Affy exon tissues: exon levels in normal tissues in human, mouse and rat (original) (raw)
Abstract
Summary: Most genes in human, mouse and rat produce more than one transcript isoform. The Affymetrix Exon Array is a tool for studying the many processes that regulate RNA production, with separate probesets measuring RNA levels at known and putative exons. For insights on how exons levels vary between normal tissues, we constructed the Affy Exon Tissues track from tissue data published by Affymetrix. This track reports exon probeset intensities as log ratios relative to median values across the dataset and renders them as colored heat maps, to yield quick visual identification of exons with intensities that vary between normal tissues.
Availability: Affy Exon Tissues track is freely available under the UCSC Genome Browser (http://genome.ucsc.edu/) for human (hg18), mouse (mm8 and mm9), and rat (rn4).
Contact: cline@soe.ucsc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The mammalian transcriptome is complex. By recent estimates, as many as 94% of human genes undergo alternative splicing (Wang, et al., 2008). Alternative splicing is consequential as well as frequent, with effects ranging from altering protein structure to targeting mRNA for early decay (Hartmann and Valcarcel, 2009). Furthermore, mammalian genomes contain an abundance of non-coding RNA genes (Chu and Rana, 2007). In short, to understand the consequences of transcription, one must look beyond overall expression levels of known genes.
Affymetrix exon arrays facilitate transcriptome analysis with probesets that measure RNA abundance for individual exons, conserved genomic regions, and blocks from syntenic alignments (see http://www.affymetrix.com/support/technical/technotes/exon_array_design_technote.pdf). These arrays have offered new insights on how transcript isoforms may be influenced by a myriad of factors including tissue type (Clark et al., 2007), genetic variation (Kwan et al., 2007; Zhang et al., 2009), differentiation (Yeo et al., 2007), and disease (Soreq et al., 2008; Thorsen et al., 2008).
Alternative splicing can arise through normal, regulated processes; or through abnormalities such as mutation, disease, and environmental stress (Yeo et al., 2005). Before one can understand the abnormal conditions, it is valuable to understand the scope of normal alternative splicing by comparing splicing patterns between normal tissues. To facilitate this, we have provided the Affy Exon Tissues tracks in the UCSC Genome Browser (Kuhn et al., 2009), depicting exon probeset intensities in normal tissues in human, mouse, and rat.
2 DESCRIPTION
The Affy Exon Tissues track consists of two parts: genomic coordinates of the exon array probesets; and a heat map indicating exon probeset intensities in normal tissues, based on data available from http://www.affymetrix.com/support/technical/sample_data/exon_array_data.affx. Briefly, normal tissues were assayed in triplicate, and were analyzed with the Affymetrix Power Tools software (http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx) to produce normalized, background-corrected probeset intensities. For each probeset, we computed its median intensity for each tissue, and then the median of these median values. For each experiment, we calculated the log ratio between the probeset intensity and this median value. For numeric stability, we added a fixed, background-level pseudocount to each observation, which also renders probesets with no expression as constant-valued. The genome browser renders these log ratios as blue–white–red (shown), green–red, or yellow–blue heat maps, with the color selection controlled via the track's details page. Additional details are provided in the Supplementary Material.
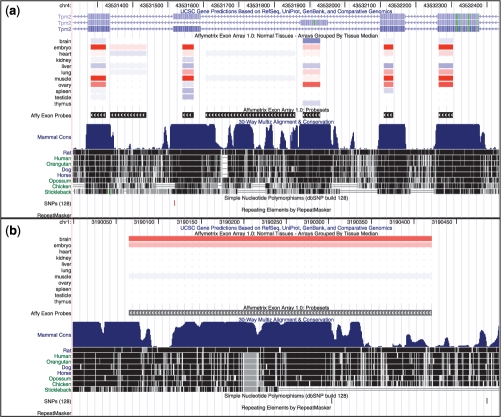
Figure 1a shows the Affy Exon Tissues track for TPM2 in mm9. The constitutive exons (those included in all transcripts) indicate that TPM2 is expressed most strongly in muscle and embryo, with some expression in ovary. TPM2 has a well-documented pattern of tissue-dependent splicing, with one isoform produced in skeletal muscle tissue and another in non-muscle tissue (Gooding and Smith, 2008). This pattern is apparent in the two mutually-exclusive exons (third and fourth from the left): one is highly-expressed (red) in muscle and embryo (a heterogeneous tissue), while the other is highly-expressed in ovary.
Fig. 1.
(a) The Affy Exon Tissues track demonstrates tissue-dependent splicing of two mutually exclusive exons (third and fourth from the left) in the mm9 TPM2 locus. Red indicates probesets that are up-regulated relative to median levels, while blue indicates down-regulated probesets. The constitutive exons (those included in all transcripts: first, second and fifth from the left) offer perspectives on overall gene expression, and indicate that the gene is up-regulated (red) in embryo, muscle and ovary. The leftmost mutually-exclusive exon appears upregulated in embryo and muscle, while the second appears upregulated in ovary. The remaining two probesets, which map to no known exons, are rendered mostly white. This indicates no variation in their expression levels, and suggests that they are not expressed in these tissues. (b) A probeset was designed for this unannotated region in mm9 based on more-speculative evidence such as genomic conservation or ab initio exon prediction. This region shows evidence of up-regulation (red) in brain, suggesting production of a brain-specific transcript.
For contrast, Figure 1b shows an unannotated conserved region on chromosome 1 in mm9. While it does not overlap with any known gene, its red (up-regulated) log intensity in brain suggests brain-specific expression. This illustrates how this data can offer insights on regions with no annotation but strong conservation.
3 CONCLUSION
The Affy Exon Tissues track displays exon probeset intensities in human, mouse, and rat tissues, including breast, cerebellum, heart, kidney, liver, muscle, pancreas, prostate, spleen, testes, and thyroid. In contrast to traditional microarray tracks such as the GNF Expression Atlas (Su et al., 2004), which provide one measure of overall expression per gene and cannot report any transcript variation, the Affy Exon Tissues track offers the ability to compare intensities of neighboring probesets and observe alternative promoter usage, polyadenylation, and splicing. Exon probeset intensities are rendered as heat maps to offer rapid visual identification of exons that vary under normal cellular conditions.
Besides the Affy Exon Tissues track, the UCSC Genome Browser currently hosts the hg18 Sestan Brain exon expression track, which contrasts exon probeset intensities between sections of the brain (Johnson et al., 2009). This set of tracks may expand further as additional datasets become available, offering further insights into transcript variation in the mammalian genomes.
ACKNOWLEDGEMENTS
Many people contributed to this track. The authors would like to thank Affymetrix in general and Alan Williams in particular for the track data. This track reflects the work of many individuals in the UCSC Genome Browser team, including Bob Kuhn, Donna Karolchik and Jim Kent. M.S.C. thanks Manuel Ares Jr, for his mentorship and encouragement.
Funding: National Human Genome Research Institute (2 P41 HG002371-06 to UCSC Center for Genomic Science, 3 P41 HG002371-06S1 ENCODE supplement to UCSC Center for Genomic Science); National Cancer Institute (Contract No. N01-CO-12400 for Mammalian Gene Collection); NIH GM-040478 (to M.S.C.)
Conflict of Interest: none declared.
REFERENCES
- Chu CY, Rana TM. Small RNAs: regulators and guardians of the genome. J. Cell Physiol. 2007;213:412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- Clark TA, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding C, Smith CW. Tropomyosin exons as models for alternative splicing. Adv. Exp. Med. Biol. 2008;644:27–42. doi: 10.1007/978-0-387-85766-4_3. [DOI] [PubMed] [Google Scholar]
- Hartmann B, Valcarcel J. Decrypting the genome's alternative messages. Curr. Opin. Cell Biol. 2009;21:377–386. doi: 10.1016/j.ceb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq L, et al. Identifying alternative hyper-splicing signatures in MG-thymoma by exon arrays. PLoS ONE. 2008;3:e2392. doi: 10.1371/journal.pone.0002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen K, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol. Cell Proteomics. 2008;7:1214–1224. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, et al. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, et al. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum. Genet. 2009;125:81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]