, Kwak E, Kong Y-Y, Lee S-K, Roeder RG, Lee JW. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. PNAS. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The Role of Chromatin during Transcription. Cell. 2007;128:707. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lowell BB, S-Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocr Rev. 2005;26:583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. Phospho-dependent protein recognition motifs contained in C/EBP family of transcription factors: in silico studies. Cell Cycle. 2006;5:2501–2508. doi: 10.4161/cc.5.21.3421. [DOI] [PubMed] [Google Scholar]
- Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison RF, Farmer SR. Role of PPARgamma in Regulating a Cascade Expression of Cyclin-dependent Kinase Inhibitors, p18(INK4c) and p21(Waf1/Cip1), during Adipogenesis. J Biol Chem. 1999;274:17088–17097. doi: 10.1074/jbc.274.24.17088. [DOI] [PubMed] [Google Scholar]
- Mueller E, Drori S, Aiyer A, Yie J, Sarraf P, Chen H, Hauser S, Rosen ED, Ge K, Roeder RG, Spiegelman BM. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor gamma isoforms. J Biol Chem. 2002;277:41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protocols. 2006;1:179. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs K-J, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPAR{gamma}:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Developmental Cell. 2007;13:580. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Surapureddi S, Zhu Y-J, Yu S, Kashireddy P, Rao MS, Reddy JK. Transcriptional Coactivator PRIP, the Peroxisome Proliferator-activated Receptor {gamma} (PPAR{gamma})-interacting Protein, Is Required for PPAR{gamma}-mediated Adipogenesis. J Biol Chem. 2003;278:25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metabolism. 2007;6:38. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Wysocka J. H3K27 Demethylases, at Long Last. Cell. 2007;131:29. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- Yu XX, Lewin DA, Forrest W, Adams SH. Cold elicits the simultaneous induction of fatty acid synthesis and {beta}-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J. 2002;16:155–168. doi: 10.1096/fj.01-0568com. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Korenberg JR, Chen X, Noya D, Rao MS, Reddy JK. Structural Organization of Mouse Peroxisome Proliferator-Activated Receptor {gamma} (mPPAR{gamma}) Gene: Alternative Promoter Use and Different Splicing Yield Two mPPAR{gamma} Isoforms. Proceedings of the National Academy of Sciences. 1995;92:7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01