Conservation of enhancer location in divergent insects (original) (raw)
Abstract
Dorsoventral (DV) patterning of the Drosophila embryo is controlled by a concentration gradient of Dorsal, a sequence-specific transcription factor related to mammalian NF-κB. The Dorsal gradient generates at least 3 distinct thresholds of gene activity and tissue specification by the differential regulation of target enhancers containing distinctive combinations of binding sites for Dorsal, Twist, Snail, and other DV determinants. To understand the evolution of DV patterning mechanisms, we identified and characterized Dorsal target enhancers from the mosquito Anopheles gambiae and the flour beetle Tribolium castaneum. Putative orthologous enhancers are located in similar positions relative to the target genes they control, even though they lack sequence conservation and sometimes produce divergent patterns of gene expression. The most dramatic example of this conservation is seen for the “shadow” enhancer regulating brinker: It is conserved within the intron of the neighboring Atg5 locus of both flies and mosquitoes. These results suggest that, like exons, an enhancer position might be subject to constraint. Thus, novel patterns of gene expression might arise from the modification of conserved enhancers rather than the invention of new ones. We propose that this enhancer constancy might be a general property of regulatory evolution, and should facilitate enhancer discovery in nonmodel organisms.
Keywords: Anopheles, dorsoventral patterning, evolution, Tribolium, cis-regulation
The maternal Dorsal nuclear gradient controls the dorsoventral (DV) patterning of the early Drosophila melanogaster embryo through the direct transcriptional regulation of ≈60–70 target genes in a concentration-dependent manner (1). Proteolytic cleavage of the Toll ligand Spatzle on the ventral side of the oocyte leads to the activation of the ubiquitous Toll receptor and the selective degradation of the Cactus inhibitor, resulting in the translocation of the Dorsal protein from the cytoplasm into the nucleus (2, 3). Thus, Toll signaling converts an extracellular gradient of processed Spatzle into a nuclear concentration gradient of the transcription factor Dorsal (4). The Dorsal gradient generates at least 3 primary thresholds of gene activity in the cellularizing embryo, leading to the specification of the mesoderm, neurogenic ectoderm, and dorsal ectoderm (5).
The subdivision of the DV axis into separate embryonic tissues depends on the differential regulation of target enhancers by distinct concentrations of the Dorsal protein. Mesodermal genes such as twist and snail have enhancers that contain low-affinity Dorsal binding sites, restricting the activation of these genes to ventral regions where there are peak levels of nuclear Dorsal (5). Neurogenic genes such as vnd, brinker, and sog possess enhancers with high-affinity Dorsal binding sites, and are consequently activated by lower concentrations of the gradient (6). These enhancers also contain Snail repressor binding sites, which inhibit the expression of neurogenic genes in the mesoderm where there are high levels of Dorsal (7, 8).
Cross-species comparisons of genes involved in insect DV patterning have made it increasingly clear that the broad Dorsal nuclear gradient is a Dipteran-specific innovation (9, 10). For example, the Dorsal gradient is narrowly restricted to ventral tissues and is transiently expressed during early embryogenesis in the flour beetle Tribolium castaneum (10). This spatial and temporal restriction in the Dorsal gradient is thought to reflect an entirely zygotic mode of Toll signaling, along with robust Dorsal feedback loops that limit Toll and Cactus expression to ventral tissues (11–13).
In most insects (but not the Diptera), Dorsal is limited to the mesoderm and perhaps the ventral-most regions of the presumptive neurogenic ectoderm (9, 10). In Tribolium, the patterning of the neurogenic ectoderm and dorsal ectoderm appears to be indirectly regulated by Dorsal, as Dorsal is involved in the formation of a broad Dpp signaling gradient emanating from the dorsal side of the embryo (14). In Drosophila, the Dpp inhibitor Sog is expressed in broad lateral stripes in response to low levels of Dorsal, where it functions to restrict Dpp signaling to the dorsal midline (8, 15). However, sog expression is limited to the ventral mesoderm of Anopheles and Tribolium embryos, and a critical transcriptional inhibitor of Dpp signaling, brinker, is altogether absent in the Tribolium embryo (14, 16). These changes in the deployment of Sog and Brinker are probably responsible for the expanded domains of Dpp signaling seen in Anopheles and Tribolium and the subsequent expansion of the amnion and serosa lineages (14, 17).
EGF signals pattern the ventral neurogenic ectoderm in Drosophila (18, 19). These signals arise from 2 separate sources. First, low levels of the Dorsal gradient directly activate the EGF signaling genes rhomboid and vein in ventral regions of the neurogenic ectoderm (6, 7). Second, the ventral midline is a source of another EGF signaling molecule, Spitz, which is released into lateral regions of the developing ventral nerve cord (19). Dorsal indirectly regulates Spitz through the activation of the midline determinant sim (6, 20). It is possible that Spitz might be sufficient to pattern the CNS in those insects where Dorsal is restricted to the mesoderm (21). Thus, the transcriptional outputs of the broad Dorsal gradient of Drosophila may be superimposed on an ancestral DV patterning system that depends on restricted expression of Dorsal and Dpp/EGF signaling relays mediated by Sog and Sim, respectively.
To explore the evolution of DV patterning mechanisms, we identified and characterized 5 different Dorsal target enhancers in Anopheles and Tribolium. Despite widespread regulatory changes, the positions of the enhancers are constant among these different insect groups. For example, the cactus enhancers of Drosophila and Tribolium are located within 3′ introns, whereas the brinker enhancers of Drosophila and Anopheles are located within the intron of a neighboring gene, Atg5. This invariance of enhancer positions reflects either orthology or functional constraint. We propose that the evolution of novel expression patterns depends on changes in conserved enhancers rather than the invention of new ones.
Results
Dorsal Target Enhancers in Anopheles.
Although not explicitly visualized, it is assumed that the Dorsal gradient in Anopheles functions in a manner similar to that seen in Drosophila. To explore this possibility, we examined the regulation of 2 Dorsal target genes in Anopheles, twist, and brinker, which are activated by high and low levels of the Dorsal gradient, respectively.
Computational analysis of an extended genomic interval encompassing the Anopheles twist locus identified a single cluster of putative Dorsal binding sites, located immediately upstream of the transcription start site (Fig. 1A). A genomic DNA fragment containing this binding-site cluster was attached to a lacZ reporter gene, and lacZ expression was examined in transgenic Drosophila embryos. Robust staining was detected in the ventral mesoderm, similar to the endogenous twist expression patterns in the early embryos of both Drosophila and Anopheles.
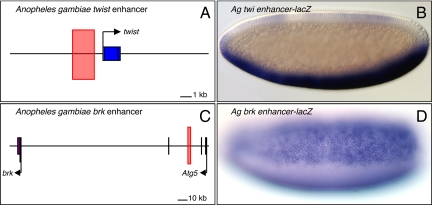
Fig. 1.
The Anopheles twist and brinker enhancers. (A and C) The locations of the Anopheles twist (A) and brinker (brk) (C) enhancers are indicated by pink rectangles. The twist enhancer is located 5′ of the gene; the brinker enhancer is ≈100 kb away in the intron of Atg5. (B and D) The Anopheles twist (twi) enhancer (B) drives lacZ reporter expression in the Drosophila mesoderm, whereas the Anopheles brinker enhancer (D) drives lacZ in the neurogenic ectoderm.
The Anopheles twist enhancer appears to correspond to the distal enhancer of the D. melanogaster twist locus, which contains separate proximal and distal enhancers in the 5′ flanking region (22, 23). The distal enhancer is responsible for sustained expression of twist in the developing mesoderm. In Drosophila virilis, the distal twist enhancer is located in the same position as in the D. melanogaster enhancer. However, the proximal enhancer is located within the second intron in D. virilis (24), and it is possible that it is located in a 3′ position in Tribolium (25). Despite this apparent movement of the proximal enhancer, the critical distal enhancer is located in comparable 5′ positions in Drosophila, Anopheles, and Tribolium.
brinker encodes a transcriptional repressor of Dpp signaling that is expressed in the presumptive neurogenic ectoderm of the early Drosophila embryo (26). A similar brinker expression pattern is observed in Anopheles embryos (16). The Drosophila gene is activated by 2 enhancers: one is located ≈10 kb 5′ of the transcription start site, and the other is located within the intron of a neighboring gene, Atg5, ≈13 kb downstream of the brinker start site (6, 27). Computational analyses identified 3 clusters of putative Dorsal binding sites in an extended region encompassing the Anopheles brinker locus. Only the cluster located within the intron of the neighboring Atg5 gene directs broad lateral stripes of lacZ expression in transgenic Drosophila (Fig. 1 C and D). Thus, it would appear that the major enhancer regulating brinker expression in the early Anopheles embryo is located in the same position as the 3′ shadow enhancer in Drosophila. This is a remarkable example of conservation in the position of a developmental enhancer because the Anopheles Atg5 gene is located nearly 100 kb from brinker and is inverted as compared with the organization in Drosophila (Fig. 1 A and C).
Dorsal Target Enhancers in Tribolium.
The preceding analysis identified twist and brinker enhancers in Anopheles that are located in the same relative positions as the corresponding enhancers in Drosophila. To determine whether this trend of fixed enhancer positions extends to even more divergent insects, we identified Dorsal target enhancers in the flour beetle T. castaneum.
Previous studies have shown that the Dorsal gradient is narrow and transient in Tribolium embryos, presumably due to a Dorsal–Cactus negative feedback loop, whereby Dorsal activates the expression of the Cactus inhibitor (10, 13). To determine whether Dorsal directly regulates cactus expression, computational methods were used to identify putative Dorsal binding sites in the vicinity of the cactus locus. The best binding-site cluster is located within the second predicted intron of the cactus transcription unit (Fig. 2C). This genomic DNA fragment mediates robust expression of a lacZ reporter gene in the presumptive mesoderm of transgenic Drosophila embryos (Fig. 2 A and E). The gap in expression in anterior regions could be due to repression by Hunchback because there is greater integration of anterior–posterior and dorsal–ventral patterning in Tribolium, as compared with Drosophila (e.g., ref. 17).
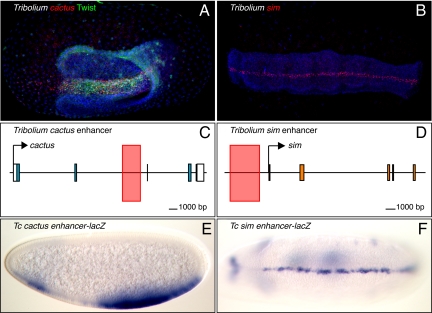
Fig. 2.
The Tribolium cactus and sim enhancers. (A) Tribolium embryo stained for cactus mRNA (red) and Twist protein to mark the mesoderm (green). (B) sim (red) is expressed in the ventral midline in beetles. DNA is stained in blue (A and B), anterior is to the left, and dorsal is up in all cases. (C and D) The locations of the Tribolium cactus (C) and sim (D) enhancers are indicated by pink rectangles. (E and F) The Tribolium cactus enhancer and the Tribolium sim enhancer drive lacZ reporter expression in transgenic Drosophila in the mesoderm (cactus) (E) and ventral midline (sim) (F).
ChIP–chip assays (11) previously identified an intronic enhancer in the Drosophila cactus gene that mediates a similar pattern of expression (summarized in Fig. 3). It is possible that the Dorsal–Cactus negative feedback loop has a stronger impact on the dynamics of the Dorsal gradient in Tribolium than Drosophila because Toll signaling is maternally deployed in Drosophila, but is strictly zygotic in Tribolium (12, 13). In any event, these studies identified comparable cactus enhancers in similar locations in Tribolium and Drosophila, despite extensive divergence in the dorsoventral patterning networks.
Fig. 3.
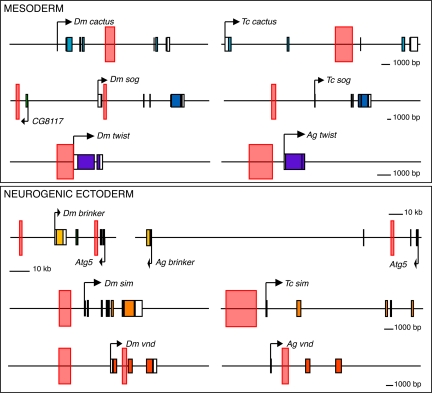
Summary of enhancer positions. Enhancers are indicated by pink rectangles. In all cases, the Drosophila locus is shown on the left and the orthologous Tribolium or Anopheles gene on the right. Abbreviations: Dm, Drosophila melanogaster; Ag, Anopheles gambiae; Tc, Tribolium castaneum.
Additional DV enhancers were identified in Tribolium to determine whether they are also located in fixed positions relative to their target genes. cactus is activated by the highest levels of the gradient, and the next gene tested, sim, is activated just beyond the limits of the cactus expression pattern where it defines the ventral midline of the developing nerve cord (28, 29). Enhancers controlling midline expression of sim are located in the immediate 5′ flanking region in both honeybees (Apis mellifera) and mosquitoes (Anopheles gambiae) (6, 21). We surveyed the Tribolium sim locus for putative Dorsal, Twist, Snail, and Sim/Tango binding sites, and identified a cluster containing all these sites upstream of the P1 promoter (Fig. 2D). A genomic DNA fragment encompassing this cluster recapitulates the sim expression pattern in the ventral midline of transgenic Drosophila embryos (Fig. 2 B and F). Thus, the sim enhancer is located in the same relative position in Drosophila, Apis, and Tribolium, despite extensive evolutionary changes in the underlying DV patterning mechanisms (21).
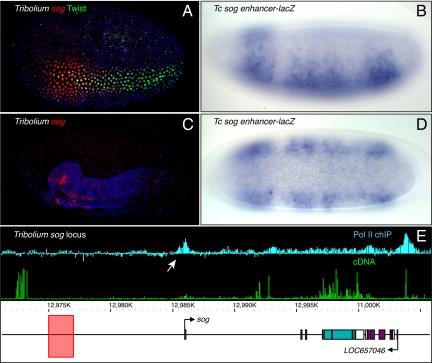
The secreted Sog inhibitor is essential for the formation of a Dpp signaling gradient (15). In Drosophila, sog is activated by low levels of the Dorsal gradient throughout the presumptive neurogenic ectoderm (8), but its expression is restricted to ventral regions of Tribolium and Anopheles embryos (14, 16). In Tribolium, sog is initially expressed in a ventral domain extending slightly beyond the mesoderm before becoming restricted to the ventral neurogenic ectoderm (Fig. 4 A and C) (14). We identified a cluster of low-affinity Dorsal sites, as well as Twist and Snail binding sites, ≈10 kb upstream of the Tribolium sog promoter (Fig. 4E). This binding cluster recapitulates the Tribolium sog pattern when expressed in transgenic Drosophila embryos (Fig. 4 A–D). As seen for the endogenous sog expression pattern, the Tribolium enhancer initially directs lacZ expression throughout the presumptive mesoderm before becoming restricted to the ventral neurogenic ectoderm and ultimately the ventral midline (Fig. 4 B and D and Fig. S1).
Fig. 4.
The Tribolium sog enhancer. (A and C) Tribolium sog is expressed first in the mesoderm and ventral neurogenic ectoderm (A) before becoming restricted to the ventral neurogenic ectoderm (C). sog is in red, Twist in green, and DNA in blue. (B and D) The Tribolium sog enhancer has the same expression pattern when driving a reporter in transgenic Drosophila. Frame E shows the location of the Tribolium sog enhancer (pink rectangle), and Pol II chIP–chip (Upper) and cDNA expression profiles (Lower) of the Tribolium sog locus. White arrow indicates the promoter.
The Drosophila sog expression pattern is controlled by 2 separate enhancers: one located within the first intron, and another (the “shadow” enhancer) located ≈20 kb upstream of the gene (see Fig. 3) (8, 27). The Tribolium enhancer is located in the same general region as the D. melanogaster sog shadow enhancer, although somewhat closer to the transcription start site. There is no evidence for an intronic sog enhancer in Tribolium. Identification of the Tribolium sog 5′ exon (14) is well supported by Pol II ChIP–chip signals at the beetle sog promoter (Fig. 4E). Syntenic relationships between sog and its neighbors are not preserved between Drosophila and Tribolium, including CG8117, which resides between the D. melanogaster sog shadow enhancer and promoter. However, this CG8117–sog linkage is not seen in divergent drosophilids outside the melanogaster group, including Drosophila virilis, which contains an experimentally verified sog enhancer located ≈30 kb upstream of the start site (30).
The previous analysis of sog regulation in Anopheles identified an enhancer located in intron 1 (16). A computational survey of putative Dorsal binding clusters identified a second enhancer located in the 5′ flanking region, similar to the position of the Drosophila shadow enhancer and 5′ enhancer in Tribolium, which directs reporter expression in the fly mesoderm (Fig. S2). It is possible that this 5′ enhancer is the major determinant of sog regulation in Drosophila, Anopheles, and Tribolium. The absence of intronic sog enhancers in some species may reflect the incomplete characterization of the locus or, alternatively, the acquisition of new enhancers in addition to the conserved 5′ enhancer.
Discussion
This study identified 5 different DV enhancers in A. gambiae and T. castaneum, representing a broad spectrum of patterning responses to the Dorsal gradient. These enhancers embody the largest collection of functionally defined regulatory DNAs engaged in a common process in divergent insects. Despite extensive differences in the DV regulatory networks of flies, mosquitoes, and beetles, the enhancers are located in similar positions relative to the promoters of the target genes they control. A constrained position is also observed for the previously identified vnd enhancer in Anopheles (31) (summarized in Fig. 3). Altogether, these 6 enhancers are located in all possible orientations, including the immediate 5′ flanking region (twist, sim), remote 5′ flanking region (sog), intron 1 (vnd), 3′ intron (cactus), and within a neighboring gene (brinker).
Given the rapid divergence of noncoding sequences due to the constant turnover of individual transcription-factor binding sites within enhancers (32), there are no arrangements of sites or even individual sites that can be thought of as orthologous in any of the pairs of enhancers described here (see Fig. S3 for binding-site arrangements). The most closely related pair of species examined in this study, Anopheles and Drosophila, last shared a common ancestor >200 million years ago. The 2 genomes have been rearranged to such an extent that only an estimated 34% of orthologous genes can be sorted into microsyntenic blocks (33). In light of this, it is somewhat surprising to see such conservation in enhancer positions. There are at least 2 possible explanations. First, the different enhancers identified in Anopheles and Tribolium might be orthologous, that is, they might derive from a common enhancer in the last shared ancestor. The position of the enhancer within the locus is apparently unchanged simply because no genomic events that would perturb its position have occurred. According to this view, selection might depend on the modification of preexisting enhancers rather than the creation of new ones, similar to the evolution of protein coding sequences (34, 35). A nonexclusive alternative explanation is that the enhancer position is functionally constrained within a genomic locus. For example, enhancers might be able to communicate with the target promoter only when located in particular positions within the higher-order structure of a complex genetic locus. Thus, enhancers might be in a constant flux of death and birth, but de novo enhancers work only when located in particular positions. The Hox gene complexes represent extreme examples of constrained enhancer organization (36). Perhaps simpler loci are subject to similar, but somewhat less stringent, organizational constraints.
The conservation of enhancer location observed in this study applies to developmental control genes. It is certainly conceivable that enhancer turnover alters the locations of regulatory DNAs, particularly in the case of housekeeping genes. An interesting example is seen for the spec genes in the sea urchin Strongylocentrotus purpuratus, which have coopted repetitive elements to function as enhancers (37). In contrast, regulatory genes engaged in interlocking network interactions, as seen for the genes considered in this study, might tend to retain old enhancers rather than invent new ones. An implication of this study is that the evolution of novel patterns of gene expression depends on the modification of existing enhancers rather than the invention of new ones. This has already been documented for the evolution of the sim expression pattern and ventral midline of divergent insects (21). We propose that the modification of constrained or orthologous enhancers will prove to be a general mechanism for the evolution of gene expression patterns.
Materials and Methods
Fly Transgenics.
T. castaneum sim, T. castaneum cactus, and A. gambiae twist enhancers were cloned into the _P_-element vector nE2G (6). A. gambiae brinker and T. castaneum sog enhancers were cloned into the ΦC31 transgenesis equivalent (eve minimal promoter, lacZ reporter) (38) provided by Michael Eisen (University of California, Berkeley). The D. melanogaster strain used for _P_-element-mediated transgenesis was _yw_67, as described previously (e.g., ref. 39). All experiments involving _P_-elements were performed with multiple independent lines. ΦC31-mediated transgenesis was carried out by using either 86Fb (40), or an AttP2 (41) pNos-integrase line, or both. Oligos are listed in Table S1.
Imaging.
Drosophila and Tribolium embryo fixation and in situ hybridizations were carried out according to methods described previously (21, 42). For RNA detection, embryos were hybridized with digoxygenin (Roche) or dinitrophenyl (Perkin–Elmer)-labeled probes and visualized colorimetrically (22) or fluorescently (42) together with the DNA stain Draq5 (Biostatus). Probes were generated with the primers listed in Table S1 and in vitro transcription. In general, probes were 2–3 kb in length, and designed to the 5′ end of the gene. The Tribolium Twist antibody was raised in rat to a nearly full-length _N_-terminal 6His-conjugated Tribolium Twist fusion protein, and was visualized with donkey anti-rat Alexa 488. Oligos are listed in Table S1.
Microarray Analysis.
Sequence for the Tribolium sog locus was taken from the Tcas_2.0 Baylor HSGC assembly. The tiled region consists of ≈100 kb on LG9 (10930000–11035000). NimbleGen designed 50-bp features covering these regions at a density of ≈1 feature per 90 bp. Chromatin for IPs was prepared from ≈15–40-h-old beetle embryos (raised at 30° C) according to ref. 43, with the exception that beetle embryos were dechorionated for 2 min in 100% bleach and were cross-linked for 10 min in formaldehyde-equilibrated hexanes. Chromatin IPs were performed according to NimbleGen's standard protocol (http://www.nimblegen.com) by using a mixture of Pol II antibodies (H14, H5, and 8WG16; Covance). Final IP samples were amplified by using the WGA-2 kit (Sigma) before being submitted to NimbleGen for labeling and hybridization. Visualization and scaling of tiling data were done with the Integrated Genome Browser (Affymetrix) and are shown as the log2 of the ratio between the IP and the input control samples. Generation of Tribolium cDNA to hybridize to the array has been described previously (44), and was carried out with ≈5–15-h-old embryos raised at 30° C.
Computational Analysis.
Anopheles and Tribolium orthologs of Drosophila genes were identified by a reciprocal BLAST–BLAST strategy. For each gene, the extended locus was scanned for clusters of binding sites by using ClusterDraw2, and the top clusters were assayed for enhancer activity in transgenic Drosophila. ClusterDraw2 has been described previously (21). The program and the motifs used in this analysis are available online at http://flydev.berkeley.edu/cgi-bin/cld/submit.cgi. Dorsal, Twist, and Snail sites have been described previously in ref. 45, and Zelda sites in refs. 46 and 47. Additionally, a table of binding sites used in this analysis is available as Table S2.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Matthew Ronshaugen and Michael Perry for insightful comments and discussions and Dave Hendrix for technical help. J. C. was funded in part by a Chang Ling Tien fellowship for the environmental sciences. This work was funded by a Moore Foundation grant and National Institutes of Health Grant GM46638.
Footnotes
The authors declare no conflict of interest.
References
- 1.Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- 2.Stein D, Nusslein-Volhard C. Multiple extracellular activities in Drosophila egg perivitelline fluid are required for establishment of embryonic dorsal-ventral polarity. Cell. 1992;68:429–440. doi: 10.1016/0092-8674(92)90181-b. [DOI] [PubMed] [Google Scholar]
- 3.Roth S, Hiromi Y, Godt D, Nusslein-Volhard C. cactus, a maternal gene required for proper formation of the dorsoventral morphogen gradient in Drosophila embryos. Development. 1991;112:371–388. doi: 10.1242/dev.112.2.371. [DOI] [PubMed] [Google Scholar]
- 4.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo-shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–R899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Stathopoulos A, Levine M. Dorsal gradient networks in the Drosophila embryo. Dev Biol. 2002;246:57–67. doi: 10.1006/dbio.2002.0652. [DOI] [PubMed] [Google Scholar]
- 6.Markstein M, et al. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- 7.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 8.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dearden PK, et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006;16:1376–1384. doi: 10.1101/gr.5108606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Handel K, Roth S. The maternal NF-kappaB/dorsal gradient of Tribolium castaneum: Dynamics of early dorsoventral patterning in a short-germ beetle. Development. 2000;127:5145–5156. doi: 10.1242/dev.127.23.5145. [DOI] [PubMed] [Google Scholar]
- 11.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxton–Kuchenmeister J, Handel K, Schmidt–Ott U, Roth S, Jäckle H. Toll homologue expression in the beetle tribolium suggests a different mode of dorsoventral patterning than in drosophila embryos. Mech Dev. 1999;83:107–114. doi: 10.1016/s0925-4773(99)00041-6. [DOI] [PubMed] [Google Scholar]
- 13.Nunes da Fonseca R, et al. Self-regulatory circuits in dorsoventral axis formation of the short-germ beetle Tribolium castaneum. Dev Cell. 2008;14:605–615. doi: 10.1016/j.devcel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 14.van der Zee M, Stockhammer O, von Levetzow C, Nunes da Fonseca R, Roth S. Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. Proc Natl Acad Sci USA. 2006;103:16307–16312. doi: 10.1073/pnas.0605154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francois V, Solloway M, O'Neill JW, Emery J, Bier E. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 1994;8:2602–2616. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- 16.Goltsev Y, et al. Evolution of the dorsal-ventral patterning network in the mosquito, Anopheles gambiae. Development. 2007;134:2415–2424. doi: 10.1242/dev.02863. [DOI] [PubMed] [Google Scholar]
- 17.van der Zee M, Berns N, Roth S. Distinct functions of the Tribolium zerknullt genes in serosa specification and dorsal closure. Curr Biol. 2005;15:624–636. doi: 10.1016/j.cub.2005.02.057. [DOI] [PubMed] [Google Scholar]
- 18.Kim IO, et al. CNS midline cells contribute to maintenance of the initial dorsoventral patterning of the Drosophila ventral neuroectoderm. J Neurobiol. 2005;62:397–405. doi: 10.1002/neu.20104. [DOI] [PubMed] [Google Scholar]
- 19.Golembo M, Raz E, Shilo BZ. The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development. 1996;122:3363–3370. doi: 10.1242/dev.122.11.3363. [DOI] [PubMed] [Google Scholar]
- 20.Chang J, Kim IO, Ahn JS, Kim SH. The CNS midline cells control the spitz class and Egfr signaling genes to establish the proper cell fate of the Drosophila ventral neuroectoderm. Int J Dev Biol. 2001;45:715–724. [PubMed] [Google Scholar]
- 21.Zinzen RP, Cande J, Ronshaugen M, Papatsenko D, Levine M. Evolution of the ventral midline in insect embryos. Dev Cell. 2006;11:895–902. doi: 10.1016/j.devcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J, Kosman D, Ip YT, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 23.Pan DJ, Huang JD, Courey AJ. Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev. 1991;5:1892–1901. doi: 10.1101/gad.5.10.1892. [DOI] [PubMed] [Google Scholar]
- 24.Pan D, Valentine SA, Courey AJ. The bipartite D. melanogaster twist promoter is reorganized in D. virilis. Mech Dev. 1994;46:41–53. doi: 10.1016/0925-4773(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 25.Handel K, Basal A, Fan X, Roth S. Tribolium castaneum twist: Gastrulation and mesoderm formation in a short-germ beetle. Dev Genes Evol. 2005;215:13–31. doi: 10.1007/s00427-004-0446-9. [DOI] [PubMed] [Google Scholar]
- 26.Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell. 1999;96:563–573. doi: 10.1016/s0092-8674(00)80660-1. [DOI] [PubMed] [Google Scholar]
- 27.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crews ST, Thomas JB, Goodman CS. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- 29.Thomas JB, Crews ST, Goodman CS. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- 30.Crocker J, Tamori Y, Erives A. Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol. 2008;6:e263. doi: 10.1371/journal.pbio.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erives A, Levine M. Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci USA. 2004;101:3851–3856. doi: 10.1073/pnas.0400611101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig MZ. Functional evolution of noncoding DNA. Curr Opin Genet Dev. 2002;12:634–639. doi: 10.1016/s0959-437x(02)00355-6. [DOI] [PubMed] [Google Scholar]
- 33.Zdobnov EM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 34.Long M. Evolution of novel genes. Curr Opin Genet Dev. 2001;11:673–680. doi: 10.1016/s0959-437x(00)00252-5. [DOI] [PubMed] [Google Scholar]
- 35.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 36.Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 37.Dayal S, et al. Creation of cis-regulatory elements during sea urchin evolution by co-option and optimization of a repetitive sequence adjacent to the spec2a gene. Dev Biol. 2004;273:436–453. doi: 10.1016/j.ydbio.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stathopoulos A, Levine M. Linear signaling in the Toll-Dorsal pathway of Drosophila: Activated Pelle kinase specifies all threshold outputs of gene expression while the bHLH protein Twist specifies a subset. Development. 2002;129:3411–3419. doi: 10.1242/dev.129.14.3411. [DOI] [PubMed] [Google Scholar]
- 40.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kosman D, et al. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 43.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shippy TD, et al. Analysis of the Tribolium homeotic complex: Insights into mechanisms constraining insect Hox clusters. Dev Genes Evol. 2008;218:127–139. doi: 10.1007/s00427-008-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 46.ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 47.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information