Coordinate Regulation of FOXO1 by miR-27a, miR-96, and miR-182 in Breast Cancer Cells (original) (raw)
Abstract
The FOXO1 transcription factor orchestrates the regulation of genes involved in the apoptotic response, cell cycle checkpoints, and cellular metabolism. FOXO1 is a putative tumor suppressor, and the expression of this gene is dysregulated in some cancers, including prostate and endometrial cancers. However, the molecular mechanism resulting in aberrant expression of human FOXO1 in cancer cells is poorly understood. We show here that FOXO1 mRNA is down-regulated in breast tumor samples as compared with normal breast tissue. Silencing of the microRNA processing enzymes, Drosha and Dicer, led to an increase in FOXO1 expression. We also identified functional and specific microRNA target sites in the FOXO1 3′-untranslated region for miR-27a, miR-96, and miR-182, microRNAs that have previously been linked to oncogenic transformation. The three microRNAs, miR-27a, miR-96 and miR-182, were observed to be highly expressed in MCF-7 breast cancer cells, in which the level of FOXO1 protein is very low. Antisense inhibitors to each of these microRNAs led to a significant increase in endogenous FOXO1 expression and to a decrease in cell number in a manner that was blocked by FOXO1 siRNA. Overexpression of FOXO1 resulted in decreased cell viability because of inhibition of cell cycle traverse and induction of cell death. We have identified a novel mechanism of FOXO1 regulation, and targeting of FOXO1 by microRNAs may contribute to transformation or maintenance of an oncogenic state in breast cancer cells.
The Forkhead Box O subfamily of transcription factors (FOXO) regulates a variety of important cellular processes including metabolism, cellular differentiation, apoptosis, and cell-cycle progression (1, 2). Acting as master cellular regulators, FOXO transcription factors can both activate and repress target gene expression (3).
The three predominant members of the FOXO family (FOXO1, FOXO3a, and FOXO4) were first implicated in tumorigenesis based on the observation that fusion proteins resulting from chromosomal breakpoints exist in certain types of cancers (2). Recent evidence suggests that FOXO proteins function as tumor suppressors based on their role in regulating cell-cycle progression and inducing apoptosis (4).
One regulatory mechanism of FOXO1 activity is through phosphorylation, primarily downstream of the insulin-stimulated phosphatidylinositol 3-kinase/AKT/protein kinase B signaling pathway, which results in nuclear exclusion (5–7). FOXO1 activity can also be regulated by acetylation (8) and ubiquitination (9, 10). In addition to insulin, FOXO1 can be down-regulated by other growth factors including estrogen (11, 12) and epidermal growth factor (13). The estrogen receptor α also complexes with phosphorylated FOXO1 and mediates its export from the nucleus. These mitogens are important for the growth and survival of breast cancer cells and may contribute to maintaining low levels of FOXO1 in breast cancer cells.
Although the activity of FOXO1 has been well characterized, very little is known about the regulation of FOXO1 expression, particularly in breast cancer. FOXO1 is down-regulated in several other cancers including endometrial carcinoma (14) and ovarian cancer (15). In addition, restoration of FOXO1 expression in endometrial carcinoma cells decreases cellular proliferation (14). Although the role of FOXO1 in tumorigenesis is not entirely clear, it has been hypothesized that the down-regulation of this gene is an important step in tumor formation. Recently, it was shown that FOXO1 is reduced in certain endometrial carcinoma cells lines as well as endometriod endometrial tumors (14). Subsequent analysis suggested that the down-regulation of FOXO1 expression was because of a post-transcriptional mechanism (14).
A novel class of small RNA molecules, microRNAs, has been implicated in the post-transcriptional regulation of thousands of mRNA transcripts resulting in decreased protein expression of target genes (16). MicroRNAs are small ∼21–25 nucleotide single-stranded RNA molecules that negatively regulate gene expression by binding to the 3′-UTR2 of a target mRNA molecule resulting in either degradation of the transcript or translational inhibition. Recent studies have shown that many microRNAs work in conjunction to fine tune protein expression on a global level (17, 18). In addition, mice harboring knockouts in microRNA genes display a variety of detrimental phenotypes such as severe immune deficiency (19, 20) and stress-induced heart defects (21), and these studies have emphasized the important role of these genes in tissue homeostasis and disease.
Here, we show that FOXO1 expression is also down-regulated in breast tumor samples compared with normal breast tissue. We hypothesized that the low levels of FOXO1 are a consequence of microRNA regulation, and we subsequently identified three microRNAs that directly target FOXO1 (miR-27a, miR-96, and miR-182) and regulate endogenous protein expression in MCF-7 breast cancer cells. Suppression of these microRNAs resulted in an increase in FOXO1 protein and in a decrease in cell number that was rescued by FOXO1 siRNA. In addition, we show that overexpression of FOXO1 in breast cancer cells resulted in decreased cell number both through inhibition of cell cycle traverse and increased cell death. This study has identified a novel mechanism for the down-regulation of FOXO1 in breast cancer samples, and this may impact the transformation of breast cells and maintenance of an oncogenic state.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF-7, T47D, MDA-MB-231, and MDA-MB-435 cells were obtained through American Type Culture Collection. MCF-7, MDA-MB-231, and MDA-MB-435 cells were cultured in Dulbecco's modified Eagle's medium/F-12 media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). T47D cells were cultured in Dulbecco's modified Eagle's medium/F-12 media supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 0.2 units/ml insulin (Sigma).
Analysis of FOXO1 Expression in Breast Cancer Samples
Breast Cancer I quantitative PCR tissue arrays were purchased from OriGene and used to assess the expression of FOXO1. This array consisted of cDNAs obtained from 48 samples ranging from stage 0 (normal) to breast tumor stage III. All the clinical information associated with each of these samples can be found on the OriGene web site (Breast Cancer Panel 1).
Luciferase Constructs
The psi-CHECKTM-2 dual luciferase reporter (Promega) was a kind gift from Dr. Henry Furneaux (University of Connecticut Health Center, Farmington, CT). All sequences were directionally cloned into plasmids cut with XhoI and NotI restriction endonucleases. Inserts were ligated into the vector and transformed into DH5α competent cells. Colonies were screened for inserts via colony PCR and standard DNA sequencing. DNA from positive clones was purified using the Qiagen Plasmid Midi kit and visualized on a 1% agarose gel to verify accurate concentrations. Sequences cloned into the psi-CHECKTM-2 vector varied depending on the nature of the assay. For the tiling assay, various regions of the FOXO1 3′-UTR ranging from 164 to 416 bp in size were amplified using gene-specific primers and cloned into the luciferase reporter assay. DNA oligonucleotides for the sensor assay were purchased from Integrated DNA Technologies (Coralville, IA) and purified by desalting. The oligonucleotides were treated with T4 polynucleotide kinase (Invitrogen), annealed, and ligated into the psi-CHECKTM-2 vector. For the deletion mutant experiments (Fig. 5D), DNA oligonucleotides matching 50 bp of endogenous FOXO1 3′-UTR sequence were annealed and inserted into the psi-CHECKTM-2 vector. Mutants were generated by synthesizing oligos missing the endogenous predicted microRNA binding site.
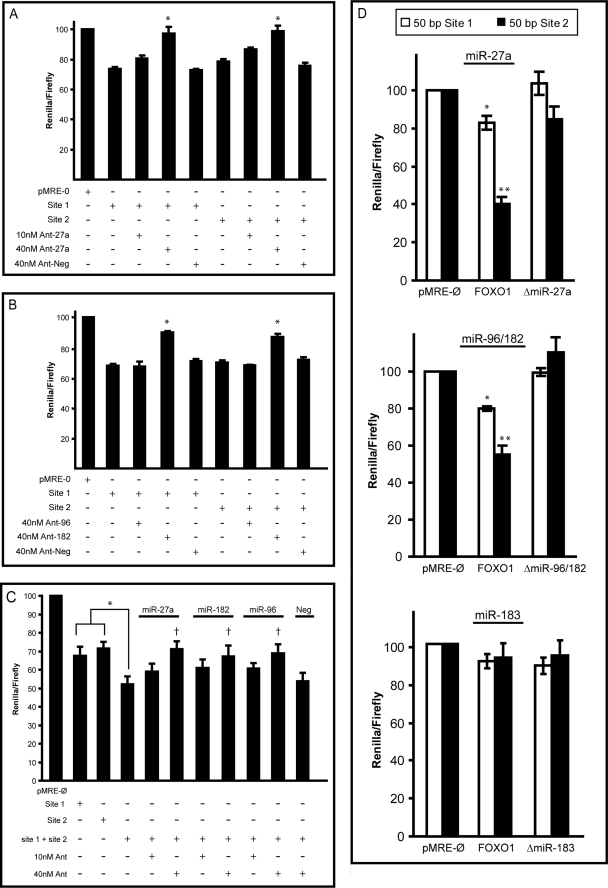
FIGURE 5.
Repression of predicted microRNA target sites in the FOXO1 3[prime]-UTR in MCF-7 cells. Short fragments of the FOXO1 3′-UTR harboring predicted microRNA sites were PCR-amplified and cloned into a dual luciferase reporter plasmid. Site 1 corresponds to nucleotides 204–492, and Site 2 corresponds to nucleotides 1907–2247 of the FOXO1 3′-UTR. Luciferase repression was conferred with Renilla activity and normalized to firefly activity. A, luciferase repression of Site 1 and Site 2 followed by treatment with antisense inhibitor against miR-27a (Ant-27a) or a scrambled inhibitor (Ant-Neg). *, p < 0.05 for inhibitor treatment versus Site alone. B, luciferase repression of Site 1 and Site 2 followed by treatment with antisense inhibitor against miR-96 (Ant-96), miR-182 (Ant-182), or a scrambled inhibitor (Ant-Neg). *, p < 0.05 for inhibitor treatment versus site alone. C, Site 1 and Site 2 were cloned into the 3′-UTR of the luciferase reporter plasmid. The vector containing Site 1 and Site 2 was treated with antisense inhibitors against the indicated microRNAs or a scrambled inhibitor. Error bars represent the S.E. of three independent experiments. *, p < 0.05; †, p < 0.05 for inhibitor treatment versus Site 1 + Site 2 alone. D, smaller sites encompassing the predicted target site and endogenous flanking sequence (50-bp total insert size) was cloned into the dual luciferase reporter plasmid. White bars represent predicted Site 1, and black bars represent predicted Site 2 for each indicated microRNA. Luciferase repression was conferred with Renilla activity and normalized to firefly activity for the empty vector (pMRE_-Ø), the endogenous site (FOXO1), or the same site with the microRNA binding site deleted (Δ_miR). Error bars represent the S.E. of three independent experiments. *, p < 0.05; **, p < 0.01 for endogenous site versus parent vector or site mutant.
Luciferase Reporter Assays
The indicated cell lines were transiently transfected with 100 ng of the indicated plasmid using Lipofectamine 2000 Transfection reagent (Invitrogen). Cells were serum-starved for 4 h followed by adding back the full media. Cells were harvested 24 h post-transfection by treatment with trypsin, lysed in 1× Passive Lysis buffer (Promega), and centrifuged at 14,000 rpm for 15 min. The supernatant was assayed for firefly and Renilla luciferase activity following the Promega protocol. Values were reported as relative Renilla to firefly luciferase activity, and significance is reported as ±S.E.
Quantitative Real-time PCR
mRNA transcripts were measured using a standard SYBR Green real-time assay. RNA was isolated using the Trizol reagent (Invitrogen), and 1 μg of total RNA was reverse-transcribed using the Superscript III enzyme (Invitrogen). Real-time PCR was then performed on cDNA in an iQ Sybr Green Supermix (Bio-Rad) with gene-specific primers (supplemental Table S1). Amplicons were analyzed using the ΔΔCt method, and data are represented as the mean of three independent experiments ±S.E.
Semiquantitative End Point Analysis of Mature MicroRNAs
The protocol for the amplification and detection of mature microRNAs using a stem-loop gene-specific reverse transcription primer was adapted from Varkonyi-Gasic et al. (40). Briefly, stem-loop primers were designed to specifically reverse transcribe the mature microRNA of interest. The cDNA was synthesized by incubating 1 μg of total RNA, 2 pmol of gene-specific primer, and 0.5 mm dNTPs at 65 °C for 5 min. The mixture was cooled on ice, 1× first strand buffer (250 mm Tris-HCl (pH 8.3), 375 mm KCl, 15 mm MgCl2), 5 mm dithiothreitol, 4 units of RNase Out, and 100 units of SuperscriptTM III reverse transcriptase (Invitrogen) were added to a reaction volume of 50 μl and incubated at 55 °C for 60 min, and then the enzyme was inactivated at 70 °C for 15 min. To ensure no DNA contamination was present, control reactions were performed as described above without the addition of SuperscriptTM III enzyme. Subsequent PCR amplification were performed by combining 1× PCR buffer (200 mm Tris-HCl (pH 8.4), 500 mm KCl), 1.5 mm MgCl2, 0.5 mm dNTPs, 0.2 μm primers, 2 μl of cDNA, 1 unit of Taq polymerase, and nuclease-free water in a reaction volume of 50 μl. Primers were designed based on the sequences of interest (supplemental Table S2). The temperature program was 95 °C, 5 min → [95 °C, 30 s → 55 °C, 30 s → 72 °C, 30 s] × 30 cycles → 72 °C 10 min. It should be noted that microRNAs expressed at lower levels (such as miR-96) required 35 cycles of PCR amplification to visualize products. PCR products were resolved on a 10% nondenaturing acrylamide gel and stained with ethidium bromide in 0.5× Tris borate-EDTA.
Antisense Inhibitors
MicroRNA expression was decreased using miRIDIANTM microRNA Hairpin Inhibitors (Dharmacon) directed against hsa-miR-27a, hsa-miR-182, hsa-miR-96, has-miR-183, hsa-miR-122, or Negative Control #1 (a non-targeting sequence). Inhibitors were resuspended in nuclease-free water at a stock concentration of 20 μm and transfected into MCF-7 cells at the indicated final concentrations using Lipofectamine 2000 (Invitrogen) in serum-free conditions.
Western Blotting
Whole cell lysates were prepared from the indicated cell lines. Protein concentrations were quantitated using a standard BCA assay, and samples were resolved on either an 8% (Dicer, Drosha) or 10% (FOXO1, GAPDH, β-actin) SDS-PAGE gel. The protein was transferred onto a nitrocellulose membrane, then blocked in 5% milk in 0.1% Tris-buffered saline-Tween 20 (TBST) at room temperature for 1 h. All primary antibodies were obtained from Abcam, Inc. and were incubated with the membrane at 4 °C overnight at the following concentrations: α-FOXO1 (1:1000), α-GAPDH (1:10,000), α-Dicer (1:1000), α-Drosha (1:2000), α-β-actin (1:2500). Membranes were washed with 1× TBST and incubated with either anti-mouse or anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody, both obtained from Santa Cruz Biotechnology. Protein expression was assessed by enhanced chemiluminescence and exposure to Biomax Light film (Eastman Kodak Co.). Image J software from the National Institutes of Health was used to quantify band intensities.
siRNA Treatments
SMARTpool siRNA targeting Dicer, Drosha, FOXO1, and GAPDH were obtained from Dharmacon and used at a final concentration of either 80 nm (Dicer and Drosha) or 100 nm (FOXO1 and GAPDH (supplemental Fig. S5)).
Trypan Blue Cell Viability Assay
MCF-7 cells were plated at 2 × 105 cells/well in 6-well culture dishes and transfected with the indicated concentrations of empty vector or expression constructs. In some experiments, cells were transfected with 40 nm antisense inhibitors to specific microRNAs alone or inhibitors plus 100 nm siRNA targeting FOXO1 or GAPDH. At the indicated time points, cells were trypsinized and stained with trypan blue, and viable cells were counted four times for each sample. Cell viability is expressed as a percentage of the empty vector or mock-treated samples. Values are expressed as the average percentage of viable cells ±S.E. of three independent experiments.
BrdUrd Assay
MCF-7 cells were plated at 1 × 106 cells/well in a 6-well culture dish and transfected with 4 μg of empty vector or expression constructs. Transfection was allowed to proceed for 30 h, then BrdUrd was added to the cells at a final concentration of 10 μm, and incorporation was carried out for 18 h. Cells were then washed and fixed in 3.7% formaldehyde and acid-washed, and staining of BrdUrd was carried out using the Ultravision Detection System kit (Thermo Scientific). Four separate fields were counted for each sample, and percentages represent the total number of BrdUrd-positive cells per treatment normalized to the total cell number. Error bars represent the S.D. of four independent experiments.
Microscopy
MCF-7 cells were grown on coverslips and transfected with the indicated plasmids. Transfection was carried out for 24 h, then cells were fixed with 4% paraformaldehyde and permeabilized with 1% Triton X-100 in phosphate-buffered saline. Cells were blocked in 3% normal goat serum (NGS), and an α-FOXO1 antibody was used at a dilution of 1:100 (Chemicon). An anti-rabbit fluorescein isothiocyanate-labeled secondary antibody was used at a dilution of 1:50. Cells were visualized using a Zeiss Pascal microscope at 40× with a NA 1.2 water immersion objective. Images were processed and analyzed using the LSM Image Browser.
Statistical Analysis
Values reported in all analysis were expressed as the mean ± S.E. However, Western blot quantifications were noted only as averages. Differences between treatments and/or groups were analyzed using an unpaired Student's t test. Statistical significance was accepted at p < 0.05.
RESULTS
FOXO1 Levels Are Decreased in Human Breast Tumor Samples
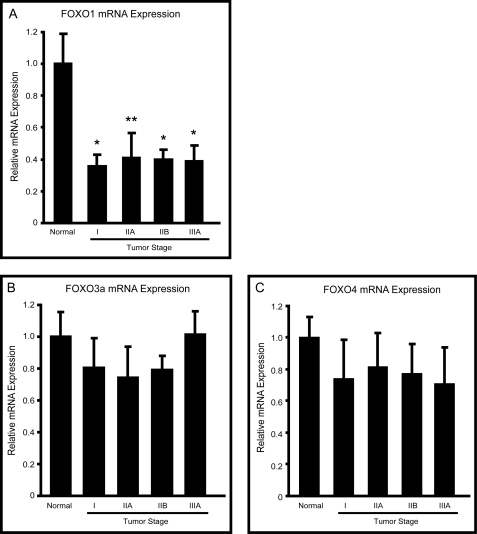
FOXO1, FOXO3a, and FOXO4 mRNA expression was measured in human breast tumor samples from a range of tumor stages as well as in normal breast tissue samples by quantitative real-time PCR. FOXO1 mRNA levels were significantly decreased by about 2-fold in all tumor samples as compared with normal breast tissue (Fig. 1A). Levels of FOXO3a and FOXO4, however, did not differ significantly between the normal and tumor samples (Fig. 1, B and C).
FIGURE 1.
FOXO1 expression in human breast tumor samples. FOXO1 (A), FOXO3a (B), and FOXO4 (C) mRNA expression was measured using gene specific primers in various breast tumor samples representing several stages and compared with normal breast tissue. Sample categories were distributed as follows: Normal (n = 7), Stage I (n = 10), IIA (n = 13), IIB (n = 7), IIIA (n = 8). All samples were normalized to β-actin expression. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
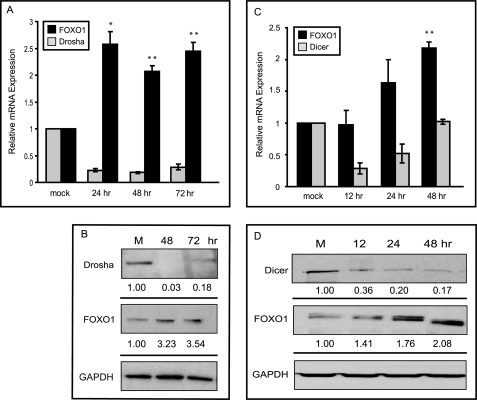
We embarked on a study of whether the lower levels of FOXO1 in breast tumors were due, in part, to post-transcriptional gene silencing for two reasons. First, FOXO1 displayed apparent post-transcriptional regulation in a previous study of uterine endometrial cancer. Second, the FOXO1 transcript harbors a very large 3′-UTR (∼3350 nucleotides), which is a region that most often confers post-transcriptional regulation through several mechanisms. MicroRNAs represent a subclass of noncoding, regulatory RNAs that play a significant role in posttranscriptional gene silencing. To test the hypothesis that microRNAs target FOXO1 mRNA in breast cancer, key effectors of the microRNA pathway were knocked down, and FOXO1 levels were assessed. Drosha is a nuclear RNase III enzyme and is required for the first step in microRNA processing, generating pre-microRNAs from primary-microRNAs. Transfection of Drosha siRNA reduced Drosha protein to nondetectable levels at 48 h, and these began to rebound at 72 h. Knockdown of Drosha yielded significantly (∼2–3-fold) increased FOXO1 mRNA and protein levels at 48 and 72 h (Fig. 2, A and B). To corroborate the findings from the Drosha knockdown, we transfected cells with siRNA to Dicer. The cytoplasmic RNase III, Dicer, is required for the last step in microRNA processing that generates a mature microRNA duplex from a precursor stem-loop form. Upon treatment of MDA-MB-231 cells with Dicer siRNA, Dicer mRNA levels fell by 12 h and rebounded by 48 h, presumably because Dicer is needed for its own knockdown (Fig. 2C). Protein levels of Dicer remained low after 48 h (Fig. 2D). This transient loss of Dicer was associated with an ∼2-fold increase (p < 0.05) in endogenous FOXO1 mRNA and protein at 24 and 48 h. These data support the possibility that microRNAs play a role either directly or indirectly in the suppression of FOXO1 expression in human breast cancers.
FIGURE 2.
Knockdown of Dicer and Drosha resulted in increased FOXO1 mRNA and protein levels. A, quantitative PCR analysis of FOXO1 and Drosha mRNA levels at the indicated time points post-transfection of Drosha siRNA in MDA-231 cells. Relative mRNA expression was normalized to RPL-P0 (B634). Error bars indicate the S.E. of three independent experiments. B, representative Western blot of Drosha and FOXO1 protein levels after treatment of MDA-231 cells with Drosha siRNA. Numbers represent the average -fold change compared with mock for three independent experiments. C, quantitative PCR analysis of FOXO1 and Dicer mRNA levels at the indicated time points post-transfection of Dicer siRNA in MDA-231 cells. Relative mRNA expression was normalized to RPL-P0 (B634). Error bars indicate the S.E. of three independent experiments. D, representative Western blot of Dicer, FOXO1, and GAPDH mRNA after transfection with Dicer siRNA in MDA-231 cells. Numbers represent the average -fold change compared with mock for three independent experiments. *, p < 0.05; **, p < 0.01. M, molecular mass standards.
MicroRNA Regulation of the FOXO1 3′-UTR
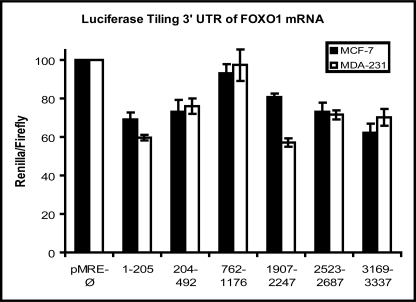
In mammals microRNAs commonly form imperfect duplexes with target sites within the 3′-UTR region of target mRNAs. The FOXO1 transcript harbors a very large 3′-UTR (∼3350 nucleotides) and, therefore, many potential microRNA binding sites. Predicted microRNA binding sites were mapped in the FOXO1 3′-UTR using target prediction databases (miRanda, Targetscan, and PicTar; supplemental Fig. S2 and Table S3). Potential microRNA sites formed six clusters within the FOXO1 mRNA 3′-UTR. To test whether these regions contained bona fide repressive elements, small sections of the FOXO1 3′-UTR were cloned into a dual luciferase reporter system so that the FOXO1 sequence was inserted into the 3′-UTR of the Renilla luciferase gene, and luciferase expression was compared with an empty vector construct in two different breast cancer cell lines, MCF-7 and MDA-MB-231 cells. The six sections of the FOXO1 3′-UTR (ranging in size from 164 to 413 base pairs) were analyzed, and several sections conferred up to 1.5-fold repression (Fig. 3). Five of the microRNA candidates (miR-27a, miR-96, miR-182, miR-183, and miR-128) were predicted to target the FOXO1 3′-UTR in two different locations (sections 204–492 and 1907–2247), and consequently, these microRNAs were selected for further detailed analysis. Some of these microRNAs are predicted to target FOXO3a (miR-27a, miR-96, miR-182, miR-128) and FOXO4 (miR-96, miR-128). However, the regulation of these isoforms was not pursued in this study as the mRNA levels of these transcripts did not vary in normal versus tumor breast tissue.
FIGURE 3.
Repression of predicted microRNA target sites in the FOXO1 3′-UTR. Short fragments of the FOXO1 3′-UTR harboring predicted microRNA sites were PCR amplified and cloned into a dual luciferase reporter plasmid. Nucleotides corresponding to the segments of the 3′-UTR tested are indicated. Luciferase repression was conferred with Renilla activity and normalized to firefly activity.
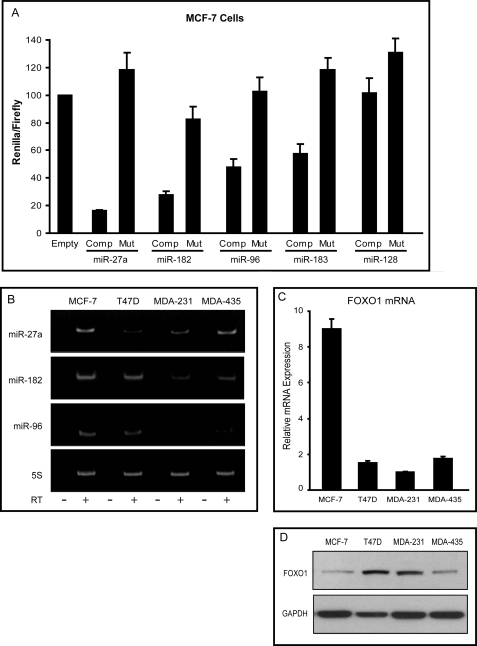
A dual luciferase reporter construct containing a target site perfectly complementary to the microRNA of interest, termed a “sensor” construct, was used to measure the endogenous activity of the candidate microRNAs. The binding of a microRNA to the target sequence provided would result in decreased luciferase activity because of the repressive nature of microRNAs in most cellular contexts. Four of the five microRNAs, as detected by the corresponding sensor constructs, displayed significant activity in MCF-7 cells: miR-27a (∼10-fold repression), miR-182 (∼3-fold repression), miR-96 (∼2-fold repression), and miR-183 (∼2-fold repression) (Fig. 4A). To ensure the specificity of these interactions, mutant constructs were synthesized that contained a 2-nucleotide change in the seed region of the target sequence (nucleotides 3 and 4). In each case the mutant construct abrogated the repressive activity. It should be noted that miR-96 and miR-182 possess identical seed sequences, so overlap in their respective activities may exist. Because miR-128 did not display activity in MCF-7 cells, it was not further studied. Additionally, later studies on miR-183 (Figs. 5 and supplemental Fig. S3) indicated that it neither interacted with the predicted miR-183 site in FOXO1 mRNA nor regulated FOXO1 protein expression. Thus, further study focused on miR-27a, miR-96, and miR-182.
FIGURE 4.
MicroRNA expression and activity in breast cancer cell lines. A, microRNA activity in MCF-7 cells was assessed using a dual luciferase reporter construct containing either a target sequence fully complementary to the microRNA (Comp) or a mutant sequence harboring a two-nucleotide change in the seed region of the target site (Mut). Activity is expressed as Renilla luciferase activity normalized to firefly luciferase activity to control for transfection efficiency. Values are represented as the average of three independent experiments ±S.E. B, semiquantitative end point PCR analysis of the expression of miR-27a, miR-182, miR-96, and 5 S ribosomal RNA in breast cancer cell lines. PCR products were resolved on non-denaturing acrylamide gels and stained with ethidium bromide. RT, reverse transcription. C, quantitative PCR analysis of FOXO1 mRNA in four breast cancer cell lines. Samples were normalized to RPL-P0 (BL34) expression. D, Western analysis of FOXO1 and GAPDH levels in breast cancer cell lines.
In addition to measuring the activity of the candidate microRNAs, the expression levels of the three most active microRNAs, miR-27a, miR-96 and miR-182, were assessed using semiquantitative end point PCR. Gene specific stem-loop reverse transcription primers were designed for each mature microRNA, and PCR amplification using gene-specific primers was carried out after cDNA synthesis. Expression of miR-27a, miR-96, and miR-182 was measured in four breast cancer cell lines (Fig. 4B), and all three microRNAs displayed robust levels in the MCF-7 cells. We also examined FOXO1 mRNA and protein levels in the four cell lines. Although levels are relative, MCF-7 cells showed a marked discrepancy between FOXO1 mRNA (Fig. 4C) and protein levels (Fig. 4D), suggesting microRNA-mediated translational repression rather than mRNA degradation as the primary mechanism of microRNA regulation. Therefore, we focused on MCF-7 cells to validate putative microRNA target sites and the effects of suppression of endogenous microRNA levels on FOXO1 expression.
As previously stated, sections harboring base pairs 204–492 (Site 1) and 1907–2247 (Site 2) of the FOXO1 3′-UTR each contained one binding site for miR-27a, miR-96, and miR-182. Thus, each construct (Site 1 or Site 2) was transfected into MCF-7 cells either alone or with antisense inhibitors to miR-27a (Fig. 5A), miR-96, or miR-182 (Fig. 5B). A non-targeting inhibitor was used as a negative control. Upon transfection of segments of the FOXO1 3′-UTR, a 1.5-fold repression was observed. In the presence of antisense inhibitors directed against miR-27a or miR-182, this repression was abrogated. It should be noted that inhibitor to miR-96 did not relieve the suppression from either site. These data indicate that the repression of endogenous FOXO1 3′-UTR sequence is specifically because of the activity of microRNAs, particularly miR-27a and miR-182.
MicroRNAs often regulate gene expression coordinately, and multiple microRNAs may act on a single target. To assess the combinatorial effect of multiple microRNA sites in the FOXO1 3′-UTR, a dual luciferase reporter construct containing a target site encompassing both active sections (Site 1 and Site 2) was created. When both sites were combined, they conferred a somewhat greater repression (∼2-fold) than each site alone (∼1.5-fold). To test if the candidate microRNAs examined specifically targeted these predicted regions, antisense inhibitors directed against the individual microRNAs (or an unrelated sequence) were co-transfected with the reporter constructs, and luciferase activity was measured in MCF-7 cells. Inhibitors against miR-27a, miR-182, and, unexpectedly, against miR-96, relieved repression, indicating that these microRNAs are responsible for the microRNA-mediated repression observed (Fig. 5C). This series of experiments was followed by insertion of 50-bp regions encompassing either the endogenous miR-27a or miR-96/182 sites into the luciferase reporter vector. Both sites conferred ∼1.5–2-fold repression of luciferase activity (Fig. 5D). Excision of the endogenous site (ΔmiR-27a or ΔmiR-96/182) abrogated repression. As mentioned above, the endogenous miR-183 site was devoid of suppressive activity in MCF-7 cells.
MicroRNAs Regulate Cell Proliferation and Survival Specifically through Their Suppression of Endogenous FOXO1 Protein
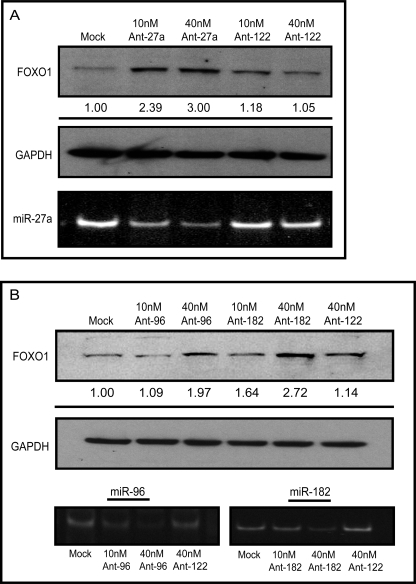
To assess the role of microRNA regulation on endogenous FOXO1 protein expression, antisense inhibitors directed against miR-27a (Fig. 6A) or miR-96 and miR-182 (Fig. 6B) were transiently transfected into MCF-7 cells and levels of FOXO1 protein were measured by Western blot. Inhibitors directed against miR-122, a liver-specific microRNA, were used as a negative control. Knockdown of the indicated microRNAs was confirmed using semiquantitative end point PCR analysis (Fig. 6, A and B, lower panels). Endogenous FOXO1 protein levels were increased by up to 3-fold in a dose-dependent manner upon knockdown of miR-27a, miR-96, and miR-182, indicating that these microRNAs coordinately regulate the expression of endogenous FOXO1. As discussed above, knockdown of inactive miR-183 had no effect of FOXO1 protein (supplemental Fig. S3). These data support the hypothesis that FOXO1 expression is repressed at the post-transcriptional level by one or more microRNAs.
FIGURE 6.
Regulation of endogenous FOXO1 levels by miR-27a, miR-96, and miR-182. MCF-7 cells were treated with antisense inhibitors targeting miR-27a (A) or miR-96 and miR-182 (B), or the unrelated microRNA miR-122 (Ant-122) at the indicated concentrations. Cells were harvested 48 h post-transfection, and FOXO1 and GAPDH levels were measured by Western analysis. Numbers represent the average -fold change compared with mock for three independent experiments. Knockdown of endogenous microRNAs by the appropriate inhibitors was verified using semi-quantitative reverse transcription-PCR.
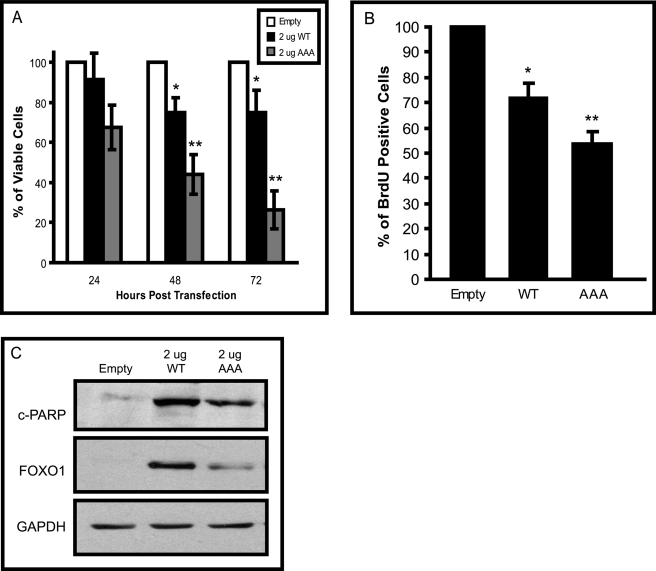
A previous study in MCF-7 cells showed that overexpression of FOXO1 decreased cell number and colony size (22). In this study we examined cell counts but also BrdUrd incorporation and cleaved poly(ADP-ribose) polymerase as assays of cell cycle and terminal cell death, respectively, in breast cancer cells overexpressing FOXO1. MCF-7 cells (which contain very low levels of FOXO1 protein) were transiently transfected with constructs expressing either wild-type FOXO1 (WT)* or a constitutively active FOXO1 mutant (AAA)* that cannot be phosphorylated and is, therefore, contained to the nucleus. Overexpression of FOXO1 was confirmed by Western analysis (supplemental Fig. S4_A_). In addition, proper localization of the FOXO1 expression constructs were confirmed using confocal microscopy (supplemental Fig. S4_B_). Cells transfected with empty vector did not express any fluorescence (data not shown). Overexpression of FOXO1 protein was detected in MCF-7 cells transfected with either a wild-type FOXO1 construct (WT) or the mutant construct (AAA). Overexpression of the wild-type FOXO1 construct resulted in protein primarily localized to the cytoplasm (presumably from regular turnover), whereas the mutant construct displayed protein primarily confined to the nucleus. Functionality of the vectors was also tested by measuring the regulation of known downstream targets of FOXO1 (p21 and SOD2) by quantitative real-time-PCR (supplemental Fig. S4, C and D, respectively). To assess if FOXO1 overexpression had a functional consequence, MCF-7 cell viability was measured by trypan blue staining. MCF-7 cells were seeded out in equal numbers and transfected with 2 μg of FOXO1 constructs or empty vector. Cells were harvested at 24-h time points and stained with trypan blue, and viable cells were counted in triplicate (Fig. 7A). Expression of wild-type FOXO1 (WT) resulted in a 25% decrease in cell viability 72 h post-transfection. A more dramatic result was observed with overexpression of the constitutively active mutant FOXO1 (AAA), which resulted in a dose dependent decrease in cell viability reaching a 70% decrease 72 h post-transfection. The percentage of viable cells for each treatment was normalized to the number of viable cells transfected with equivalent concentrations of empty vector.
FIGURE 7.
Restoring FOXO1 expression in MCF-7 cells results in decreased cell proliferation and induction of apoptosis. A, MCF-7 cells were seeded at equal densities and transfected with the indicated amount of DNA. Empty vector (Empty), wild-type FOXO1 (WT), and constitutively active mutant (AAA). Viable cells were counted after staining with trypan blue at the indicated time points post-transfection. B, BrdUrd incorporation in MCF-7 cells transfected with empty vector, wild-type FOXO1 (WT), or mutant FOXO1 (AAA). C, Western blot of cleaved poly(ADP-ribose) polymerase (c-PARP; top panel) after transfection with the indicated vectors. Middle panel, overexpression of FOXO1 was confirmed. The Western blot is a representative analysis from three independent experiments. All error bars represent the S.D. from four independent experiments. *, p < 0.05; **, p < 0.01.
To further expand these studies, BrdUrd staining was conducted to observe the effects of FOXO1 overexpression on cell proliferation. MCF-7 cells were transfected with either empty vector, wild-type FOXO1 (WT), or mutant FOXO1 (AAA), and the percentage of BrdUrd-positive cells was assessed 24 h post-transfection (Fig. 7B). MCF-7 cells overexpressing wild-type FOXO1 (WT) had ∼30% decrease in proliferation, whereas cells overexpressing the mutant FOXO1 protein (AAA) had an ∼50% decrease in cell proliferation. To evaluate whether the observed decrease in cell viability was also because of the induction of apoptosis, cleaved poly(ADP-ribose) polymerase expression was measured after overexpression of FOXO1 in MCF-7 cells (Fig. 7C). A significant increase in cleaved poly(ADP-ribose) polymerase was observed after transfection of either wild-type FOXO1 or mutant FOXO1, and overexpression of these constructs was confirmed in these samples. Levels of GAPDH were assessed as a loading control and remained unchanged after overexpression of FOXO1. These results show that the restoration of FOXO1 in MCF-7 breast cancer cells results in reduced cell number because of a decrease in proliferation and induction of apoptosis. This phenotype is further exacerbated when a FOXO1 constitutively active mutant is overexpressed.
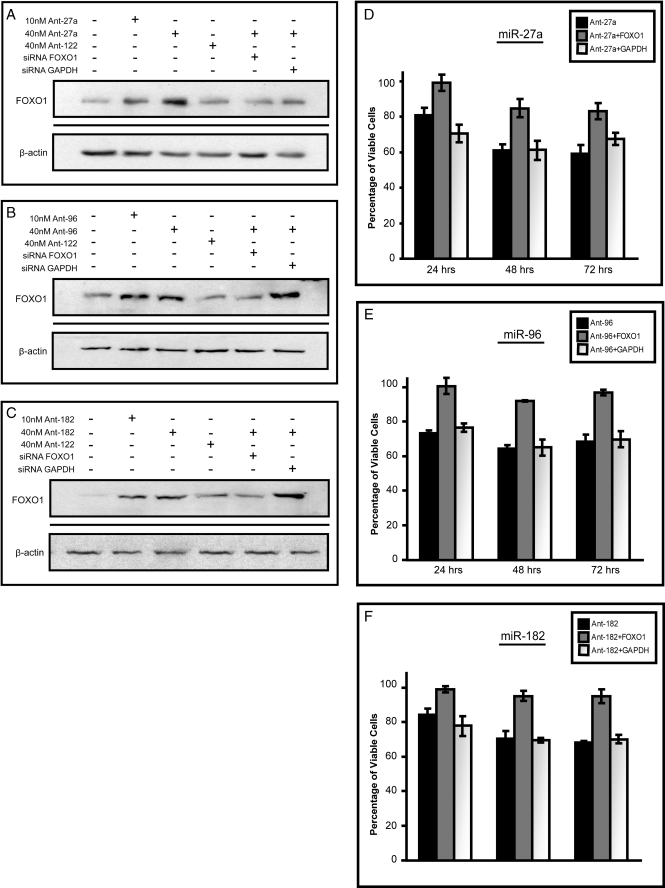
Next, we examined whether the down-regulation of miR-27a, miR-96, or miR-182 reduced cell number in a manner that could be “rescued” by siRNA to FOXO1. Again, antisense inhibitors against either miR-27a, miR-96, or miR-182, but not miR-183, increased endogenous FOXO1 protein levels in MCF-7 cells (Fig. 8, A–C, supplemental Fig. S3_A_), and this was associated with a ∼30–40% reduction in cell number by 72 h relative to mock-transfected controls (Fig. 8, D–F). Co-transfection with FOXO1 siRNA, but not GAPDH siRNA, significantly blunted the effects of the antisense inhibitors against miR-27a, miR-96, and miR-182. These data provide direct evidence that the specific suppression of FOXO1 contributes to the growth-promoting actions of these three microRNAs in MCF-7 cells.
FIGURE 8.
Inhibiting miR-27a, miR-96, and miR-182 results in decreased cell viability because of increased FOXO1 expression. A–C, Western blot analysis of FOXO1 expression after the treatment of MCF-7 cells with antisense inhibitors (Ant) to miR-27a (A), miR-96 (B), or miR-182 (C) or inhibitors plus siRNA to either FOXO1 or GAPDH. Treatments were carried out for 48 h. β-Actin was used as a loading control. D–F, MCF-7 cells were plated at 2.5 × 105 cells per well and treated with either 40 nm antisense inhibitors (Ant) to miR-27a (D), miR-96 (E), or miR-182 (F) or inhibitors plus siRNA targeting FOXO1 or GAPDH. At each time point wells were trypsinized, stained with trypan blue, and counted four separate times using a hemacytometer. Bars represent the average number of viable cells ±S.E. of three independent experiments.
DISCUSSION
The FOXO gene family encodes tumor-suppressive transcription factors that regulate multiple aspects of cell cycle traverse and survival. With respect to estrogen-dependent breast cancer, it should also be noted that FOXO1 heterodimerizes with estrogen receptor α and inhibits its transcriptional activity. A previous study showed that the overexpression of FOXO1 in MCF-7 cells resulted in decreased cell number and colony formation (22). In the present study we extended these findings by showing that overexpression of FOXO1 strongly inhibited proliferation and induced cell death in breast cancer MCF-7 cells. The dysregulation of cell cycle and evasion of apoptosis play a pivotal role in the development of cancer, and changes in FOXO1 expression and/or activity likely contribute to tumor progression. Although numerous studies have addressed the mechanisms by which FOXO1 protein activity and subcellular localization have been reported, little is known about the regulation of FOXO1 expression levels.
The levels of FOXO1 proteins are depressed in a variety of cancers, including prostate cancer, glioblastoma, and endometrial carcinoma (14, 23–25). Goto et al. (14) found that FOXO1 mRNA is ∼6-fold less in endometriod endometrial tumors compared with normal cycling endometrium. This group also measured the mRNA levels of the two other ubiquitously expressed FOXO family members, FOXO3a and FOXO4, and found that the levels of these messages remained unchanged between endometrial tumors and normal tissue. Similarly, in this study we have shown that FOXO1 mRNA is also down-regulated >2-fold in breast tumor samples compared with normal breast tissue. In contrast, the expression of the other FOXO family members (FOXO3a and FOXO4) that are expressed in breast did not vary significantly between normal and tumor breast tissue. FOXO3a and FOXO4 expression may change in response to specific conditions. For example, paclitaxel has been shown to induce FOXO3a in taxane-sensitive MCF-7 cells, resulting in the increased expression of the pro-apoptotic protein, Bim (26).
The mechanism by which FOXO1 expression is suppressed in breast or any other cancer has not been established. Goto et al. (14) addressed this question in endometriod endometrial carcinoma using two endometrial carcinoma cell lines, HEC-1B cells and Ishikawa cells. Ishikawa cells express FOXO1 at barely detectable levels, and these were not increased by proteasome inhibition. The authors also found that the low levels of FOXO1 in Ishikawa cells were not because of promoter methylation or a decrease in promoter activity. Subsequently, they measured the rate of FOXO1 mRNA turnover in each cell line and discovered that cells that expressed low levels of FOXO1 protein (Ishikawa) had a much faster rate of mRNA decay (half-life of ∼1.8 h) compared with the HEC-1B cells (half-life of ∼4.5 h), which express higher levels of FOXO1 protein. Based on this evidence, the authors concluded that the down-regulation of FOXO1 in endometrial carcinoma cells was because of some form of post-transcriptional regulation.
One centrally important mode of post-transcriptional regulation is the repression of mRNA transcripts by microRNAs. Therefore, we hypothesized that microRNAs may play a role in maintaining low levels of FOXO1 in breast cancer cells. We examined the potential involvement of microRNAs in three general ways. First, we knocked down two centrally important enzymes in microRNA biogenesis. Drosha excises the stem-loop microRNA precursor from within the longer transcribed primary-microRNA form. Dicer subsequently removes the hairpin from the precursor structure, yielding a microRNA:microRNA* duplex from which one strand is incorporated in the microRNA-induced silencing complex to repress the target mRNA molecule. In this study the knockdown of either Drosha or Dicer led to a significant, severalfold increase in the FOXO1 mRNA and protein levels in the MDA-MB-231 breast cancer cell line. This finding provides evidence that microRNAs are important for the suppression of endogenous FOXO1 expression in some forms of breast cancer but does not indicate whether FOXO1 mRNA is, in fact, a direct target of one or more microRNAs.
Second, we gained experimental evidence of putative microRNA target sites within the long FOXO1 mRNA 3′-UTR. In examining the human FOXO1 3′-UTR for potential microRNA target sites, we observed several clusters of target sites. As some microRNAs display cell-specific expression and not all predicted sites are functional, it was not surprising to observe that only some of these fragments conferred significant repression. However, two of these segments, termed Site 1 (nucleotides 1–205) and Site 2 (nucleotides 1907–2247) conferred a significant degree of repression, and further analysis of these sites led to the finding that miR-27a, miR-96, and miR-182 were predicted to target each site. Of note, miR-96 and miR-182 are transcribed from the same polycistronic microRNA cluster and have identical seed sequences. Thus, miR-96 and miR-182 are likely to have similar levels, and would be predicted to potentially share many of their targets. Additionally, functional compensation of target repression may occur in the absence of one of the microRNAs.
Analysis of miR-27a, miR-96, and miR-182 levels and activity in four breast cancer cell lines revealed that MCF-7 cells displayed the most robust expression. Interestingly, this was correlated with discordant mRNA and protein levels. Further analysis of these sites in the luciferase reporter assay showed that antisense inhibitors against miR-27a, miR-182, or miR-96 specifically blocked the repressive action of these sites. It should be noted that in this case, luciferase repression was not relieved to the level of the empty vector, and this could be because of overlap of coordinate microRNA regulation of these sequences. We also observed significantly greater repression when both sites were combined, as opposed to that conferred by each site alone. Other studies have demonstrated the coordinate regulation of a single mRNA transcript by multiple microRNAs. Krek et al. (27), the group that developed PicTar in a mammalian system, tested the prediction that the Mtpn gene was coordinately regulated by let-7, miR-124, and miR-375 using Western blot analysis and a functional luciferase assay. They observed that the exogenous addition of each individual microRNA in MIN6 mouse pancreatic cells was sufficient to regulate protein expression of Mtpn and repress luciferase activity of a construct harboring the Mtpn 3′-UTR. Furthermore, the greatest amount of luciferase repression was observed after the addition of all three microRNAs simultaneously, reinforcing the additive effect of combinatorial regulation by multiple microRNAs (27). In the present study we also observed significantly more repression from two cluster sites than by either site alone.
Third, we demonstrated that the microRNAs identified above regulate endogenous FOXO1 expression. In MCF-7 cells treated with increasing doses of antisense inhibitors, it was shown that knockdown of either miR-27a, miR-96, or miR-182, but not the unrelated miR-122, significantly increased endogenous FOXO1 levels by severalfold. Although FOXO/Daf-16 has been shown to repress the expression of a specific microRNA (lin-4), our findings provide the first evidence of the direct regulation of endogenous FOXO1 expression by specific microRNAs (28).
Previous to this study, miR-27a was implicated in breast cancer as an oncogenic microRNA. Mertens-Talcott et al. (29) found that miR-27a is highly expressed in breast cancer cells, and inhibition of this microRNA using antisense molecules in MDA-MB-231 cells decreased cell proliferation. This group also found that antisense RNA directed against miR-27a decreased the percentage of cells in S phase and increased the percentage of cells in the G2-M phase. Interestingly, the authors suggest that miR-27a targets genes involved in regulating the G2-M phase of the cell cycle and identify one potential target (Myt-1) (29). In a separate study, Scott et al. (30) reported that a pro-apoptotic dose of the inhibitor of histone deacetylases, LAQ824, rapidly decreases miR-27a levels in breast cancer SKBr3 cells. miR-27a was one of four microRNAs significantly up-regulated in renal cell carcinoma compared with normal kidney (31) and acts as an oncogene in gastric adenocarcinoma (32). miR-27 has also been linked to hepatic stellate proliferation, in which it targets the retinoid X receptor (33). These findings provide supporting evidence that miR-27a can act as an oncogenic microRNA and warrant further study of miR-27a regulation of downstream targets of FOXO1 to better characterize the involvement of miR-27a in maintaining a state of uncontrolled cell growth.
Although miR-96 and miR-182 have not been studied previously with respect to breast cancer, previous work has shown that miR-96 and miR-182 are dysregulated in human disease, including a variety of cancers. Loscher et al. (34) showed that miR-96 and miR-182 are down-regulated in a mouse model of retinitis pigmentosa. These microRNAs are contained in a gene cluster harboring miR-96, miR-182, and miR-183, and this cluster is frequently amplified in advanced human melanoma (35) and melanoma cell lines (36). It has also been shown that miR-96 and miR-182 are overexpressed in colorectal cancer (37), classic Hodgkin lymphoma tumors (miR-182) and cell lines (miR-96) (38), and chronic myeloid leukemia cells (39). A recent study implicates miR-182 in the promotion of melanoma metastasis. Overexpression of miR-182 in melanoma cell lines resulted in enhanced oncogenic properties, such as anchorage-independent growth and increased colony formation on soft agar as well as invasion and metastasis in vitro (36). Furthermore, this study identified a FOXO family member, FOXO3, as a direct target of miR-182. These studies implicate miR-182 and miR-96 as oncogenic microRNAs because of their frequent overexpression in cancers and the observation that their identified target genes are involved in the regulation of cell proliferation and apoptosis.
In summary, this study demonstrated that of the three ubiquitously expressed FOXO family members, FOXO1 is selectively down-regulated in breast cancer as compared with normal tissue. We also demonstrate that FOXO1 expression is regulated by multiple microRNAs (miR-27a, miR-96, and miR-182) that have previously been implicated in oncogenesis. These microRNAs directly target various regions of the 3′-UTR to repress endogenous expression of FOXO1. Blockade of these microRNAs led to restoration of FOXO1 expression. The restoration of FOXO1 expression in MCF-7 cells resulted in reduced cell number, decreased cell cycle traverse, and increased cell death. This effect was further pronounced when a constitutively active mutant FOXO1 protein was overexpressed. Additionally, suppression of FOXO1 restoration by siRNA specifically blocked the anti-proliferative effects of microRNA down-regulation, thereby functionally linking microRNA expression, FOXO1 expression, and cell proliferation and/or viability. These findings indicate that antisense targeting of miR-27a, miR-96, and miR-182 along with monitoring of microRNA and FOXO1 levels may be of therapeutic and/or prognostic value in breast cancer.
Supplementary Material
Supplemental Data
Acknowledgments
We greatly appreciate the gift of FOXO1 wild-type (WT) and mutant (AAA) overexpression constructs from Dr. Terry G. Unterman (Dept. of Medicine, University of Illinois, Chicago, IL).
2
The abbreviations used are:
UTR
untranslated region
siRNA
small interfering RNA
GAPDH
glyceraldehyde-3-phosphate dehydrogenase
BrdUrd
bromodeoxyuridine
NGS
normal goat serum
WT
wild type
AAA
constitutively active FOXO1 mutant.
REFERENCES
- 1.Gross D. N., van den Heuvel A. P., Birnbaum M. J. (2008) Oncogene 27,2320–2336 [DOI] [PubMed] [Google Scholar]
- 2.Fu Z., Tindall D. J. (2008) Oncogene 27,2312–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obsil T., Obsilova V. (2008) Oncogene 27,2263–2275 [DOI] [PubMed] [Google Scholar]
- 4.Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) Cell 128,309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rena G., Guo S., Cichy S. C., Unterman T. G., Cohen P. (1999) J. Biol. Chem. 274,17179–17183 [DOI] [PubMed] [Google Scholar]
- 6.Guo S., Rena G., Cichy S., He X., Cohen P., Unterman T. (1999) J. Biol. Chem. 274,17184–17192 [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Gan L., Pan H., Guo S., He X., Olson S. T., Mesecar A., Adam S., Unterman T. G. (2002) J. Biol. Chem. 277,45276–45284 [DOI] [PubMed] [Google Scholar]
- 8.Perrot V., Rechler M. M. (2005) Mol. Endocrinol. 19,2283–2298 [DOI] [PubMed] [Google Scholar]
- 9.Matsuzaki H., Daitoku H., Hatta M., Tanaka K., Fukamizu A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,11285–11290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H., Regan K. M., Wang F., Wang D., Smith D. I., van Deursen J. M., Tindall D. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazumdar A., Kumar R. (2003) FEBS Lett. 535,6–10 [DOI] [PubMed] [Google Scholar]
- 12.Lengyel F., Vértes Z., Kovács K. A., Környei J. L., Sümegi B., Vértes M. (2007) Steroids 72,422–428 [DOI] [PubMed] [Google Scholar]
- 13.Jackson J. G., Kreisberg J. I., Koterba A. P., Yee D., Brattain M. G. (2000) Oncogene 19,4574–4581 [DOI] [PubMed] [Google Scholar]
- 14.Goto T., Takano M., Albergaria A., Briese J., Pomeranz K. M., Cloke B., Fusi L., Feroze-Zaidi F., Maywald N., Sajin M., Dina R. E., Ishihara O., Takeda S., Lam E. W., Bamberger A. M., Ghaem-Maghami S., Brosens J. J. (2008) Oncogene 27,9–19 [DOI] [PubMed] [Google Scholar]
- 15.Goto T., Takano M., Hirata J., Tsuda H. (2008) Br. J. Cancer 98,1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel D. P. (2004) Cell 116,281–297 [DOI] [PubMed] [Google Scholar]
- 17.Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) Nature 455,64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Nature 455,58–63 [DOI] [PubMed] [Google Scholar]
- 19.Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Science 316,604–608 [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Science 316,608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. (2007) Science 316,575–579 [DOI] [PubMed] [Google Scholar]
- 22.Zhao H. H., Herrera R. E., Coronado-Heinsohn E., Yang M. C., Ludes-Meyers J. H., Seybold-Tilson K. J., Nawaz Z., Yee D., Barr F. G., Diab S. G., Brown P. H., Fuqua S. A., Osborne C. K. (2001) J. Biol. Chem. 276,27907–27912 [DOI] [PubMed] [Google Scholar]
- 23.Dong X. Y., Chen C., Sun X., Guo P., Vessella R. L., Wang R. X., Chung L. W., Zhou W., Dong J. T. (2006) Cancer Res. 66,6998–7006 [DOI] [PubMed] [Google Scholar]
- 24.Li R., Erdamar S., Dai H., Wheeler T. M., Frolov A., Scardino P. T., Thompson T. C., Ayala G. E. (2007) Hum. Pathol. 38,1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seoane J., Le H. V., Shen L., Anderson S. A., Massagué J. (2004) Cell 117,211–223 [DOI] [PubMed] [Google Scholar]
- 26.Sunters A., Fernández de Mattos S., Stahl M., Brosens J. J., Zoumpoulidou G., Saunders C. A., Coffer P. J., Medema R. H., Coombes R. C., Lam E. W. (2003) J. Biol. Chem. 278,49795–49805 [DOI] [PubMed] [Google Scholar]
- 27.Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37,495–500 [DOI] [PubMed] [Google Scholar]
- 28.Baugh L. R., Sternberg P. W. (2006) Curr. Biol. 16,780–785 [DOI] [PubMed] [Google Scholar]
- 29.Mertens-Talcott S. U., Chintharlapalli S., Li X., Safe S. (2007) Cancer Res. 67,11001–11011 [DOI] [PubMed] [Google Scholar]
- 30.Scott G. K., Mattie M. D., Berger C. E., Benz S. C., Benz C. C. (2006) Cancer Res. 66,1277–1281 [DOI] [PubMed] [Google Scholar]
- 31.Gottardo F., Liu C. G., Ferracin M., Calin G. A., Fassan M., Bassi P., Sevignani C., Byrne D., Negrini M., Pagano F., Gomella L. G., Croce C. M., Baffa R. (2007) Urol. Oncol. 25,387–392 [DOI] [PubMed] [Google Scholar]
- 32.Liu T., Tang H., Lang Y., Liu M., Li X. (2009) Cancer Lett. 273,233–242 [DOI] [PubMed] [Google Scholar]
- 33.Ji J., Zhang J., Huang G., Qian J., Wang X., Mei S. (2009) FEBS Lett. 583,759–766 [DOI] [PubMed] [Google Scholar]
- 34.Loscher C. J., Hokamp K., Wilson J. H., Li T., Humphries P., Farrar G. J., Palfi A. (2008) Exp. Eye Res. 87,529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin W. M., Baker A. C., Beroukhim R., Winckler W., Feng W., Marmion J. M., Laine E., Greulich H., Tseng H., Gates C., Hodi F. S., Dranoff G., Sellers W. R., Thomas R. K., Meyerson M., Golub T. R., Dummer R., Herlyn M., Getz G., Garraway L. A. (2008) Cancer Res. 68,664–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura M. F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S., Zakrzewski J., Blochin E., Rose A., Bogunovic D., Polsky D., Wei J., Lee P., Belitskaya-Levy I., Bhardwaj N., Osman I., Hernando E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106,1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bandrés E., Cubedo E., Agirre X., Malumbres R., Zárate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzó M., García-Foncillas J. (2006) Mol. Cancer 5,29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navarro A., Gaya A., Martinez A., Urbano-Ispizua A., Pons A., Balagué O., Gel B., Abrisqueta P., Lopez-Guillermo A., Artells R., Montserrat E., Monzo M. (2008) Blood 111,2825–2832 [DOI] [PubMed] [Google Scholar]
- 39.Agirre X., Jiménez-Velasco A., San José-Enériz E., Garate L., Bandrés E., Cordeu L., Aparicio O., Saez B., Navarro G., Vilas-Zornoza A., Pérez-Roger I., García-Foncillas J., Torres A., Heiniger A., Calasanz M. J., Fortes P., Román-Gómez J., Prósper F. (2008) Mol. Cancer Res. 6,1830–1840 [DOI] [PubMed] [Google Scholar]
- 40.Varkonyi-Gasic E., Wu R., Wood M., Walton E. F., Hellens R. P. (2007) Plant Methods 3,12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data