The Ubiquitin-Specific Peptidase USP15 Regulates Human Papillomavirus Type 16 E6 Protein Stability (original) (raw)
Abstract
Proteomic identification of human papillomavirus type 16 (HPV16) E6-interacting proteins revealed several proteins involved in ubiquitin-mediated proteolysis. In addition to the well-characterized E6AP ubiquitin-protein ligase, a second HECT domain protein (HERC2) and a deubiquitylating enzyme (USP15) were identified by tandem affinity purification of HPV16 E6-associated proteins. This study focuses on the functional consequences of the interaction of E6 with USP15. Overexpression of USP15 resulted in increased levels of the E6 protein, and the small interfering RNA-mediated knockdown of USP15 decreased E6 protein levels. These results implicate USP15 directly in the regulation of E6 protein stability and suggest that ubiquitylated E6 could be a substrate for USP15 ubiquitin peptidase activity. It remains possible that E6 could affect the activity of USP15 on specific cellular substrates, a hypothesis that can be tested as more is learned about the substrates and pathways controlled by USP15.
Human papillomaviruses (HPVs) are associated with several human cancers, most notably human cervical cancer, the second most common cancer among women worldwide (43). Papillomaviruses cause proliferative squamous epithelial lesions, and more than 100 HPV types have been described (14). The HPV types associated with mucosal squamous epithelial lesions have been further classified into high- or low-risk types based on the propensity for the lesions with which they are associated to progress to cancer. Among the high-risk HPV types, HPV type 16 (HPV16) and HPV18 account for approximately 70% of cervical cancers (43). The high-risk HPV types carry two genes, the E6 and E7 genes, which have oncogenic properties and are always expressed in HPV-positive cancers. E6 and E7 interfere with the p53 and retinoblastoma (pRB) tumor suppressor pathways, respectively, and contribute directly to cell cycle alterations, protection from apoptosis, and transformation (14). The dysregulated expression of the E6 and E7 oncoproteins is an important step in the progression from a preneoplastic stage to cancer in HPV-infected cells and is often a consequence of the integration of the viral genome into the host chromosome.
The interaction between E6 and p53 is mediated by the E3 ubiquitin ligase E6AP (15). E6, p53, and E6AP form a complex in which E6 directs the ligase activity of E6AP to p53, thereby targeting p53 for ubiquitin-mediated degradation (36). E6, however, has a number of other cellular partners and other functions. For instance, the C terminus of the high-risk E6 protein contains a PDZ binding motif (20, 25) that mediates the interaction with several PDZ domain-containing proteins, including discs large (Dlg), Scribble (Scrib), the MAGI family of proteins, MUPP1, and PATJ (9, 10, 29). Some of these proteins are also targeted for degradation in an E6AP-dependent manner (22, 29). While the major mechanism of oncogenesis revolves around E6's ability to inhibit the proapoptotic effects of p53, recent work involving the PDZ domain proteins indicates that these interactions are also important to the oncogenic potential of E6 (38, 41). Furthermore, E6 has been reported to bind a number of other cellular proteins, including but not limited to Bak, CBP/p300, c-Myc, E6TP1, hADA3, IRF3, MCM7, PTPH1, and TNF-R1 (7, 8, 17, 23, 24, 32, 35, 39, 40). The importance of the binding of several of these proteins with regard to the transformation or other functions of E6 remains to be established. E6 itself is thought to be targeted for degradation by an ubiquitin-proteasome pathway (18), although how E6 protein stability is regulated has not been well studied.
Many of the E6 binding partners have been identified using purified bacterially expressed E6 fusion proteins and cell lysates from various cell types or using yeast two-hybrid screenings. While some of these interactions with E6 have been validated, the physiologic relevance of a number of proposed E6 targets remains undetermined. In an effort to identify E6-interacting proteins, perhaps under more physiologic conditions, we employed tandem affinity purification (TAP) using tagged HPV16 E6 stably expressed in the HPV16-positive cervical cancer cell line SiHa. We have discovered several new interacting proteins, including an interaction between E6 and the cellular deubiquitylating enzyme (DUB) USP15. USP15 is not targeted for degradation by E6, but we found that USP15 stabilizes E6 protein levels, suggesting that E6 may itself be a target for USP15 DUB activity.
MATERIALS AND METHODS
Plasmids.
The HPV16 gene E6 was cloned into the pOZN retroviral expression plasmid (30), incorporating a FLAG-hemagglutinin (HA) tag on the E6 N terminus by using the XhoI and NotI sites to generate pOZN16E6 (plasmid p5132). The USP15 gene was cloned out of plasmid pXW107 (3) and into pcDNA-4C (Invitrogen), with Xpress and His tags fused to the N terminus by using the NotI and XhoI sites to generate pcDNA-USP15 (plasmid p5953). The USP5 gene was cloned out of pTAP-USP5 (provided by J. W. Harper, Harvard Medical School, Boston, MA) and into pcDNA-4C, with Xpress and His tags fused to the N terminus by using the PCR-engineered BamHI and XbaI sites to generate pcDNA-USP5 (plasmid p6042). The catalytically inactive USP15 was made by mutating the cysteine at position 269 into an alanine by using site-directed mutagenesis to generate pcDNA-USP15C269A (plasmid p5954). The codon-optimized HPV16 E6 construct (16E6) was produced by GenScript Corp. (NJ) and cloned into the pOZN vector using the XhoI and NotI sites to generate pOZN16E6Opt (plasmid p5955). IRF3 was cloned out of pcDNA-IRF-3 (19) and into pOZN using the PCR-engineered XhoI and NotI sites to generate pOZN-IRF3 (plasmid p5035). Plasmids LXSH, L(E6)SH, and L(E7)SH were derived from the analogous LXSN vectors (11), originally provided by D. Galloway (Fred Hutchinson Cancer Research Center, Seattle, WA).
Cell culture.
C33A, Caski, U2OS, HeLa, and SiHa cells as well as C33A cells stably expressing L(E7)SH and either pOZN, pOZN16E6, or pOZN16E6Opt constructs were maintained in Dulbecco modified Eagle medium with 10% fetal calf serum, penicillin (10 U/ml), and streptomycin (100 μg/ml). C33A and U2OS cells stably transfected with the LXSH parental plasmid, or with the expression plasmid L(E6)SH or L(E7)SH, were selected and maintained in Dulbecco modified Eagle medium with 10% fetal calf serum and penicillin-streptomycin supplemented with 250 μg/ml hygromycin B. Primary human foreskin keratinocytes were maintained in keratinocyte-SFM media (Invitrogen) supplemented with bovine pituitary extract, epidermal growth factor, and amphotericin B (Fungizone).
Immunoprecipitations (IPs).
For TAP, SiHa-pOZN or SiHa pOZN16E6 cells were harvested from six to eight 500-cm2 plates and lysed in 1% Triton X-100 and 1% NP-40 buffer. After 30 min on ice, lysates were sonicated once for 10 s at 35% and centrifuged at 4°C to obtain the soluble fraction. Equivalent amounts of total protein, as determined by Pierce BCA protein assay (catalog no. 23225), were immunoprecipitated with anti-FLAG-agarose beads (Sigma), rotating at 4°C overnight. Beads were washed four times with 100 mM NaCl buffer, and bound proteins were eluted four times at 15 min each with 250 μg/ml FLAG peptide. The eluates were then immunoprecipitated with anti-HA-agarose beads (Sigma), rotating for 4 h at 4°C. Beads were again washed four times with 100 mM NaCl buffer. Bound proteins were eluted twice at 20 min each with glycine-HCl, pH 2.5. After pH adjustment, proteins were acetone precipitated overnight and resuspended in 1× loading buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and colloidal blue staining.
For single IPs, cells were lysed in 1% Triton X-100-1% NP-40 lysis buffer, as described above, incubated for 3 to 4 h with anti-HA-agarose beads, washed four times with 100 mM NaCl buffer, and resuspended in loading buffer for SDS-PAGE.
Antibodies.
All antibodies used in this study were obtained from the following commercial sources, except for the 4C6 mouse monoclonal antibody to HPV16 E6 (5, 28), generated by Etienne Weiss (University of Strasbourg) and provided through Arbor Vita Corporation (Sunnyvale, CA): actin (1501; Chemicon), E6AP (sc-25509; Santa Cruz), HA-horseradish peroxidase (Roche), IκBα (9242; Cell Signaling), p53 (OP43; Calbiochem), ubiquitin (sc-9133; Santa Cruz), USP15 (H00009958-M01; Abnova), and Xpress (R910-25; Invitrogen).
Plasmid transfections.
All DNA plasmid transfections were performed using FuGene transfection reagent as per the manufacturer's instructions at a FuGene/DNA ratio of 2:1.
Quantitative real-time reverse transcriptase PCR.
Total RNA was isolated from cell pellets using the NucleoSpin RNA II kit (Clontech, Palo Alto, CA), according to the manufacturer's instructions. The concentration of each sample was determined by UV spectrophotometry. To remove residual DNA contamination, purified RNAs were treated with DNase using the Turbo DNA-free kit (Ambion), which includes a DNase inactivation and removal step. Equal amounts of RNA were reverse transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative real-time PCR was performed in an Applied Biosystems ABI 7500 fast sequence detection system using the TaqMan fast universal PCR master mix (Applied Biosystems), oligonucleotide primers, and TaqMan dual-labeled probes (5′FAM and 3′Black Hole quencher or 5′FAM and 3′Iowa Black quencher; Integrated DNA Technologies, Coralville, IA). HPV16 E6Opt primers and probes were designed using Primer Express software (Applied Biosystems). The sequences are as follows: HPV16 E6OPT FWD, 5′ CCTGCTGATTCGCTGTATTAATTG 3′; HPV16 E6Opt REV, 5′ CCAGGTGCCGCTGTTTCT 3′; and HPV16 E6OPT PROBE, 5′ CAGAAGCCCCTGTGCCCCGA 3′. The relative amounts of cDNA in each sample were calculated based on a standard curve prepared using serial dilutions of one reference cDNA. The change in transcription of the gene following small interfering RNA (siRNA) treatment was calculated by comparison of that to the sample transfected with E6OPT DNA only. To ensure that equal amounts of cDNA were included in each reaction, samples were analyzed with primers and TaqMan probes specific to the cellular housekeeping gene encoding glucose-6-phosphate dehydrogenase (G6PD). The sequences for G6PD (42) primers and probes have been previously published.
Cycloheximide determination of E6 protein half-life.
C33A cells were transfected, as described above. At 48 hours posttransfection, cultures were treated with 40 μg/ml of cycloheximide for the times indicated previously. Cells were lysed under denaturing conditions (2% SDS), followed by SDS-PAGE and Western blot analysis.
In vitro degradation assay.
Cell lysates were mixed with purified bacterially expressed GST-16E6, ATP, and ubiquitin and incubated at 30°C for 90 min. SDS-PAGE and Western blotting were performed using anti-p53 and anti-USP15 antibodies.
siRNA knockdown experiments.
siRNA oligonucleotides against E6 (5′-AGAGAUCAGUUGUCUCUGGUU-3′) (sequence provided by Kenneth Alexander, University of Chicago), as well as the following control siRNAs, were purchased from Dharmacon: siGLO (D-001630-02), siCONT (nontargeting siRNA #1, D-001210-01), and siBRD4 (D-004937-02). siUSP15-134 and siUSP15-308 were purchased from Ambion's predesigned Silencer siRNA collection, catalog no. 105134 and 113308, respectively. siRNA was transfected into the C33A(E7)-NE6Opt cell line using DharmaFECT 2 transfection reagent, as per the manufacturer's instructions; cells were lysed in 2% SDS lysis buffer containing 50 mM Tris, pH 6.8, at the indicated time points posttransfection. siRNA was transfected into Caski cells using Oligofectamine (Invitrogen), as per the manufacturer's instructions; cells were lysed in 2% SDS lysis buffer containing 50 mM Tris, pH 6.8, at the indicated time points posttransfection.
RESULTS
TAP of HPV16 E6.
To generate a cell line suitable for TAP of protein complexes containing HPV16 E6, we established cell lines using SiHa, an HPV16-positive cervical cancer cell line. These cells were stably transfected with a retroviral expression construct containing a dual FLAG-HA-tagged HPV16 E6 gene driven by a retroviral murine sarcoma virus long terminal repeat promoter, allowing low-level stable expression of the tagged E6 protein. We demonstrated that the FLAG-HA-tagged E6 was functional for E6AP binding and p53 degradation in C33A cells, as well as for telomerase activation in human foreskin keratinocytes (data not shown).
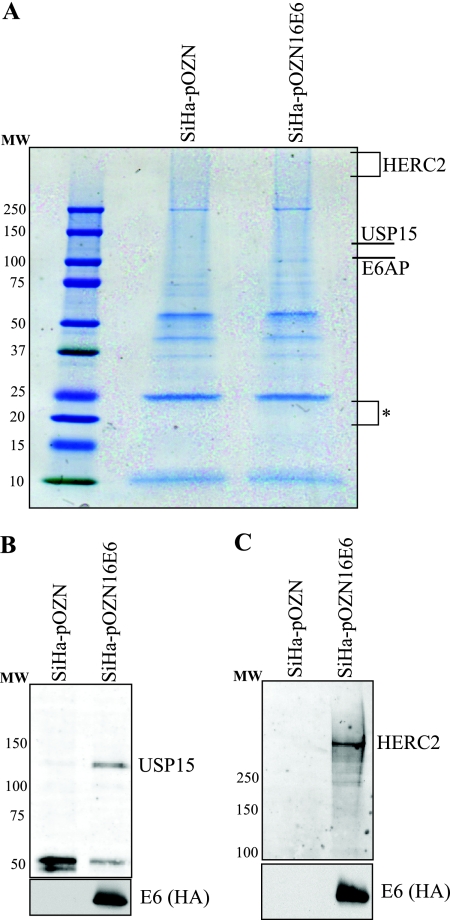
Lysates from HPV16 E6-expressing stable cells and from control cells were subjected to TAP. Proteins that precipitated in complex with tagged E6 but not from cells transduced with the empty pOZN vector were stained with Coomassie blue and identified by mass spectrometry (Fig. 1A). E6 was not observed following Coomassie blue staining, due to the low level of expression and possibly due in part to its small size. Using targeted ion mass spectrometry, E6 was identified in the appropriate region of the gel. After eliminating nonspecific proteins that were present in both experimental and vector control samples, we limited our analysis to those proteins for which two or more peptides were identified (see Table S1 in the supplemental material). The most peptides were detected for E6AP. We did not detect p53 or Scrib, both proteins that are reported to bind E6, likely because these proteins are substrates for degradation via their interactions with E6 and E6AP. We identified several novel E6-interacting proteins. Our results show that the DUB USP15 interacts with HPV16 E6, and this interaction was confirmed following tandem IP of tagged E6, followed by Western blotting against endogenous USP15 in the tagged E6-expressing SiHa cells (Fig. 1B). In addition, we identified the 528-kDa protein HERC2, which contains a HECT-like domain and is a putative E3 ligase (16). This interaction was also confirmed by IP-Western blotting (Fig. 1C). A number of other proteins were also identified as potential E6-interacting partners (see the supplemental material). For this study, we have focused on the interaction between USP15 and HPV16 E6.
FIG. 1.
USP15 and HERC2 interact with HPV16 E6. (A) SiHa cells stably transfected with either parental plasmid pOZN or the dual-tagged E6 expression plasmid pOZN16E6 were lysed under mild conditions, and complexes containing E6 were tandem affinity purified. Mass spectrometry analysis revealed specific interaction of E6 with E6AP, USP15, and HERC2. The asterisk indicates the region of the gel analyzed by targeted ion mass spectrometry for the identification of HPV16 E6. Molecular weight (MW) standards are listed to the left of each gel. (B) The dual-tagged HPV16 E6 was tandem immunoprecipitated, followed by Western blotting to confirm the E6-USP15 interaction. (C) The dual-tagged HPV16 E6 was tandem immunoprecipitated, followed by Western blotting to confirm the E6-HERC2 interaction.
HPV16 E6-mediated degradation of USP15.
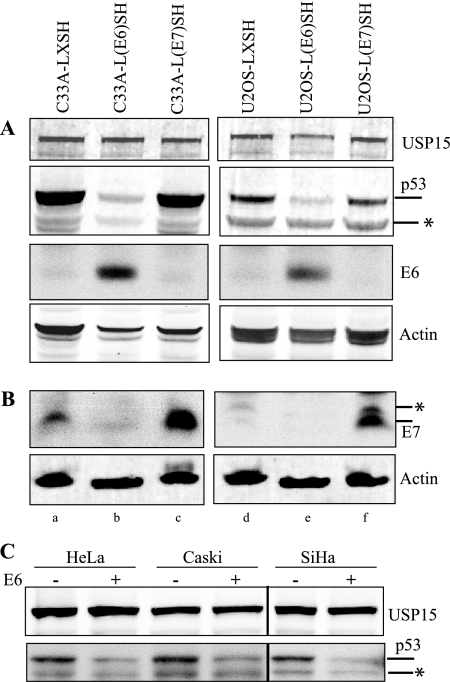
Since a number of proteins that interact with HPV16 E6 are targeted for ubiquitin-mediated degradation, we explored the possibility that HPV16 E6 might mediate USP15 proteolysis. We compared the levels of endogenous USP15 in the C33A and U2OS cell lines stably expressing HPV16 E6 or, as controls, HPV16 E7 or vector alone. The steady-state level of USP15 was not affected by either E6 or E7 expression (Fig. 2A). In this experiment, p53 served as a positive control; its level was low in the E6-positive cell lines compared to that in cells expressing HPV E7 or vector alone. E7 expression was also confirmed by Western blotting (Fig. 2B). We also performed an in vitro cell free degradation assay using various cell lysates either with or without the addition of purified E6 and did not observe any E6-mediated degradation of USP15 (Fig. 2C). Degradation of p53 served as a positive internal control. We therefore conclude that HPV16 E6 expression does not lead to USP15 degradation.
FIG. 2.
HPV16 E6 does not promote the degradation of USP15. (A) Endogenous USP15 expression was analyzed in C33A and U2OS cell lines stably transfected with empty parental vector LSXH (lanes a and d, respectively), HPV16 E6 (lanes b and e, respectively), or HPV16 E7 (lanes c and f, respectively) expression vector. Degradation of p53 was used as a positive control for E6 activity. Confirmation of E6 expression is also shown. The asterisk specifies a nonspecific band below the p53 band. (B) Confirmation of E7 expression was detected in a separate blot using the HPV16 E7 monoclonal antibody (8C9). Actin is included as a loading control. The asterisk specifies a nonspecific band seen in the E7 Western blot that runs just above the specific E7 band. (C) USP15 degradation was assessed in an in vitro degradation assay using cell extracts with or without the addition of purified GST-16E6. Degradation of p53 was used as a positive control. The asterisk specifies a nonspecific band below the p53 band.
USP15 overexpression causes an increase in HPV16 E6 protein levels.
In order to more readily detect HPV16 E6, we created a codon-optimized version of E6. While the amino acid sequence of E6 remained unchanged, the nucleotide changes introduced ablated the splice donor site at nucleotide 226, thereby eliminating the possible synthesis of the E6 proteins. Cells transfected with the codon-optimized construct (E6Opt) expressed about three times more E6 protein than cells transfected with the genomic sequence. The codon-optimized expressed E6 protein retained its ability to bind E6AP (see Fig. 4A).
FIG. 4.
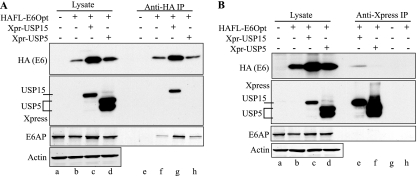
HPV16 E6 and USP15 interact in C33A cells. Epitope-tagged versions of E6 and USP15, or USP5 as a control, were cotransfected into C33A cells. At 48 hours posttransfection, cells were lysed under mild conditions; anti-HA IPs for E6 (A) and anti-Xpress (Xpr) IPs for USP15 and USP5 (B) were performed. Western blotting to confirm expression, pulldown, and specific and reciprocal coimmunoprecipitation of HPV16 E6 and USP15 but not USP5 is shown. Endogenous E6AP coimmunoprecipitation with E6 served as an internal positive control. Transfection efficiency for this experiment was approximately 70%.
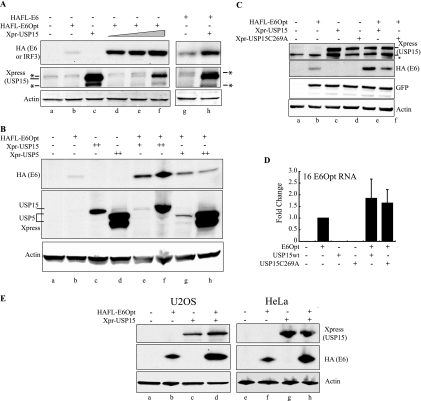
Next, we characterized the interaction between HPV16 E6 and USP15 by coexpressing E6Opt with His-Xpress-tagged USP15 in C33A cells. We observed higher levels of E6 protein in cells that had been cotransfected with USP15 expression vector than in cells transfected with E6Opt alone (Fig. 3A, compare lanes b and d). E6 protein levels increased in a dose-dependent manner with increasing USP15 expression (Fig. 3A, lanes d to f). We confirmed that the levels of the non-codon-optimized E6 protein were increased by coexpression of USP15 (Fig. 3A, lanes g and h). To assess the specificity of USP15 in increasing E6 levels, we performed similar cotransfection experiments with another DUB, USP5 (also known as isopeptidase T). We observed an increase of three- to fivefold in E6 protein levels in cells expressing USP5 (Fig. 3B, compare lanes e and f versus lanes g and h), but this increase was less than the increase of 8- to 11-fold observed with USP15 expression. This increase could be due to the general ability of USP5 to disassemble polyubiquitin chains (33).
FIG. 3.
Overexpression of USP15 increases HPV16 E6 protein and RNA levels. (A) C33A cells were cotransfected with a constant amount of codon-optimized 16E6 (E6Opt) along with increasing amounts of Xpress-tagged USP15 (Xpr-USP15) (lanes d to f). E6 levels were analyzed by SDS-PAGE and Western blotting with the indicated antibodies at 48 h posttransfection. The experiment was also performed using the original non-codon-optimized E6 construct (lanes g and h) as shown. Transfection efficiency for this experiment was approximately 70%. The asterisks indicate nonspecific bands. (B) C33A cells were cotransfected with E6Opt and either USP15 or USP5, as indicated. Lysates taken at 48 h posttransfection were analyzed by SDS-PAGE and Western blotting for E6 levels. Transfection efficiency for this experiment was approximately 70%. (C) C33A cells were cotransfected with E6Opt and either USP15wt or the USP15(C269A) catalytic mutant, as indicated. Lysates taken at 48 h posttransfection were analyzed by SDS-PAGE and Western blotting for E6 levels. GFP is shown for each sample as a transfection control; transfection efficiency for this experiment was approximately 50%. The asterisk indicates a nonspecific band. (D) C33A or U2OS cells were transfected with the indicated constructs and harvested at 48 h posttransfection. Total cellular RNA was reverse transcribed and analyzed by quantitative real-time PCR to measure E6Opt transcript levels. The graph indicates the average of five independent experiments, each normalized to G6PD transcript levels. Error bars indicate one standard deviation from the means of the combined experiments. (E) U2OS and HeLa cells were transfected with E6Opt and USP15 alone or in combination, as indicated. Lysates taken at 48 h posttransfection were analyzed by SDS-PAGE and Western blotting for E6 levels. U2OS transfection efficiency was approximately 60%. HeLa transfection efficiency was approximately 80%.
To determine whether the effect on the E6 levels requires USP15 activity, we generated a catalytically inactive form of USP15 by mutating the active cysteine residue at position 269 (12). In cotransfection experiments with this construct, we observed an increase of 1.3-fold in E6 levels relative to the green fluorescent protein (GFP) internal control with the mutant compared to an increase of 2.4-fold seen with USP15wt (Fig. 3C). Since some of the increase in E6 expression when USP15 is overexpressed could be due to an increase in RNA levels, we measured HPV16 E6Opt RNA levels after cotransfection with a USP15 expression vector. RNA was isolated from C33A or U2OS cells transfected with 16E6Opt, USP15, and USP15(C269A) expression vectors. The samples were reverse transcribed into cDNA, and the relative amount of the E6Opt transcript present in each sample was measured by quantitative real-time PCR. The average of five independent experiments is shown (Fig. 3D). We observed an increase in the E6Opt transcript in the presence of overexpressed USP15 that was variable but by approximately twofold compared to that of the sample with E6Opt alone. Introducing the catalytically inactive USP15(C269) expression vector resulted in a similar increase in E6Opt transcript levels. We concluded that an increase in the abundance of the E6Opt transcript does contribute somewhat to the USP15-mediated increase in E6Opt protein levels. Other ways in which USP15 causes this increase, including lengthening the half-life of E6Opt, are discussed below. Since the initial observation in this study was that the HPV16 E6 and USP15 proteins interact, we focused subsequent experiments on understanding the mechanism of increased protein stability.
To rule out a cell type-dependent effect of USP15 on E6Opt, we performed similar cotransfection experiments in the following two additional cell lines: HPV18-positive HeLa cells and HPV-negative U2OS cells. In each of these cell lines, the E6Opt protein levels were significantly higher in the presence of transfected USP15 (Fig. 3E).
We next confirmed that E6 binds to USP15 but not to USP5. Following cotransfection in C33A cells, IP of E6Opt specifically coprecipitated USP15 but not USP5 (Fig. 4A). E6Opt could be observed in IPs with USP15 but not with USP5 (Fig. 4B). In these experiments, E6 retained the ability to coimmunoprecipitate E6AP (Fig. 4A, lanes e to h), suggesting that USP15 binding to E6 does not block E6 binding to E6AP. Furthermore, E6AP was not detected after USP15 IP (Fig. 4B, lanes e to h), indicating that E6AP did not mediate the binding of E6 to USP15.
USP15 stabilizes HPV16 E6 protein.
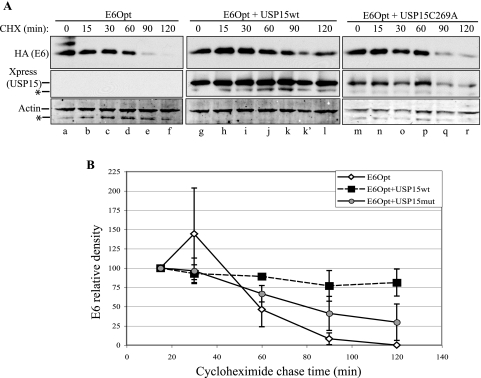
To examine whether USP15 affected the stability of HPV16 E6, we examined the half-life of E6 in the presence or absence of cotransfected USP15 using a cycloheximide chase in C33A cells. Lysates were immunoblotted for HA (E6), Xpress (USP15), and actin. The densities of E6 and actin were determined relative to the 15-min time point, and ratios were calculated to account for differences in loading (Fig. 5A). Using data from three independent experiments (Fig. 5B), we observed that E6Opt expressed alone had a half-life of 68 min. The levels of E6Opt in the presence of USP15wt never dropped below 50% of the level at baseline; however, we were able to extrapolate a half-life of 278 min. In the presence of the catalytically inactive USP15(C269A), the half-life of E6Opt was 87 min, similar to the half-life of E6Opt alone. We concluded that USP15 overexpression stabilizes 16E6 protein and that the catalytic activity of USP15 is required for this effect.
FIG. 5.
Coexpression of USP15 increases 16E6 steady-state levels by extending the half-life of E6. (A) C33A cells were transfected with E6 alone or together with USP15wt or the catalytically inactive USP15(C269A). At 48 hours posttransfection, cells were treated with 40 μg/ml cycloheximide to inhibit de novo translation. Cells were lysed under denaturing conditions at the time points indicated, and lysates were analyzed by SDS-PAGE and Western blotting to monitor E6 protein levels. Lanes k and k′ represent one sample for which a broken gel well (lane k) allowed some sample to spill into the adjacent lane (lane k′); for densitometry measurements, values from the two lanes were combined. Transfection efficiency for this experiment was approximately 80%. The asterisks indicate nonspecific bands. (B) Cumulative data from three experiments are shown.
USP15 protein knockdown by siRNA.
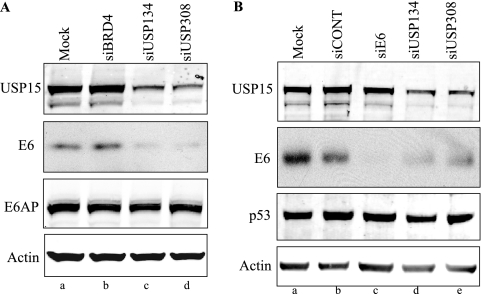
Since the overexpression of USP15 increased the steady-state levels of E6, we investigated whether the knockdown of USP15 by siRNA would decrease E6 protein levels. We used two siRNAs that targeted different regions of USP15 to knock down USP15 in C33A cells stably expressing 16E6Opt. Treatment with each of the USP15 siRNAs resulted in lower expression levels of USP15 and a reduction of E6Opt protein levels (Fig. 6A, lanes c and d) compared to those of cells treated with a control siRNA directed to BRD4 (Fig. 6A, lane b), a protein that is not known to interact with either E6 or USP15. A similar result was obtained in HeLa cells stably expressing the tagged protein (data not shown). Since the reduction in E6 protein levels was observed with both of the siRNAs that target USP15, off-target effects causing the reduction in E6 protein levels are unlikely.
FIG. 6.
USP15 suppression by RNA interference reduces steady-state levels of 16E6 in stably transfected and HPV16-positive cell lines. (A) C33A cells expressing E7 were stably transfected with the parental plasmid pOZN or pOZN16E6Opt and mock transfected (lane a) or transiently transfected with the siBrd4 control (lane b) or the USP15 siRNAs (lanes c and d). At 72 hours posttransfection, cells were lysed under denaturing conditions and analyzed by SDS-PAGE and Western blotting. (B) HPV16-positive Caski cells were mock transfected (lane a) or transiently transfected with the siCONT control (lane b), siE6 that targets the endogenous E6/E7 mRNA (lane c), or the USP15 siRNAs (lanes d and e). At 96 h posttransfection, cells were lysed under denaturing conditions and analyzed by SDS-PAGE and Western blotting.
We also investigated whether the knockdown of USP15 by siRNA would affect the levels of endogenous E6 detectable in the HPV16-positive Caski cell line. At 96 h posttransfection, we observed a decrease in E6 protein levels in cells in which USP15 had been knocked down (Fig. 6B, lanes d and e). A siRNA directed at the E6/E7 mRNA in Caski cells served as a positive control (Fig. 6B, lane c).
Since E6 targets the ubiquitylation and proteolysis of p53, we examined whether the lower levels of E6 resulting from the USP15 knockdown at 96 h affected p53 levels in these cells. We did not observe any significant increase in p53 levels with either of the siRNAs directed against USP15 or that directed against E6 (Fig. 6B). The stability of p53, despite the lower levels of E6 from the USP15 knockdown, suggests that even low levels of E6 can direct the enzymatic ubiquitylation of p53 and that perhaps we had not reached the threshold of E6 levels necessary to inhibit this activity in our siRNA knockdown experiments. This hypothesis and alternatives are discussed further below.
DISCUSSION
There are more than 90 human DUBs (2). The largest family of these enzymes includes the ubiquitin-specific proteases, characterized by prototypic Cys and His box motifs. DUBs play important roles in a wide variety of cellular processes. These include protecting substrates from K48-linked ubiquitin-mediated degradation by the proteasome and altering signaling and localization of proteins through the removal of K63-linked ubiquitin moieties. USP15 was first characterized a decade ago (3); however, only recently have some of its functions and targets been elucidated.
In this study, we report that the HPV16 E6 oncoprotein and USP15 interact and demonstrate that USP15 is involved in regulating E6 protein stability. Overexpression of USP15 stabilizes E6, and siRNA-mediated knockdown of USP15 results in decreased levels of E6 protein. The catalytic activity of USP15 was required for increased E6 protein levels, suggesting that ubiquitylated E6 may itself be a target for USP15 deubiquitylation.
There are previous reports in the literature of viral proteins interacting with cellular DUBs. For instance, USP7 (or HAUSP) interacts with the herpes simplex virus type I immediate early protein ICP0 (4, 6) as well as the Epstein-Barr virus protein EBNA1 (13). USP7 is known to regulate the cellular turnover of p53 (26), suggesting that these herpesviruses may be able to manipulate the apoptotic status of infected cells through interaction with USP7. Binding of USP11 has been reported to stabilize HPV16 E7 through deubiquitylation, extending its half-life and enhancing the downstream degradation of Rb (27). In addition, HPV16 and HPV18 E6 proteins have been implicated in the ubiquitylation and degradation of the CYLD DUB, resulting in the sustained hypoxia-induced activation of NF-κB (1). That study demonstrated CYLD in complex with E6 but did not provide evidence of a direct interaction between E6 and the CYLD DUB (1).
No link between USP15 and cancer has yet been established. USP15 was found to be active in various tumor cell lines, including those derived from human cervix, colon, lung, brain, and kidney cancers, as well as in some human lymphomas (31). A subsequent study compared the activities of a variety of USPs from primary cervical carcinoma tissue and matched normal cervical tissue; no consistent up- or downregulation of USP15 activity was detected in samples from 27 patients (34).
In our study, we examined whether the decrease in E6 following USP15 knockdown resulted in increased p53 levels. We did not observe any effect up to 96 h after introduction of the siRNAs. The lack of effect could be due to the previously observed phenomenon that when E6 is removed as the major regulator of p53 in HPV-positive cells, the “natural” regulators MDM2, COP1, and Pirh2 resume control of p53 levels (21). Alternatively, the reduction in E6 levels might not have been sufficient to affect the overall cellular E6/E6AP ubiquitylation activity. It is also possible that the E6-USP15 interaction has functional consequences in a pathway other than that defined for p53.
Identification of the substrate targets for USP15 is ongoing. USP15 associates with the COP9 signalosome (CSN) (12), a multiprotein complex involved in regulating the ubiquitin-proteasome pathway predominantly though interaction with cullin-based E3 ligases. In this context, USP15 has been shown to deubiquitylate the E3 ligase Rbx1, presumably to protect Rbx1 from autoubiquitylation (12). More recently, IκBα was shown to be deubiquitylated by USP15 in association with the CSN after tumor necrosis factor alpha-mediated stimulation of the NF-κB pathway (37). Our preliminary data suggest that E6 may affect the ability of USP15 to break down ubiquitin chains, but no link to any of the aforementioned pathways has been established (our unpublished data).
Future efforts will focus on understanding the normal cellular role of USP15 and how its function may be affected by the presence of E6, as well as evaluating a larger subset of known E6-interacting proteins to determine if USP15 may be involved in their regulation. We were unable to establish cell lines with either stable overexpression or stable knockdown by short hairpin RNA of USP15, indicating that USP15 likely plays a vital role within the cell and that its expression levels are tightly regulated. Insights into the relevance for carcinogenesis will also be gained by investigating whether both high-risk and low-risk HPV E6 proteins can interact with and are stabilized by USP15.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank John Asara from the BIDMC Mass Spectrometry Core for useful discussion and troubleshooting advice. We also thank Kenneth Alexander for providing the endogenous E6/E7 siRNA sequence.
This work was supported by grants from the National Cancer Institute to P.M.H. (RO1 CA050661 and R37 CA64888) and the following NIH-sponsored training grants: Viral-Host Interactions in Cancer training grant to R.M.V. (T32 CA09031), the Pathobiology of Cancer training grant to J.A. (T32 CA09382), and an individual NRSA training grant to E.A.W. (F32 AI080075).
Footnotes
▿
Published ahead of print on 24 June 2009.
REFERENCES
- 1.An, J., D. Mo, H. Liu, M. S. Veena, E. S. Srivatsan, R. Massoumi, and M. B. Rettig. 2008. Inactivation of the CYLD deubiquitinase by HPV E6 mediates hypoxia-induced NF-kappaB activation. Cancer Cell 14394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek, K. H. 2003. Conjugation and deconjugation of ubiquitin regulating the destiny of proteins. Exp. Mol. Med. 351-7. [DOI] [PubMed] [Google Scholar]
- 3.Baker, R. T., X. W. Wang, E. Woollatt, J. A. White, and G. R. Sutherland. 1999. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics 59264-274. [DOI] [PubMed] [Google Scholar]
- 4.Boutell, C., and R. D. Everett. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 27836596-36602. [DOI] [PubMed] [Google Scholar]
- 5.Courtete, J., A. P. Sibler, G. Zeder-Lutz, D. Dalkara, M. Oulad-Abdelghani, G. Zuber, and E. Weiss. 2007. Suppression of cervical carcinoma cell growth by intracytoplasmic codelivery of anti-oncoprotein E6 antibody and small interfering RNA. Mol. Cancer Ther. 61728-1735. [DOI] [PubMed] [Google Scholar]
- 6.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 161519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippova, M., H. Song, J. L. Connolly, T. S. Dermody, and P. J. Duerksen-Hughes. 2002. The human papillomavirus 16 E6 protein binds to TNF R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 27721730-21739. [DOI] [PubMed] [Google Scholar]
- 8.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 185487-5496. [DOI] [PubMed] [Google Scholar]
- 10.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 195270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hetfeld, B. K., A. Helfrich, B. Kapelari, H. Scheel, K. Hofmann, A. Guterman, M. Glickman, R. Schade, P. M. Kloetzel, and W. Dubiel. 2005. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr. Biol. 151217-1231. [DOI] [PubMed] [Google Scholar]
- 13.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 27829987-29994. [DOI] [PubMed] [Google Scholar]
- 14.Howley, P. M., and D. R. Lowy. 2007. Papillomaviruses, p. 2299-2354. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 15.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP: a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji, Y., M. J. Walkowicz, K. Buiting, D. K. Johnson, R. E. Tarvin, E. M. Rinchik, B. Horsthemke, L. Stubbs, and R. D. Nicholls. 1999. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum. Mol. Genet. 8533-542. [DOI] [PubMed] [Google Scholar]
- 17.Jing, M., J. Bohl, N. Brimer, M. Kinter, and S. B. Vande Pol. 2007. Degradation of tyrosine phosphatase PTPN3 (PTPH1) by association with oncogenic human papillomavirus E6 proteins. J. Virol. 812231-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kehmeier, E., H. Ruhl, B. Voland, M. C. Stoppler, E. Androphy, and H. Stoppler. 2002. Cellular steady-state levels of “high risk” but not “low risk” human papillomavirus (HPV) E6 proteins are increased by inhibition of proteasome-dependent degradation independent of their p53- and E6AP-binding capabilities. Virology 29972-87. [DOI] [PubMed] [Google Scholar]
- 19.Kirshner, J. R., A. Y. Karpova, M. Kops, and P. M. Howley. 2005. Identification of TRAIL as an interferon regulatory factor 3 transcriptional target. J. Virol. 799320-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 9411612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koivusalo, R., A. Mialon, H. Pitkanen, J. Westermarck, and S. Hietanen. 2006. Activation of p53 in cervical cancer cells by human papillomavirus E6 RNA interference is transient, but can be sustained by inhibiting endogenous nuclear export-dependent p53 antagonists. Cancer Res. 6611817-11824. [DOI] [PubMed] [Google Scholar]
- 22.Kuballa, P., K. Matentzoglu, and M. Scheffner. 2007. The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J. Biol. Chem. 28265-71. [DOI] [PubMed] [Google Scholar]
- 23.Kukimoto, I., S. Aihara, K. Yoshiike, and T. Kanda. 1998. Human papillomavirus oncoprotein E6 binds to the C-terminal region of human minichromosome maintenance 7 protein. Biochem. Biophys. Res. Commun. 249258-262. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, A., Y. Zhao, G. Meng, M. Zeng, S. Srinivasan, L. M. Delmolino, Q. Gao, G. Dimri, G. F. Weber, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 225801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 749680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416648-653. [DOI] [PubMed] [Google Scholar]
- 27.Lin, C. H., H. S. Chang, and W. C. Yu. 2008. USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. J. Biol. Chem. 28315681-15688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masson, M., C. Hindelang, A. P. Sibler, G. Schwalbach, G. Trave, and E. Weiss. 2003. Preferential nuclear localization of the human papillomavirus type 16 E6 oncoprotein in cervical carcinoma cells. J. Gen. Virol. 842099-2104. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 208244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370430-444. [DOI] [PubMed] [Google Scholar]
- 31.Ovaa, H., B. M. Kessler, U. Rolen, P. J. Galardy, H. L. Ploegh, and M. G. Masucci. 2004. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. USA 1012253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 185061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes-Turcu, F. E., J. R. Shanks, D. Komander, and K. D. Wilkinson. 2008. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 28319581-19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolen, U., V. Kobzeva, N. Gasparjan, H. Ovaa, G. Winberg, F. Kisseljov, and M. G. Masucci. 2006. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol. Carcinog. 45260-269. [DOI] [PubMed] [Google Scholar]
- 35.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. The human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 122061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquination of p53. Cell 75495-505. [DOI] [PubMed] [Google Scholar]
- 37.Schweitzer, K., P. M. Bozko, W. Dubiel, and M. Naumann. 2007. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 261532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonson, S. J., M. J. Difilippantonio, and P. F. Lambert. 2005. Two distinct activities contribute to human papillomavirus 16 E6's oncogenic potential. Cancer Res. 658266-8273. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, M., and L. Banks. 1999. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 801513-1517. [DOI] [PubMed] [Google Scholar]
- 40.Veldman, T., X. Liu, H. Yuan, and R. Schlegel. 2003. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 1008211-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson, R. A., M. Thomas, L. Banks, and S. Roberts. 2003. Activity of the human papillomavirus E6 PDZ-binding motif correlates with an enhanced morphological transformation of immortalized human keratinocytes. J. Cell Sci. 1164925-4934. [DOI] [PubMed] [Google Scholar]
- 42.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 781817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.zur Hausen, H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384260-265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]