Unexpected expression of α- and β-globin in mesencephalic dopaminergic neurons and glial cells (original) (raw)
Abstract
The mesencephalic dopaminergic (mDA) cell system is composed of two major groups of projecting cells in the substantia nigra (SN) (A9 neurons) and the ventral tegmental area (VTA) (A10 cells). A9 neurons form the nigrostriatal pathway and are involved in regulating voluntary movements and postural reflexes. Their selective degeneration leads to Parkinson's disease. Here, we report that gene expression analysis of A9 dopaminergic neurons (DA) identifies transcripts for α- and β-chains of hemoglobin (Hb). Globin immunoreactivity decorates the majority of A9 DA, a subpopulation of cortical and hippocampal astrocytes and mature oligodendrocytes. This pattern of expression was confirmed in different mouse strains and in rat and human. We show that Hb is expressed in the SN of human postmortem brain. By microarray analysis of dopaminergic cell lines overexpressing α- and β-globin chains, changes in genes involved in O2 homeostasis and oxidative phopshorylation were observed, linking Hb expression to mitochondrial function. Our data suggest that the most famed oxygen-carrying globin is not exclusively restricted to the blood, but it may play a role in the normal physiology of the brain and neurodegenerative diseases.
Keywords: astrocytes, hemoglobin, oligodendrocytes, oxidative phosphorylation, Parkinson
Dopaminergic neurons (DA) are an anatomically and functionally heterogeneous group of cells involved in a wide range of neuronal network activities and behavior. Among them, mesencephalic DA (mDA) are the major source of dopamine in the brain. They present two main groups of projecting cells: the A9 neurons of the substantia nigra (SN) and the A10 cells of the ventral tegmental area (VTA).
A9 neurons form the nigrostriatal pathway and are involved in regulating voluntary movements and postural reflexes. Their selective degeneration leads to Parkinson's disease (PD), and the loss of DA synapses in the striatum is believed to be primary cause for the disruption of the ability to control movements (1). A10 cells constitute the mesocorticolimbic pathway, playing a fundamental role in reward and attention. Their abnormal function has been linked to schizophrenia, attention deficit, and addiction, whereas they are relatively spared in PD (2).
The description of the repertory of genes of mDA neurons may provide crucial information on their physiology and on the mechanisms of cell-type specific dysfunction. Interestingly, in previous gene expression profiling experiments, mDA cell groups presented a limited number of differentially expressed genes with A9-enriched transcripts mainly related to energy metabolism and mitochondrial function (3–5).
A crucial requirement for metabolically active aerobic cells is a steady supply of oxygen. To this purpose, hemoglobin (Hb)-like molecules occur widely in organisms ranging from bacteria to human (6). Vertebrate Hb is the oxygen- and carbon dioxide-carrying protein in cells of erythroid lineage and is responsible for oxygen delivery to the respiring tissues of the body. Additional vertebrate heme-containing proteins with structural homology to globin chains include cytoglobin, mostly described in connective tissues (7), and neuroglobin, broadly expressed in the brain (8).
Surprisingly, Hb chains have been recently detected in nonerythroid cells including macrophages, alveolar cells, eye's lens, and mesangial cells of the kidney (9–12).
By a combination of different gene expression platforms with laser capture microdissection (LCM), we have identified the transcripts of Hb α, adult chain 1 (Hba-a1), and Hb β, adult chain 1 (Hbb-b1) in A9 neurons. Interestingly, Hb immunoreactivity (Hb-IR) decorated the large majority of A9 cells, whereas it stained only <5% of A10 neurons. Furthermore, we detected Hb expression in almost all oligodendrocytes and cortical and hippocampal astrocytes and proved that this pattern of expression was conserved in mammals. Importantly, A9 DA neurons from human postmortem brain showed Hb expression.
By gene expression analysis of mouse dopaminergic cell lines stably transfected with α- and β-chains, we observed changes in genes involved in O2 homeostasis and oxidative phosphorylation, suggesting a link between Hb and mitochondrial activity.
These results open a scenario for a role for Hb in brain physiology and PD pathogenesis.
Results
Identification of α- and β-Globin Transcripts by Expression Analysis of A9 DA Neurons.
To study the cellular physiology of A9 DA neurons, we determined their gene expression profiles with two independent techniques: cDNA microarrays and a nanoscale version of the cap analysis of gene expression (nanoCAGE). To this purpose we took advantage of transgenic mice that selectively express green fluorescent protein (GFP) in catecholaminergic cells under the control of tyrosine hydroxylase (TH) gene promoter (TH-GFP mice) (13). In this mouse line we can identify the majority of mDA neurons for their GFP labeling. Furthermore, we can distinguish A9 neurons from A10 for their anatomical localization. Thus, LCM and pressure catapulting were used to harvest A9 neurons after fixation with a zinc fix-based method that assured the preservation of both tissue morphology and RNA integrity. RNA was then used as template in two different gene expression approaches.
In the cDNA microarray experiment, RNA was processed with a μMACS amplification kit (Miltenyi), labeled, and used as a target to monitor gene expression on a custom-made cDNA microarray platform. All of the experiments were performed in three biological replicates. The complete description of the transcripts expressed in A9 neurons will be presented elsewhere. Interestingly, among the genes expressed in A9 cells, transcripts for the α- and β-chains of mouse Hb were identified.
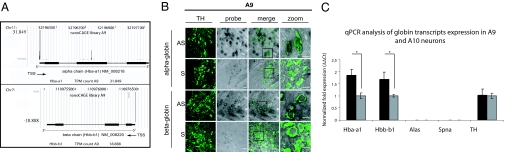
For the nanoCAGE transcriptome analysis, 2000 A9 cells were harvested, full-length cDNAs were produced and, after cleavage with a class IIS restriction endonuclease, 5′ end tags were purified and sequenced by using the second generation of sequencers. Finally, transcription start sites (TSS) were identified by mapping tags to the genome (15). This kind of analysis unveiled TSS on the mouse chromosomes 11 and 7 in the genomic regions corresponding to the 5′ end of Hba-a1 (NM_008218.2) and Hbb-b1 (NM_008220.3) transcripts (Fig. 1A). In Table S1 a list of nanoCAGE tags in A9 neurons is provided for these two loci.
Fig. 1.
Expression of α- and β-globin transcripts in A9 and A10 neurons. (A) NanoCAGE tracks visualization concerning Hbb-a1 (Chr.11) and Hbb-b1 (Chr.7) gene loci. A Genome Browser view is presented. A9 library is indicated, and the tags per million (TPM) for each gene are reported. The structure of Hbb-a1 and Hbb-b1 transcripts is depicted at the bottom. Transcriptional start sites (TSS) are indicated at the 5′ untranscribed region of each transcript. (B) In situ hybridization of α- and β-globin transcripts on A9 DA neurons: ventral midbrain slices were processed with antisense (AS) and sense (S) probes for the two globins transcripts. DA neurons were visualized by immunohistochemistry using an anti-TH antibody (green). The overlay (merge) shows colocalization of the transcripts of α- and β-globin in the cytoplasm of A9 neurons. Probes synthesis from sense transcripts were used as negative controls. The zoom offers magnifications of the area in the boxes of the overlay images. (Scale bars: 20 μm.) (C) qPCR starting from 500 LCM-isolated neurons from A9 (black column) and A10 (grey column) regions of the ventral midbrain of TH-GFP mice. TH, α-globin, and β-globin transcripts were amplified and the absence of blood contamination was evaluated by using primers for Alas and Spna. α- and β-transcripts are more expressed in A9 neurons (≈2-fold), four biological replicas, P < 0.05.
Therefore, cDNA microarray data and CAGE tags distribution suggested that A9 neurons express transcripts for α- and β-chains of Hb.
To estimate potential blood contamination, we monitored the expression of several erythrocyte-specific transcripts: no expression was detected for Rhag, Gypa, Alas2, Spna1, and Epb4.2.
Validation of the Expression of α- and β-Globin Transcripts in A9 DA Cells.
We then validated the expression of α- and β-globin transcripts in A9 DA neurons by two independent approaches.
We studied the precise distribution of α- and β-globin transcripts in the mouse midbrain by in situ hybridization. Experimental procedures and antisense RNA probes were first tested on bone marrow as a positive control for globin expression (Fig. S1). Brains were fixed by using conventional methods after extensive perfusion with PBS to minimize blood contamination. As shown in Fig. 1B, antisense probes gave specific and reproducible signals in A9 neurons that were identified by anti-TH immunoreactivity and anatomical localization. Interestingly, a specific labeling for the antisense probe was also evident in A10 neurons (Fig. S2). Higher magnification of the merged images showed colocalization of the antisense signals for each Hb chains in the cytosol of TH-positive A9 and A10 DA cells.
We then took advantage of LCM to collect 500 TH-GFP-positive neurons for each mDA cell group. After RNA extraction and cDNA synthesis, quantitative PCR (qPCR) amplification was carried out to detect the expression of α- and β-globin. As visualized in Fig. 1C, the qPCR showed that α- and β-globin transcripts were 2-fold more expressed in A9 than in A10 neurons.
Hb-IR Is Present in DA Neurons of the SN.
To characterize Hb expression at protein level, we took advantage of a commercial antibody produced against highly purified total mouse Hb (Cappel). For the high homology among proteins of the globin family and the well-described expression of atypical globins in neurons (7, 8), we first verified the repertory of immunoreactivity of the Cappel antibody in immunofluorescence experiments (Fig. S3_a_). We transfected HEK cells with expression vectors carrying myc-tagged globin isoforms (Hba-a1, Hbb-b1, Hba-x, Hbb-y, Hbb-bh1, and Hbq1) and atypical globins (Cygb, Ngb, and Mg). Cappel antibody reactivity was then monitored in parallel with anti-myc staining. A strong signal was obtained for β-globin chain. A weaker labeling was also detected when the Hbb-y embryonic chain was expressed. Importantly, no cross-reaction was present with any of the atypical globins normally expressed in neurons.
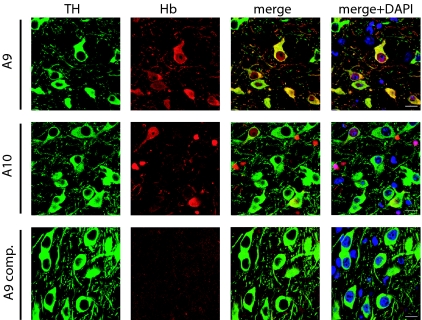
Therefore, an immunohistochemical analysis of the mouse mesencephalon was carried out. To avoid strong reactivity from blood cells, brains were extensively washed in PBS during perfusion before fixation. Immunohistochemistry on mouse midbrain revealed a complex pattern of protein expression (Fig. 2). In the SN, 65.8 ± 6.1% of A9 DA neurons were presenting Hb-IR. On the contrary, a very limited number of DA neurons in the A10 region were stained (2.9 ± 0.82%). Interestingly, Hb-IR in mDA neurons was localized in both the cytoplasm and the nucleus. Importantly, the specificity of the antibody was further verified by competition assays using mouse spleen extracts on midbrain sections (Fig. 2). Experiments performed on brains without perfusion showed a strong labeling of red blood cells that were clearly different in size, location, and number from DA neurons (Fig. S4_a_). To further confirm Hb-IR, we then used a second commercial antibody against purified mouse Hb (ICL Labs). First, we proved that its chain specificity pattern is limited to the detection of both β- and α-chains and that no cross-reaction with other typical or atypical globins was observed (Fig. S3_b_). Then we confirmed an extensive overlap of Hb-IR for DA neurons between Cappel and ICL antibodies (Fig. S4_b_).
Fig. 2.
Hb protein is expressed in A9 and A10 DA neurons of mouse brain. A double immunohistochemistry analysis using anti-Hb (red), anti-TH (green), and DAPI (blue) is presented: nearly 70% of A9 but only 3% of A10 neurons were double labeled for Hb and TH (merge). Hb staining is present in the nucleus, except for the nucleolus, and in the cytoplasm. Adsorption of the anti-Hb antibody with spleen extract completely prevents Hb staining (A9 comp). (Scale bar: 20 μm.)
Because nanoCAGE data shows that both α- and β-chains synthesis involve the same TSS used in blood, we investigated whether the major transcription factors implicated in controlling primitive and definitive erythroid lineages may be involved in mDA cells transcription. Notably, the expression of Gata family members has been also described in the midbrain and hindbrain (16, 17). Interestingly, as shown in Fig. S4_c_, several DA neurons (≈50%) were decorated for both Gata-1 and Hb staining.
Additional Hb-IR Cells Are Present in Distinct Areas Throughout the Brain.
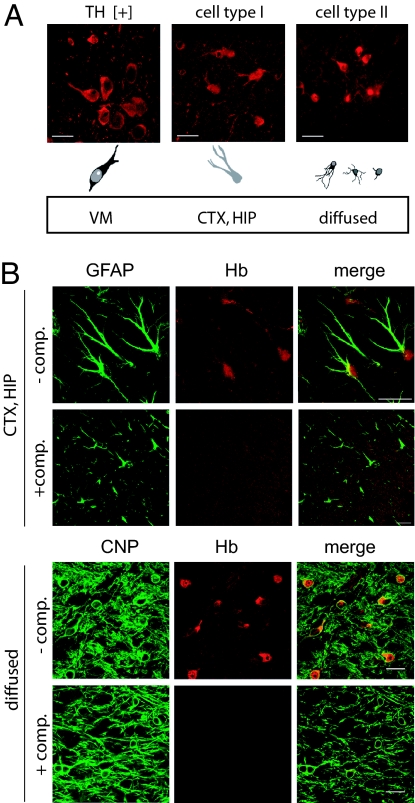
Together with mDA neurons (Fig. 3A Left), two additional TH-negative cell types were labeled by the Cappel antibody in brain sections.
Fig. 3.
Hb protein is expressed in different regions of the mouse brain. (A) (Upper) Immunohistochemistry using anti-Hb antibody on mouse brain revealed different Hb-IR cells: large neurons located in the ventral midbrain, positive for TH (TH+); cell type I large cells located in the cortex (CTX) and hippocampus (HIP); and cell type II small cells, widely diffused in all of the brain regions tested and presenting a strong Hb-IR. (Scale bars: 20 μm.) (Lower) A schematic representation of the morphologies of Hb-IR cells is presented. (B) Double immunohistochemistry using anti-Hb antibody (red) together with astrocytes and oligodendrocytes markers (GFAP and CNP, green). (Upper) In the cortex (CTX) and in the hippocampus (HIP), Hb–IR cell type I colocalizes with GFAP staining. (Lower) Hb-IR cell type II colocalizes with the oligodendrocytes marker CNP. Adsorption of the anti-Hb antibody with spleen extract completely prevents Hb staining (+ comp). (Scale bars: 20 μm.)
Cell type I are large cells located in the cortex and the hippocampus, faintly labeled in the cytoplasm and the nucleus (approximate diameter: 15–18 μm) (Fig. 3A Center).
Cell type II are small cells widely diffused in all of the brain regions analyzed, strongly labeled in the thin cytoplasm and the nucleus (approximate diameter: 7–10 μm) (Fig. 3A Right).
To identify these cells, we carried out extensive double immunohistochemistry on mouse midbrain slices with the Cappel antibody and antibodies specific for different neuronal and glial cell populations (NG2, Iba-1, NeuN, GFAP, and CNP). As shown in Fig. 3B, we found a specific and reproducible Hb-IR in a subpopulation (73.2 ± 4.8%) of hippocampal and cortical astrocytes labeled with anti-GFAP antibody and in a large fraction (>99%) of mature oligodendrocites, characterized by the expression of CNP.
Primary Cultures of Hb-IR Neuronal and Glial Cell Populations.
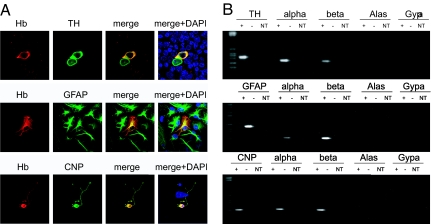
We studied globin expression at the mRNA and protein levels in vitro on primary cultures obtained from dissociated mouse ventral midbrain, cortex, or hippocampus. Immunofluorescence experiments confirmed the expression pattern observed in vivo: Hb-IR was found in a subpopulation of TH-positive DA neurons, cortical GFAP-positive astrocytes, and the large majority of CNP-positive oligodendrocytes (Fig. 4A).
Fig. 4.
Primary cultures of DA neurons, astrocytes, and oligodendrocytes: immunofluorescence and RT-PCR. (A) Immunofluorescence on primary cultures of DA neurons, cortical and hippocampal astrocytes, and oligodendrocytes. (Magnification: 63×.) Specific cell population markers (green) and Hb staining (red) are shown. (B) RT-PCR results obtained from 2,000 single FACS-sorted cells. α- and β-globin transcripts and the population-specific markers (TH, GFAP, and CNP, respectively) were amplified (+). The absence of blood contamination was evaluated by using primers for Alas and Gypa. Negative controls, retrotranscriptase free (−), and no-template control samples (NT) are presented. RNA extracted from blood was used as positive control (Fig. S9).
Taking advantage of TH-GFP, GFAP-GFP (Jackson Laboratory), and CNP-GFP transgenic mice lines (18), we next validated the expression of α- and β-chains transcripts after resorting to FACS for purifying, respectively, mDA neurons, astrocytes, and oligodendrocytes. After enzymatic digestion and mechanical trituration of dissected regions, the cell suspension was sequentially panned on four Bandeiraea Simplicifolia lectin I-coated dishes (19). This step minimized endothelial, microglial, and red blood cell contamination of the preparation. Then, the FACS procedure was applied and GFP-positive cells were collected. FACS-purified cell culture showed an elevated enrichment of the cells of interest (98% for mDA, 96% for astrocytes, and 97.8% for oligodendrocytes). RNA was then extracted from 2,000 GFP-positive cells for each cell type and, after RT-PCR amplification, the specific amplicons of α- and β-globins were observed in TH-, GFAP-, and CNP-enriched cells (Fig. 4B). The identity of PCR products was confirmed by cloning and sequencing.
Hb-IR Pattern Is Conserved in Mammals.
We then addressed whether the characteristic pattern of globin expression described for C57BL/6J line is conserved in different genetic backgrounds. Immunohistochemistry of BALB/cJ, FVB/NJ, and CD-1 mouse strains was carried out by showing the same morphological and topographical organization of Hb-IR cells. Furthermore, Hb-IR in mDA neurons, cortical and hippocampal astrocytes, and oligodendrocytes was confirmed as early as postnatal day 6. Importantly, other rodents, like Rattus norvegicus, presented the same pattern of Hb-IR, as shown in Fig. S5_a_ for the adult mesencephalon.
We then analyzed the SN of human postmortem brains by two different antibodies: a subset of TH-positive neurons was Hb-immunoreactive, proving that Hb expression in the brain is conserved from mouse to human (Fig. S5_b_).
Hb Overexpression on Mouse DA Cell Line MN9D Changes the Expression of Genes Involved in O2 Homeostasis and Mitochondrial Oxidative Phosphorylation.
We took advantage of the MN9D dopaminergic cell line to address the function of Hb in DA neurons. RT-PCR demonstrated that transcripts for α- and β-chains of mouse Hb were indeed expressed. RNA from mouse blood was used as positive control. By using a specific antibody against mouse Hb, protein expression was detected by Western blot analysis although Hb level was very low. By resorting to immunoprecipitation, a clear band of 17 kDa was specifically enriched from cell lysates (Fig. S6).
As Hb is likely to act as heterotetramer of two different subunits, we took advantage of pBUDCE 4.1 vector to overexpress both mouse globin chains in a series of stably transfected MN9D cell lines (Fig. S7). The expression of α- and β-chains was verified by qPCR and immunocytochemistry, and the presence of α/β heterodimers was confirmed by coimmunoprecipitation experiments (Fig. S7).
We then took advantage of the Affymetrix platform to interrogate the GeneChip Mouse Genome 430A 2.0 Array for gene expression differences between control and globin chain stable cell lines (see Materials and Methods for details). The experiment was carried out with three biological replicas.
A total of 4,617 genes was found to be differentially expressed with a fold change >1.2. A total of 2,057 were up-regulated in α- and β-chains over-expressing clones, and 2,560 were down-regulated. qPCR confirmed microarrays data for all 14 genes tested for validation. A complete list of genes is provided in Table S2.
By applying Ingenuity software, two major pathways were affected: O2 homeostasis and oxidative phopshorylation (Fig. 5). Other changes were observed in genes involved in oxidative stress, iron metabolism, and nitric oxide (NO) synthesis (Table S3).
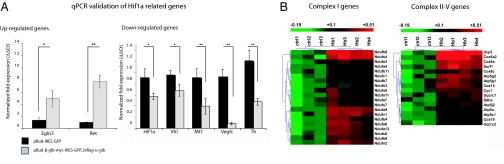
Fig. 5.
Array analysis of Hb overexpressing mouse dopaminergic cell line reveals changes in the expression of genes involved in O2 homeostasis and mitochondrial oxidative phosphorylation. (A) Genes involved in Hif1a pathway are presented. qPCR experiments of selected genes, up-regulated (Left) and down-regulated (Right), validate array data. (B) Genes involved in mitochondrial oxidative phosphorylation pathway. Heat maps of genes components of complex I (Left) and complexes II–V (Right) are presented.
O2 homeostasis mainly occurs through the activity of Hif1a, a transcription factor whose expression is decreased in globin-overexpressing cells (Fig. 5) (20). Its physiological activity is regulated by Egln3, a mediator of Hif1a hydroxylation, and Vhl that targets Hif1a for degradation. The overexpression of α- and β-chains decreased Vhl mRNA whereas it strongly increased Egln3 transcripts. Interestingly, the expression of TH and Ret, two targets of Hif1a, was also changed (Fig. 5).
Importantly, genes involved in mitochondrial oxidative phosphorylation were increased upon overexpression of Hb chains. A total of 36 of 78 genes that encode for subunits of mitochondrial complex I–V were up-regulated. This induction occurred mainly in complex I (20 genes of 46) and to a lesser extent in complex II (1 gene), III (3 genes), IV (6 genes), and V (5 genes). Interestingly, the mitochondrial, proton carrier, uncoupling protein 2, was also strongly up-regulated.
Discussion
The first descriptions of globins in the nervous tissue date back to the 19th century. More recently, globin-like molecules have been detected in neurons of various invertebrates (6). In the bivalve mollusc Tellina alternate, neural excitability is sustained as long as oxygen can be delivered by a neural globin (21). In Aplysia, a gastropod mollusc, the firing activity of the neural ganglia is proportional to the degree of oxygenation of the neural globin. Natural variation in a neural globin in Caenorhabditis elegans strains has been linked to changes in electrophysiological responses and sensory behaviors (22).
Recently, neuroglobin has been identified in mammalian brains where it is probably involved in the hypoxia response (8, 23).
Here, we show that α- and β-chains transcripts of Hb and Hb-IR are present in a subpopulation of DA neurons, cortical and hippocampal astrocytes, and all mature oligodendrocytes.
We observed the expression of Hb transcripts in DA cells by using four different approaches: cDNA microarrays, nanoCAGE, RT-PCR, and in situ hybridization. Furthermore, we took advantage of two different methods (LCM and FACS) to isolate a pure population of DA neurons.
Interestingly, when analyzing Hb-IR we found that Hb protein expression does not fully overlap with transcript distribution: the large majority of A10 DA cells and a small number of A9 neurons showed mRNA expression but not Hb-IR. There are thus at least two potential explanations for this discrepancy: the level of Hb expression in those DA cells is lower and below antibody sensitivity and/or Hb protein expression may be regulated at posttranscriptional level. Interestingly, our in situ hybridization data may suggest the expression of mRNA for Hb chains in hippocampal neurons as recently proposed (24, 25).
Globin RNAs and protein expression overlap in hippocampal and cortical astrocytes and almost all mature oligodendrocytes. Globin mRNAs have been detected as differentially expressed between acutely purified and cultured oligodendrocytes (11) and during regeneration of the sciatic nerve (26). Here, we observed globin staining in the oligodendrocytes of all of the brain regions including striatum, corpus callosum, and medulla oblongata. We also found Hb-immunoreactive cells in perinatal pups. In the adult, no NG2-positive cells were Hb-immunoreactive, thus Hb expression seems restricted to mature oligodendrocytes.
Although Hb function in the brain remains to be investigated in vivo, here we have provided some interesting cues by using MN9D cells, a mouse dopaminergic cell line that represents a well-accepted in vitro model to study dopaminergic cell physiology and dysfunction (27, 28).
By carrying out a gene expression analysis of MN9D stably transfected with α- and β-chains we found that Hb expression acts on the main elements of O2 homeostasis. This observation was not surprising because Hb may function as an oxygen storage and transport molecule. It is well known that both hyperoxia and hypoxia can be detrimental to cellular physiology in the nervous system (29). Brain Hb may then act as storage of oxygen to provide a homeostatic mechanism in anoxic conditions, which is especially important for A9 DA neurons that have an elevated metabolism with a high requirement for energy production.
Extending this model to other Hb-expressing cells in the brain, the widespread distribution of oligodendrocytes and their localization adjacent to neuronal cells may provide a net of oxygen-storage cells. In hypoxia conditions, oxygen may then be released and provide to the neighboring neurons some highly needed relief for the maintenance of the aerobic metabolism.
Interestingly, 46% of genes that encode for subunits of mitochondrial complex I–V were induced in the stable cell lines overexpressing Hb chains. It is well known that oxygen tension regulates mtDNA-encoded complex I gene expression (30) and high oxygen concentration induces mitochondrial biogenesis (31). Complex I plays a central role in PD because deficits in its subunits and activity have been consistently detected in the SN of PD patients (32). Furthermore, in PD animal models administration of the toxic metabolite MPP+ and the pesticides rotenone and paraquat cause dopaminergic degeneration in part by mitochondrial complex I inhibition. Therefore, these gene expression data may suggest Hb as a central player in the control of mitochondrial function in normal and pathological conditions.
High mitochondrial activity is usually linked to oxidative stress, which may be especially detrimental for A9 neurons because they are normally under intense oxidative stress caused by the production of hydrogen peroxide via autoxidation and/or monoamine oxidase (MAO)-mediated deamination of dopamine and the subsequent reaction of accessible ferrous iron to generate highly toxic hydroxyl radicals (33).
Hemoglobin may indeed play homeostatic roles as both an antioxidant and a regulator of iron metabolism. In rat mesangial cells Hb carries out an antioxidant function (12). According to our gene expression data, this ability may be mediated by well-known detoxifying agents from cellular free radicals (Table S3).
Mitochondrial oxidative phosphorylation, oxidative stress, and iron deposits are all important components of PD pathogenesis (34, 35). Significantly, here we proved the expression of Hb in A9 DA neurons of human postmortem brain. Interestingly, a functional polymorphism in the gene for the Hb-binding protein haptoglobin has been shown influencing susceptibility for idiopathic PD (14).
The establishment of a series of transgenic mice with cell type-specific globin gene knockout in DA neurons, astrocytes, and oligodendrocytes will provide an essential tool for unveiling Hb function in the brain. It is of note that to our knowledge no loxP mouse line for globin genes is currently available: the unexpected expression of this old protein in the brain will soon change this surprising shortfall.
Materials and Methods
Animal Procedures.
All of the experiments involving the use of animals were performed in accordance with guidelines of the international and Italian ethical committees and under the supervision of local veterinary services.
LCM of mDA Neurons from TH-GFP Mice.
Eight- to 12-week-old mice were deeply anesthetized and extensively perfused transcardially with TBS followed by 1× zinc fixative (BD) diluted in RNase free water (Ambion). Brains were removed and postfixed in 1× zinc fixative for 8 h at + 4 °C. The region containing the SN was isolated, included in freezing medium Neg-50 (Richard Allan Scientific), and frozen on dry ice for 10 min. The frozen block was brought into cryostat (Microm International) and left at −21 °C for 30 min. Coronal sections of midbrain (14 μm) were cut with a clean blade and transferred on Superfrost Plus glass slides (Menzel-Glaser Menzel). Glass slides were air-dried for 5 min; selections of single DA were marked with a LCM microscope, microdissected, collected in adhesive caps (Zeiss), and immediately processed.
Dissociation and FACS.
To isolate DA, astrocytes, and oligodendrocytes, transgenic mice including TH-GFP (13), GFAP-GFP (Jackson Laboratories), and CNP-GFP mice (18) were used, respectively. For DA and cortical or hippocampal astrocytes P4-P8 pups were used. Oligodendrocytes were collected by using P13/P20 animals. Solitary DA were prepared as described (see SI Text). A similar procedure was followed for the dissociation of cortical and hippocampal astrocytes and oligodendrocytes.
A cell strainer with 70-μm nylon mesh was used to obtain a single-cell suspension (BD Falcon) before sorting. 7-Amino-actinomycin d (7-AAD) (Beckman–Coulter) was added to the cell suspension to exclude dead cells. Subpopulation of cells expressing GFP emission was isolated with a high-speed cell sorter (MoFlo). Sorting parameters for the three different populations are visualized in Fig. S8.
See SI Text for detailed description of experimental procedures.
Supplementary Material
Supporting Information
Acknowledgments.
We thank the members of S.G.'s laboratory for thought-provoking discussions and help; Dr. Helena Krmac and Dr. Dario Motti for technical help; Prof. Marco Pierotti and Drs. Manuela Gariboldi, and Loris de Cecco for cDNA slide preparation; Michael J. Zigmond and Dr. Juliann Jaumotte (University of Pittsburgh, Pittsburgh, PA) for providing MN9D cells; Prof. Antonello Mallamaci, Prof. Mauro Giacca, Dr. Lorena Zentilin, and Drs. Remo Sanges, Elia Stupka, and Andrea Lunardi for technical help and discussion; Drs. Marco Stebel and Cristina de Grassi for mouse colony handling and breeding; Mr. Tullio Bigiarini and Ms. Elisa Puppato for technical help; and Dr. Yoshinori Imai (National Institute of Neuroscience, Tokyo) for anti-Iba-1. This work was supported by a career developmental award from The Giovanni Armenise-Harvard Foundation and a Research Grant from the Michael J. Fox Foundation (to S.G.) and a RIKEN presidential grant (to P.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE16192).
References
- 1.Hirsch E, Graybiel AM, Agid YA. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson's disease. Nature. 1988;334:345–348. doi: 10.1038/334345a0. [DOI] [PubMed] [Google Scholar]
- 2.Meyer-Lindenberg A, et al. Reduced prefrontal activity predicts exaggerated striatal dopaminergic function in schizophrenia. Nat Neurosci. 2002;5:267–271. doi: 10.1038/nn804. [DOI] [PubMed] [Google Scholar]
- 3.Grimm J, Mueller A, Hefti F, Rosenthal A. Molecular basis for catecholaminergic neuron diversity. Proc Natl Acad Sci USA. 2004;101:13891–13896. doi: 10.1073/pnas.0405340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung CY, et al. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14:1709–1725. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene JG, Dingledine R, Greenamyre JT. Gene expression profiling of rat midbrain dopamine neurons: Implications for selective vulnerability in parkinsonism. Neurobiol Dis. 2005;18:19–31. doi: 10.1016/j.nbd.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Vandergon TL, Riggs CK, Gorr TA, Colacino JM, Riggs AF. The mini-hemoglobins in neural and body wall tissue of the nemertean worm, Cerebratulus lacteus. J Biol Chem. 1998;273:16998–17011. doi: 10.1074/jbc.273.27.16998. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt M, et al. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J Biol Chem. 2004;279:8063–8069. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- 8.Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Zeng M, Stamler JS. Hemoglobin induction in mouse macrophages. Proc Natl Acad Sci USA. 1999;96:6643–6647. doi: 10.1073/pnas.96.12.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton DA, Rao KM, Dluhy RA, Baatz JE. Hemoglobin is expressed by alveolar epithelial cells. J Biol Chem. 2006;281:5668–5676. doi: 10.1074/jbc.M509314200. [DOI] [PubMed] [Google Scholar]
- 11.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishi H, et al. Hemoglobin is expressed by mesangial cells and reduces oxidant stress. J Am Soc Nephrol. 2008;19:1500–1508. doi: 10.1681/ASN.2007101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawamoto K, et al. Visualization, direct isolation, and transplantation of midbrain dopaminergic neurons. Proc Natl Acad Sci USA. 2001;98:6423–6428. doi: 10.1073/pnas.111152398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa-Mallen P, et al. The functional polymorphism of the hemoglobin-binding protein haptoglobin influences susceptibility to idiopathic Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147:216–222. doi: 10.1002/ajmg.b.30593. [DOI] [PubMed] [Google Scholar]
- 15.Valen E, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2008;19(2):255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 17.Ogilvy S, et al. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol Cell Biol. 2007;27:7206–7219. doi: 10.1128/MCB.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan X, et al. Expression of the green fluorescent protein in the oligodendrocyte lineage: A transgenic mouse for developmental and physiological studies. J Neurosci Res. 2002;70:529–545. doi: 10.1002/jnr.10368. [DOI] [PubMed] [Google Scholar]
- 19.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp FR, Bernaudin M. HIF1 and oxygen sensing in the brain. Nat Rev Neurosci. 2004;5:437–448. doi: 10.1038/nrn1408. [DOI] [PubMed] [Google Scholar]
- 21.Kraus DW, Colacino JM. Extended oxygen delivery from the nerve hemoglobin of Tellina alternata (Bivalvia) Science. 1986;232:90–92. doi: 10.1126/science.232.4746.90. [DOI] [PubMed] [Google Scholar]
- 22.McGrath PT, et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Jin K, Mao XO, Zhu Y, Greenberg DA. Neuroglobin is up-regulated by and protects neurons from hypoxic-ischemic injury. Proc Natl Acad Sci USA. 2001;98:15306–15311. doi: 10.1073/pnas.251466698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schelshorn DW, et al. Expression of hemoglobin in rodent neurons. J Cereb Blood Flow Metab. 2009;29:585–595. doi: 10.1038/jcbfm.2008.152. [DOI] [PubMed] [Google Scholar]
- 25.He Y, et al. Effects of cerebral ischemia on neuronal hemoglobin. J Cereb Blood Flow Metab. 2009;29:596–605. doi: 10.1038/jcbfm.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setton-Avruj CP, et al. Presence of α-globin mRNA and migration of bone marrow cells after sciatic nerve injury suggests their participation in the degeneration/regeneration process. Exp Neurol. 2007;203:568–578. doi: 10.1016/j.expneurol.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Choi HK, et al. Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 1991;552:67–76. doi: 10.1016/0006-8993(91)90661-e. [DOI] [PubMed] [Google Scholar]
- 28.Choi WS, et al. Two distinct mechanisms are involved in 6-hydroxydopamine- and MPP+-induced dopaminergic neuronal cell death: role of caspases, ROS, and JNK. J Neurosci Res. 1999;57:86–94. doi: 10.1002/(SICI)1097-4547(19990701)57:1<86::AID-JNR9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Diringer MN. Hyperoxia: Good or bad for the injured brain? Curr Opin Crit Care. 2008;14:167–171. doi: 10.1097/MCC.0b013e3282f57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piruat JI, Lopez-Barneo J. Oxygen tension regulates mitochondrial DNA-encoded complex I gene expression. J Biol Chem. 2005;280:42676–42684. doi: 10.1074/jbc.M507044200. [DOI] [PubMed] [Google Scholar]
- 31.Gutsaeva DR, Suliman HB, Carraway MS, Demchenko IT, Piantadosi CA. Oxygen-induced mitochondrial biogenesis in the rat hippocampus. Neuroscience. 2006;137:493–504. doi: 10.1016/j.neuroscience.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 32.Mann VM, et al. Brain, skeletal muscle and platelet homogenate mitochondrial function in Parkinson's disease. Brain. 1992;115:333–342. doi: 10.1093/brain/115.2.333. [DOI] [PubMed] [Google Scholar]
- 33.Youdim MB, Lavie L. Selective MAO-A and B inhibitors, radical scavengers and nitric oxide synthase inhibitors in Parkinson's disease. Life Sci. 1994;55:2077–2082. doi: 10.1016/0024-3205(94)00388-2. [DOI] [PubMed] [Google Scholar]
- 34.Berg D, Hochstrasser H. Iron metabolism in Parkinsonian syndromes. Movement Disorders. 2006;21:1299–1310. doi: 10.1002/mds.21020. [DOI] [PubMed] [Google Scholar]
- 35.Youdim MB, Ben-Shachar D, Yehuda S. Putative biological mechanisms of the effect of iron deficiency on brain biochemistry and behavior. Am J Clin Nutr. 1989;50(Suppl):607–615. doi: 10.1093/ajcn/50.3.607. discussion 615–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information