Alternating Access of the Putative Substrate-Binding Chamber in the ABC Transporter MsbA (original) (raw)
. Author manuscript; available in PMC: 2010 Oct 30.
Published in final edited form as: J Mol Biol. 2009 Aug 26;393(3):574–585. doi: 10.1016/j.jmb.2009.08.051
Summary
MsbA is an ATP binding cassette (ABC) transporter from E. coli involved in trafficking lipid A across the inner membrane. ABC transporters harness the free energy of ATP binding and hydrolysis to drive the uphill translocation of substrates against their concentration gradients. A model of protein motion coupling energy input to work was inspired by crystallographic snapshots of MsbA. Central to this model is a switch in the accessibility of a transmembrane chamber, implicated in substrate binding, from an inward- to an outward-facing configuration. Here, we used spin labeling and electron paramagnetic resonance (EPR) spectroscopy to systematically explore rearrangements in MsbA structure during the ATP hydrolysis cycle. Spin label accessibility and local dynamics were determined in liposomes for the nucleotide-free intermediate and the transition state of ATP hydrolysis. The changes in the EPR parameters between these two intermediates fit a global pattern consistent with alternating access of the chamber. In the transition state of ATP hydrolysis, spin labels on the cytoplasmic side report increased dynamic restrictions and reduced water accessibility while those on the extracellular side report increased water penetration. Furthermore, spin label mobility and accessibility as well as their changes are consistent with those expected based on the crystal structures. The reversal in chamber hydration is likely to reduce the free energy of amphipathic substrate binding and promote translocation across the membrane.
Keywords: ABC transporter, electron paramagnetic resonance (EPR), site-directed spin labeling, MsbA
Cellular homeostasis requires the energetically uphill transport of a wide range of molecules across membranes. Active transporters couple vectorial translocation of molecules against their concentration gradients to thermodynamically favorable processes such as ATP hydrolysis or ions moving down their electrochemical gradients. In prokaryotes, ATP energy powers the largest class of transporters, the ATP-binding cassette (ABC) transporters, to traffic a structurally and chemically diverse spectrum of molecules1. Bidirectional transport is accomplished by two distinct subclasses of ABC transporters referred to as importers and exporters 2. Both are invariably characterized by the presence of a conserved module, the ABC or nucleotide binding domain (NBD), that harnesses ATP energy for biological work. Substrate specificity is encoded in highly divergent transmembrane domains (TMD) that also provide the passageway in the membrane. Although importers and exporters have a common molecular architecture consisting minimally of two NBDs and two TMDs, the organization and packing of these four domains in the functional unit differ significantly2. Bacterial ABC exporters are homodimers of molecules consisting of fused ABC and TMD whereas importers assemble from four independent subunits. A cognate protein delivers the substrate to ABC importers while exporters pick up their substrates from the inner leaflet of the membrane.
Transport entails the transduction of ATP energy to alternately expose the transport pathway to opposite sides of the membrane 3. Crystallographic glimpses of ABC transporters 4–10 and analysis by spin labeling and EPR spectroscopy11–13 have led to models of the underlying molecular motion. The exporter-centric model is primarily based on studies of MsbA, the putative lipid A flippase from E. coli14. MsbA crystallizes in three different conformations: two inward-facing and one outward-facing10. In all three structures, the TMD consists of two wings each containing helices from both monomers. The cross-over helices hold the dimer together and impart a remarkable degree of conformational flexibility. The nucleotide-free (apo) structure is defined by a V-shaped chamber open to the cytoplasm and the inner leaflet of the bilayer. In this structure, referred to as the open apo, the two NBDs are disengaged and separated by 50Å. In contrast, when bound to the non-hydrolyzable ATP analog, AMPPNP, the structure is tightly packed at the cytoplasmic side where the NBDs form the canonical dimer15. The TMD wings point away from each other in the outer leaflet of the bilayer resulting in an outward-facing conformation. The open-closed crystallographic transformation implies extensive molecular motion involving dissociation and association of the ATP dimer sandwich, swapping and repacking of helices in the transmembrane domain and rearrangements in the extracellular loops such that the inward and outward openings are mediated by different sets of helices. While the AMPPNP-bound closed conformation has been observed in two crystal structures of an MsbA homolog, Sav18664, the rearrangements implied by the MsbA structures remain unique and at variance with the structural analysis of ABC importers captured in different intermediates. The importers structures suggest more subtle changes mediate the inward- to outward- facing transition wherein the two NBDs remain in contact throughout the cycle6.
We have used spin labeling and EPR spectroscopy16 to obtain a complementary perspective on the nature and amplitude of MsbA conformational motion in a native-like environment without conformational selectivity subjected by lattice forces. In addition to testing conflicting models of transport, our analysis seeks to place the crystal structures in the context of the ATPase cycle. In previous work 12, mobilities and accessibilities of spin labels along helices 2, 5 and 6 reported substantial changes in local environments upon ATP binding consistent with a transition from an inward- to an outward-facing conformation.
While a cursory, qualitative comparison of the EPR and the corrected crystal structures resolve earlier incongruencies brought about by incorrect helix assignment 17, verification of the conformational changes deduced from the crystal structures requires a more detailed quantitative comparison. In this manuscript and the accompanying report18, we expand the spin labeling analysis to develop a global perspective on the dynamic events driving the transport cycle. Here, residue accessibility and mobility along helices 3 and 4 and adjacent intracellular and extracellular segments are determined in the apo and high energy post-hydrolysis intermediates. Spin labels were introduced along the intracellular coupling helices, IH1 and IH2, that transmit the signal of ATP binding and hydrolysis. Spin labels along helix 3 which contact the transmembrane chamber are optimally located to report on the changes in accessibility during the ATPase cycle.
Results
ATPase activity of spin labeled mutants
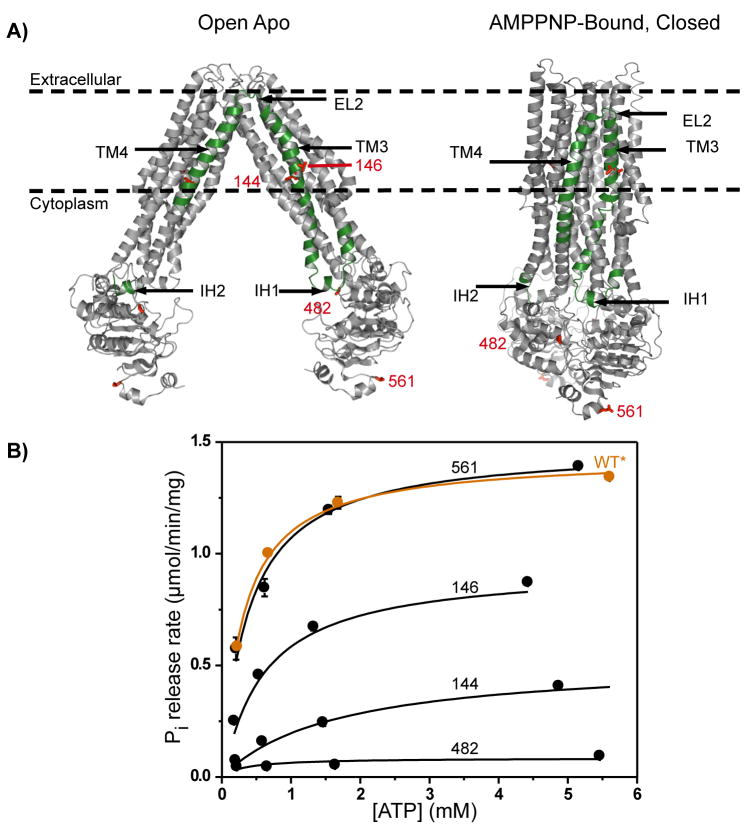
The spin label scan, highlighted in Figure 1 panel A, consists of residues in TM2 (104–111), IH1(112–120) and IH2(214–221), TM3(121–161), EL2 (162–166), TM4(167–193) and NBD (394, 482, 561). Following labeling and reconstitution, the ATPase activity of each spin labeled mutant was analyzed as described in the methods section and illustrated for representative sites in Figure 1B. The range of Vmax and Km reported in Table 1 suggests site-specific perturbation by the cysteine substitution and the subsequent labeling. Based on these values, the spin labeled mutants can be divided into three broadly defined classes. The first class consists of sites where the substitution severely reduces the catalytic efficiency. Notable in this class are residues in IH2 (supplementary table 1) where no ATPase activity is detected following reconstitution in liposomes. IH2 contacts the NBD of the opposite subunit contributing critical interactions for the swapped dimer. Indeed, sharp components in the corresponding EPR spectra suggest the presence of a population of locally or globally misfolded transporters (Supplemental Figure 1). Loss of ATPase activity is also observed at catalytic sites such as site 482 located in the LSGGQ motif of the NBD15. Therefore, motifs of nucleotide binding and catalysis in the NBD were not pursued in the spin label scan.
Figure 1.
A) Ribbon representation of the open and closed structures. Highlighted in green are the segments in the monomer subjected to cysteine substitution and spin labeling. The coordinates for the AMPPNP-bound structures were obtained from the protein data bank, PDB ID is 3B60. The model of the open-apo with side chains was provided by Dr. Geoffrey Chang, Scripps Research Institute. B) ATPase activity of MsbA cysteine-less (WT*) and representative mutants depicting three classes of activity ranging from unperturbed, intermediates and catalytically inactive.
Table 1.
ATPase activity of MsbA mutants
| Location | Mutant | Residue | Vmax (μmol/min/mg) | Km (mM) |
|---|---|---|---|---|
| TM1 | 24 | K | 1.48±0.32 | 0.85±0.55 |
| 28 | I | 1.47±0.19 | 0.30±0.16 | |
| 42 | T | 1.55±0.12 | 0.26±0.09 | |
| 43 | F | 1.42±0.01 | 0.31±0.01 | |
| 103 | R | 0.20±0.02 | 0.32±0.12 | |
| 104 | L | 0.13±0.03 | 1.12±0.59 | |
| 105 | F | 0.20±0.06 | 0.14±0.15 | |
| 106 | G | 0.35±0.04 | 0.54±0.18 | |
| 107 | H | 0.25±0.03 | 2.38±0.41 | |
| 108 | M | 0.24±0.06 | 1.97±1.13 | |
| 109 | M | 0.18±0.03 | 1.95±0.63 | |
| 110 | G | 0.27±0.01 | 1.53±0.17 | |
| 111 | M | 0.34±0.04 | 0.60±0.20 | |
| IH1 | 113 | V | 0.21±0.06 | 0.99±0.73 |
| 114 | S | 0.21±0.05 | 0.76±0.24 | |
| 115 | F | 1.08±0.02 | 1.21±0.06 | |
| 116 | F | 0.16±0.06 | 0.33±0.03 | |
| 117 | D | 0.56±0.02 | 1.90±0.14 | |
| 118 | K | 0.42±0.03 | 1.27±0.25 | |
| 119 | Q | 1.42±0.04 | 0.80±0.06 | |
| 120 | S | 0.29±0.02 | 1.48±0.29 | |
| TM3 | 121 | T | 0.49±0.07 | 2.27±0.80 |
| 122 | G | 0.35±0.02 | 0.71±0.13 | |
| 123 | T | 0.34±0.05 | 3.18±0.91 | |
| 124 | L | 0.30±0.01 | 3.48±0.22 | |
| 125 | L | 0.29±0.03 | 2.97±0.54 | |
| 126 | S | 0.67±0.01 | 0.37±0.02 | |
| 127 | R | 0.40±0.01 | 1.41±0.06 | |
| 128 | I | 0.13±0.04 | 2.96±0.54 | |
| 129 | T | 0.42±0.01 | 1.42±0.07 | |
| 130 | Y | 0.92±0.05 | 0.68±0.11 | |
| 131 | D | 0.37±0.02 | 2.04±0.26 | |
| 132 | S | 0.87±0.08 | 2.06±0.26 | |
| 133 | E | 0.42±0.02 | 1.83±0.15 | |
| 136 | A | 0.81±0.02 | 1.06±0.05 | |
| 137 | S | 0.74±0.03 | 1.15±0.14 | |
| 138 | S | 0.59±0.01 | 1.17±0.06 | |
| 139 | S | 0.83±0.04 | 1.37±0.16 | |
| 140 | S | 1.63±0.21 | 0.57±0.25 | |
| 141 | G | 0.27±0.03 | 1.32±0.39 | |
| 142 | A | 1.48±0.03 | 0.27±0.02 | |
| 143 | L | 1.01±0.18 | 1.95±0.81 | |
| 144 | I | 0.52±0.04 | 1.46±0.29 | |
| 145 | T | 0.31±0.03 | 0.11±0.05 | |
| 146 | V | 0.98±0.03 | 0.58±0.05 | |
| 147 | V | 0.34±0.02 | 1.42±0.16 | |
| 148 | R | 0.17±0.02 | 0.28±0.17 | |
| 149 | E | 0.18±0.02 | 1.20±0.37 | |
| 150 | G | 0.30±0.06 | 0.96±0.56 | |
| 151 | A | 0.72±0.06 | 0.49±0.13 | |
| 152 | S | 0.56±0.03 | 0.84±0.15 | |
| 153 | I | 0.78±0.05 | 1.49±0.20 | |
| 154 | I | 0.32±0.01 | 0.52±0.06 | |
| 155 | G | 0.65±0.105 | 0.86±0.38 | |
| 156 | L | 0.6±0.02 | 1.22±0.11 | |
| 157 | F | 1.03±0.04 | 1.17±0.12 | |
| 158 | I | 0.16±0.01 | 1.05±0.11 | |
| 159 | M | 1.01±0.04 | 1.69±0.15 | |
| 160 | M | 1.40±0.02 | 0.42±0.02 | |
| 161 | F | 1.95±0.03 | 0.43±0.02 | |
| EL2 | 162 | Y | 0.27±0.01 | 0.69±0.07 |
| 163 | Y | 0.15±0.01 | 1.07±0.15 | |
| 164 | S | 0.45±0.01 | 0.23±0.02 | |
| 165 | W | 1.77±0.03 | 0.42±0.02 | |
| 166 | Q | 1.77±0.01 | 0.37±0.01 | |
| TM4 | 167 | L | 0.25±0.01 | 0.76±0.07 |
| 168 | S | 0.43±0.03 | 0.42±0.02 | |
| 169 | I | 0.48±0.02 | 1.34±0.13 | |
| 170 | I | 0.53±0.01 | 1.39±0.09 | |
| 171 | L | 0.48±0.04 | 0.68±0.16 | |
| 172 | I | 0.49±0.02 | 1.29±0.13 | |
| 173 | V | 0.66±0.15 | 0.74±0.06 | |
| 174 | L | 0.61±0.04 | 1.20±0.17 | |
| 175 | A | 0.46±0.04 | 0.46±0.05 | |
| 176 | P | 0.35±0.01 | 1.01±0.10 | |
| 177 | I | 0.56±0.05 | 1.60±0.29 | |
| 178 | V | 0.59±0.02 | 2.03±0.15 | |
| 179 | S | 1.27±0.04 | 0.46±0.05 | |
| 180 | I | 0.36±0.01 | 1.23±0.11 | |
| 181 | A | 0.32±0.02 | 0.69±0.12 | |
| 182 | I | 1.27±0.05 | 0.81±0.09 | |
| 183 | R | 0.67±0.07 | 1.61±0.43 | |
| 184 | V | 1.76±0.06 | 0.42±0.05 | |
| 185 | V | 1.81±0.06 | 0.29±0.09 | |
| 186 | S | 0.41±0.04 | 1.05±0.26 | |
| 187 | K | 0.35±0.02 | 1.37±0.17 | |
| 188 | R | 0.20±0.15 | 2.46±1.07 | |
| 189 | F | 0.37±0.04 | 1.32±0.33 | |
| 190 | R | 0.54±0.01 | 1.38±0.08 | |
| 191 | N | 0.67±0.02 | 1.21±0.11 | |
| 192 | I | 0.29±0.02 | 1.43±0.18 | |
| 193 | S | 0.13±0.01 | 0.20±0.05 | |
| NBD | 394 | D | 1.41±0.07 | 0.47±0.10 |
| 482 | S | 0 | 0 | |
| 561 | T | 1.48±0.07 | 0.39±0.07 | |
| WT* | 1.48±0.09 | 0.33±0.09 |
Less severe perturbations are observed in the second class consisting predominantly of tertiary contact sites where the destabilization reflects the free energy cost of introducing the spin label. Reduced ATP affinity and hydrolysis rates are consistent across the IH1, and residues of TM3 at the dimer interface of the closed structure (e.g. 116,125 and 141). This is not unexpected given the repacking required to accommodate the spin label molar volume in buried regions. One notable exception in this class are exposed Gly residues (106,110,122,150) where the substitution consistently reduced Vmax. The third class consists of exposed residues in the membrane or in solution where wild-type Vmax and Km are observed. It includes NBD residues 394 and 561 and most of the lipid-facing residues in helices 3 and 4 and ECL2.
Spin label accessibility in the apo intermediate
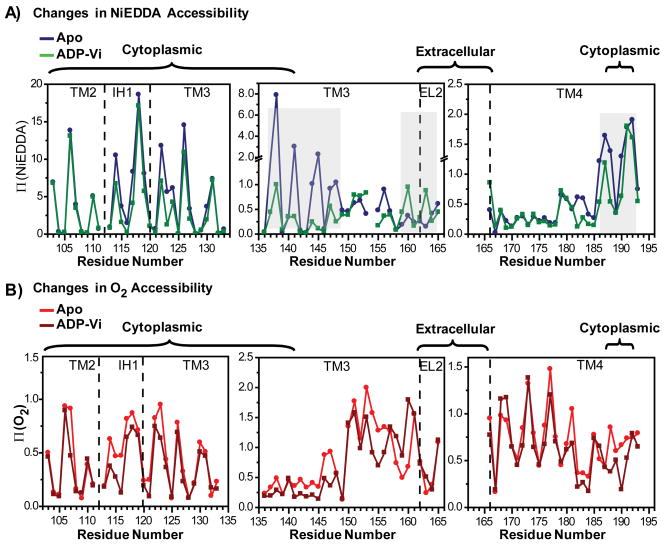
Collision frequencies with molecular O2 and NiEDDA were determined in the apo intermediate for spin labeled mutants reconstituted in liposomes. The sequence-specific patterns of the accessibility parameter Π (Figure 2) reflects the helical character of the backbone varying with 3.6 periodicity across IH1 and long stretches of helices 3 and 4. The accessibility values at the maxima reflect the differential solubility of the two relaxants in the water and bilayer phases and are also shaped by the tertiary contacts of the helices. Thus, large collision frequencies with NiEDDA are observed at solvent exposed sites in IH1 consistent with its expected location well beyond the membrane/water interface. The accessibilities become progressively smaller as the spin label approaches the bilayer and helix 3 weaves in and out of tertiary contacts with helices 1 and 2. Small Π(NiEDDA) values are observed in transmembrane regions reflecting the low solubility of this reagent in lipids. The uniformly low NiEDDA accessibility of EL2 places this loop in a lipid environment. Π(NiEDDA) gradually increases towards the cytoplasmic side as helix 4 emerges at the membrane/water interface.
Figure 2.
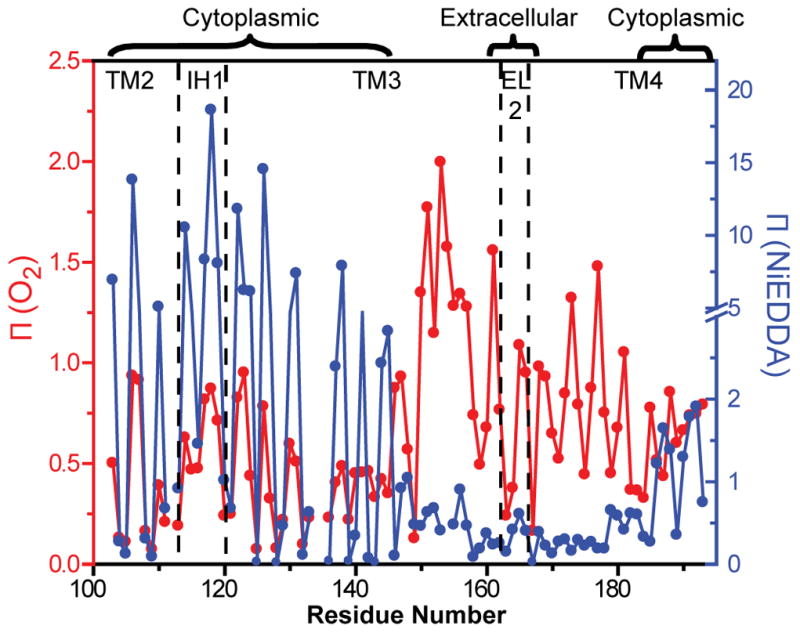
Accessibility profiles of Apo MsbA. Red Π (O2), blue (NiEDDA). Both parameters show periodic variation as a function of residue number. Chamber-facing residues of helix 3 report large accessibility to NiEDDA. In contrast, EL2 appears to be in the lipid bilayer. The dashed lines indicate helix and loop boundaries on the basis of the crystal structure assignment.
In contrast, O2 accessibilities increase in amplitude towards the middle of the bilayer in a manner consistent with the O2 concentration gradient19. The lipid-exposed face of a TM helix is defined by the Π (O2) maxima while the minima delineate the packed interface with other helices. This motif is generally observed in Figure 2 except in the 150–158 region where Π (O2) values are uniformly high. This indicates tertiary interactions with other TM helices are loose or absent. In both crystal structures, this region of helix 3 is not tightly packed presumably allowing significant access to O2. The high and periodic Π (O2) along EL2 suggest an ordered helical backbone buried in the membrane consistent with its low NiEDDA accessibility. A signature pattern of O2 accessibility is observed along TM4 reflecting its peripheral location in the crystal structure.
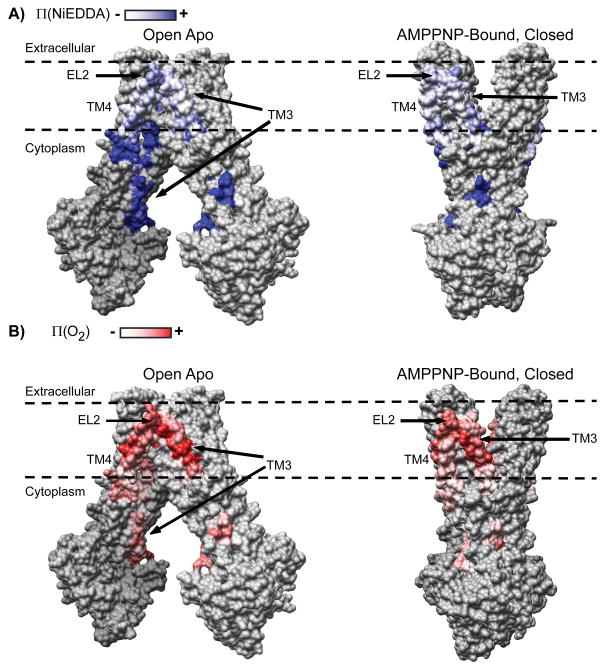
To obtain a qualitative perspective on the compatibility between the EPR data and the crystal structures, the accessibility values were mapped onto a surface representation of both MsbA structures (Figure 3). Residues with high Π (NiEDDA) are located on the cytoplasmic side in the open structure facing the chamber. In contrast, the same residues, such as 141 and 144, are buried at the TMD interface in the AMPPNP-closed structure. Thus, disruption of this interface is required to achieve the Π (NiEDDA) values observed in the apo intermediate. Mapping of the O2 accessibility data highlights the gradient of accessibility along the lipid facing surface of TM3 and TM4 and illustrates the membrane location of EL2 in the apo intermediate.
Figure 3.
A) Π (NiEDDA) mapped onto a surface rendering of the open and closed structures highlighting regions of water penetration in the chamber. B) Π (O2) mapped onto a surface rendering of the open and closed structures. The dashed lines represent the boundaries of the lipid bilayer determined from current and previous results.
Spin label accessibility in the ADP-Vi intermediate
To determine the accessibility changes in the TMD during the transport cycle, we measured spin label exposure to NiEDDA and O2 in a post ATP-hydrolysis intermediate trapped by Vanadate (Vi). This is the highest energy intermediate in the ATPase cycle of ABC transporters. Vi acts as γ-phosphate analog and, along with ADP, forms a complex mimicking the transition state of ATP hydrolysis20.
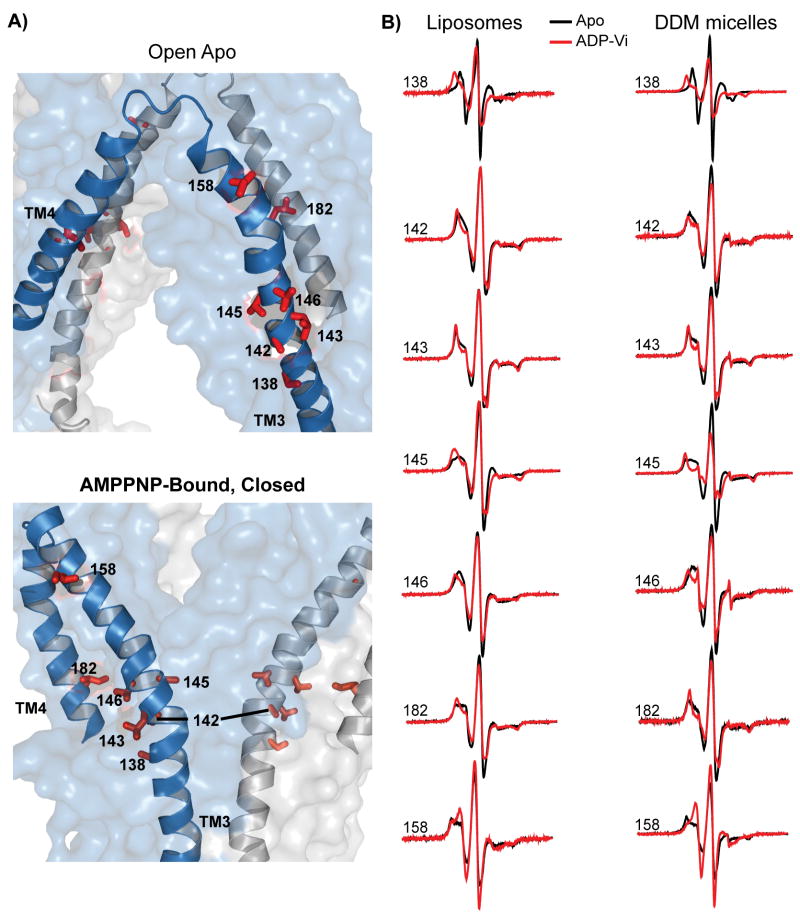
Formation of the ADP-Vi intermediate leads to opposite changes in Π (NiEDDA) on the two sides of helix 3. Whereas a decrease in accessibility is reported at chamber-facing residues in the cytoplasm, an increase is reported on the extracellular side, particularly at residues near EL2. In the absence of movement of this lipid-buried loop towards the extracellular side as demonstrated by the consistently high oxygen accessibility, the increase in NiEDDA accessibility at residues such as 160 and 163 (Figure 4) can only be due to an increase in water penetration well into the hydrophobic regions of the outer membrane leaflet.
Figure 4.
Changes in accessibility upon transition to the ADP-Vi intermediate. The dashed lines are as in Figure 2. Shading highlights regions of changes in Π (NiEDDA). These are clustered at the ends of helix 3 and in EL2. Changes in Π (O2) near the middle of helix 3 reflects the bending of this helix in the open/closed transformation (Figure 8).
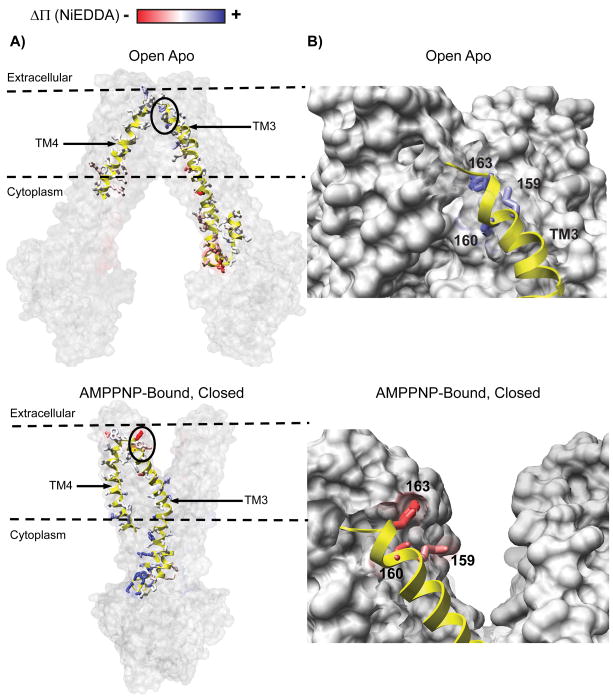
Mapping the changes in Π (NiEDDA) onto a ribbon representation of the open and closed structures reveals the overall pattern of accessibility changes as an alternation in the hydration profile at the interface of the two TMD wings (Figure 5A). For the open-apo structure, residues that report reduced NiEDDA accessibility cluster on the cytoplasmic side while they are on the extracellular side in the closed structure. A close-up view of the extracellular side highlights changes in accessibility of residues 159, 160 and 163 (Figure 5B). This is consistent with the position of EL2 in the closed structure bordering the extracellular opening. Overall, there is a general agreement between the EPR accessibilities changes induced by the transition between the apo and ADP-Vi intermediates and the inward-outward transition of the chamber in the crystal structures.
Figure 5.
A) Changes in NiEDDA accessibility, ΔΠ (NiEDDA), mapped onto a surface rendering of the open and closed structures. For clarity, ribbon representation is only shown for sites investigated in Figure 3. The opposite color distributions in the open and closed structures confirm the alternating access of the transmembrane chamber. B) Close up view of the extracellular region of helix 3 and EL2 showing their locations in the two structures. The transition predicts an increase in NiEDDA accessibility (blue) relative to the open state due to direct contact with the outward facing cavity in the closed structure.
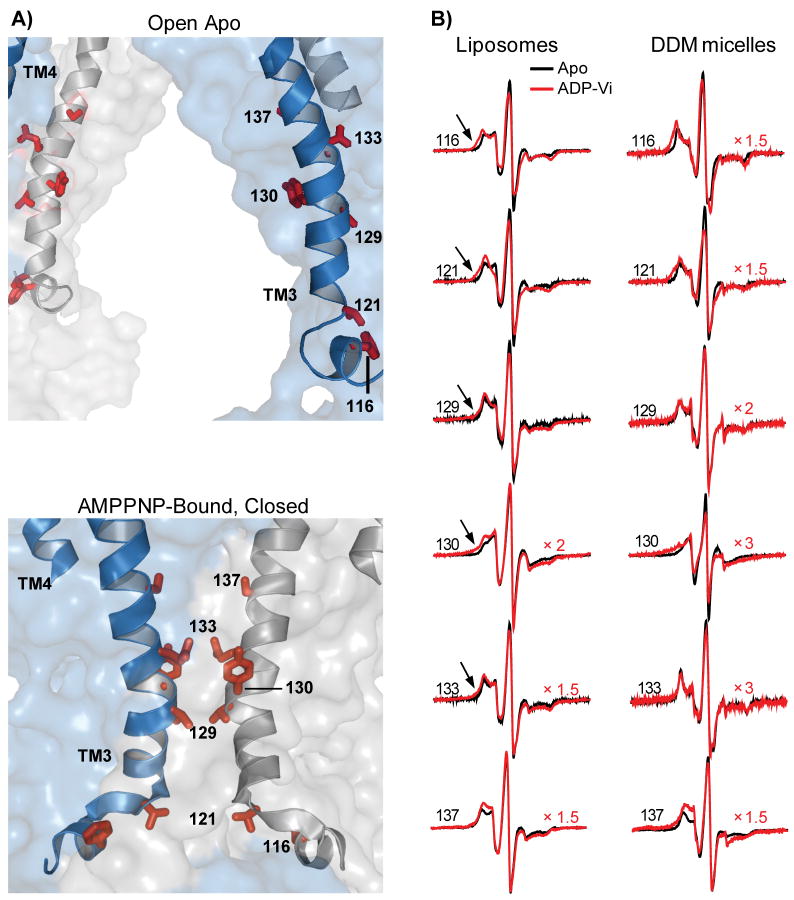
Spin label mobility in the open-closed transition
The transition to the ADP-Vi intermediate alters the dynamics of spin labels and changes their relative distances in the dimer. Starting in the cytoplasm and continuing through the TMD, spin labels at the interface report motional restrictions indicating a more sterically crowded environment21 in the post-hydrolysis intermediate. This is illustrated in Figure 6 for selected sites in IH1, helices 3 and 4 in detergent micelles and liposomes. In contrast, the pattern of mobility changes at the extracellular side reflects increased spin label dynamics. Site-specific lineshape changes are also observed at residues near helix-helix interfaces (e.g. site 158) consistent with the extensive repacking predicted by the crystal structures.
Figure 6.
A) Close up view of MsbA open and closed structures. The sites shown are expected to have different environments in the two structures. B) EPR lineshapes at representative sites in the apo and ADP-Vi intermediates showing changes in spin label local environments. Residues at the cytoplamic side report increased steric restrictions in the ADP-Vi intermediate consistent with the predicted environments in the closed conformation.
Evidence of dipolar coupling, spectral splitting and/or broadening22,23, are observed in the EPR lineshape upon formation of the ADP-Vi intermediate. Distinct spectral features (arrows in Figure 7) and/or reduction in intensity are observed at sites on the cytoplamsic side of helix 3. While these features indicate that the spin labels are brought closer by the formation of the ADP-Vi intermediate, the extent of broadening is less than expected based on relative distances in the closed crystal structure. The most likely origin of this discrepancy is perturbation and/or repacking induced by the spin labels. At sites where spin labels are predicted to be in van der Waals contacts, the catalytic efficiency is significantly impaired (Table 1). In the accompanying paper18 the magnitude of the distance is reported to be consistent with that predicted from the crystallographic transformation. However, at some of these sites there is a distinct population of spin labels that retain the distance characteristic of the apo intermediate. The consequence of this heterogeneity is an attenuation of the expected dipolar broadening observed by the CW-EPR lineshape analysis.
Figure 7.
A) Close up view of MsbA open and closed structures. The sites shown are expected to have close proximity (< 20Å) in the closed conformation. B) EPR lineshapes at representative sites in the apo and ADP-Vi intermediates. Residues at the cytoplamic side report increased steric restrictions in the ADP-Vi intermediate consistent with their environments in the closed conformation. The spectra in the ADP-Vi intermediate were scaled to highlight details of the lineshape.
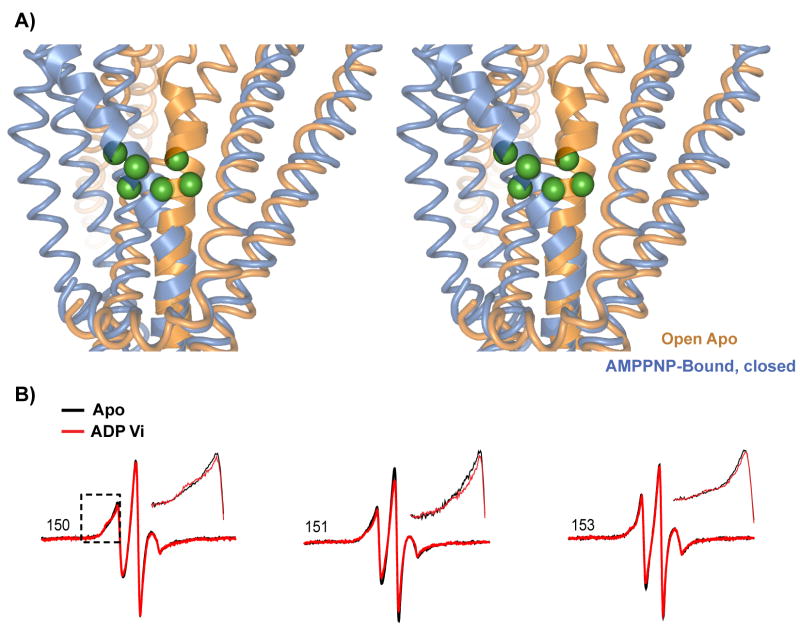
The EPR lineshapes also identify a highly dynamic region in helix 3. It corresponds to the segment of high Π (O2) discussed above. In addition to confirming the lack of strong tertiary contacts by this helix, the lineshapes at the surface sites 150, 151 and 153 are similar to those observed at loops21 suggesting a dynamic backbone. Comparison of the monomer structure in the open and closed conformations reveals that helix 3 undergoes a bending motion starting at the 150–154 turn (Figure 8). This motion, facilitated by the flexible backbone, is likely responsible for the change in Π (O2) upon ADP-Vi trapping (Figure 4B).
Figure 8.
A) Comparison of MsbA monomers in the open and closed conformations demonstrating the bending of helix 3. The NBDs have been removed to simplify the representation. The two structures were aligned by using helices 1 and 2. The highlighted α-carbons are for residues 150, 151, and 153. B) EPR lineshapes at sites in the 150–155 stretch of residues. The mobility if the spin label most likely reflects the flexibility required for the bending of the helix. The inset is an expanded view of the low field resonance line.
Discussion
How do ABC exporters move their substrates across membranes? Substrate binding from the inner leaflet of the bilayer or directly from the cytoplasm triggers the cycle. Although speculative at this point, the chamber at the interface of the two MsbA TMD wings is considered to be the substrate binding site24,25. Similar to other multidrug binding sites 26–28, this chamber has hydrophobic, aromatic and polar amino acids where multiple ligands can bind through different interactions with distinct sets of amino acids. Chamber accessibility to the inner leaflet of the bilayer and the cytoplasm allows efficient substrate access. The relatively large size, needed to accommodate the putative physiological substrate, lipid A, fits with the general theme of large and loose multidrug binding sites.
For amphipathic substrates, the high dielectric environment of MsbA’s interior chamber stabilizes their polar regions while more hydrophobic areas are favorably solvated by direct access to the bilayer. Unlike binding-protein dependent importers, MsbA displays high basal ATPase activity. Because the Km for ATP is well below the cellular concentration, the transporter only transiently samples the apo intermediate. Thus, a highly flexible open conformation may be critical for efficient substrate binding.
ATP binding and hydrolysis in the NBD trigger a series of conformational changes that promote substrate movement across the transport pathway. Our previous studies of MsbA12 along with functional data on homologous ABC exporters demonstrate that ATP binding provides the power stroke for ABC exporters. ATP binding occludes p-gp and LmrA29 cytoplasmic binding sites thereby lowering the substrate apparent affinity. The structural basis of this effect is the closing of the TM chamber manifested by a reduction in NiEDDA accessibilities of chamber-facing spin labels reported along five of the six helices of MsbA. In this model, the ATP-induced reversal in chamber polarity reduces the free energy of amphipathic substrate binding and promotes translocation to more hydrated binding sites presumably at the extracellular side.
Together, these results demonstrate that the transition to the ADP-Vi intermediate switches TMD accessibility from inward- to outward-facing. Overall, changes in EPR parameters in liposomes are consistent with the crystallographic transformation between apo-open and AMPPNP-closed conformations. At critical sites, spin label mobility exposure to NiEDDA can be rationalized from the expected local environments in each structure. Thus, these two structures are adequate models of the rearrangements induced during the ATPase cycle. The nature and amplitude of the underlying conformational motion is extensively mapped in the accompanying paper.
Materials and Methods
Materials
MTSSL (1-oxyl-2,2,5,5-tetramethylpyrrolinyl-3-methyl)-methanethiosulfonate spin label was obtained from Toronto Research Chemicals. n-Dodecyl-α-D-maltoside (α-ddm) was purchased from Anatrace Inc. Soybean polar lipid extract was obtained from Avanti Polar Lipids (Alabama, USA).
Expression and purification of MsbA mutants
MsbA cysteine mutants were generated by the QuikChange kit (Stratagene) as previously described12. All MsbA mutants were transformed into E. coli BL21 (DE3) cells and expressed in minimal media A30. Recombinant MsbA proteins were extracted with 1% α-dodecyl maltoside (α-ddm) from isolated membrane pellets and the supernatant loaded on a nickel affinity column (Ni-NTA, Qiagen) After elution from the Ni-NTA column in a buffer containing 0.015% w/v α-ddm, the protein was immediately labeled by addition of 20-fold molar excess MTSSL and incubated at room temperature for 4–6 hours followed by storage on ice overnight. Labeled proteins were further purified by size exclusion chromatography (SEC) on Superdex 200 column to remove free spin labels in an SEC buffer containing 50mM Hepes, 50mM NaCl, 0.015% w/v α-ddm at pH 7.5. Purified MsbA mutants were reconstituted in destabilized liposomes at the molar ratio of 1:1250 (protein: lipids). The mixture was incubated for 2 hours at 4°C. Subsequently, the detergent was removed by three additions of Bio-beads at 80mg/ml concentration. The recovered proteoliposome pellet was resuspended in buffer containing 50 mM Hepes, 50 mM NaCl, 2 mM MgCl2 at pH 7.5. To reconstitute MsbA trapped in the ADP-Vi intermediate, the protein was incubated in SEC buffer containing 10 mM ATP, 10 mM MgCl2 and 5 mM Vanadate (Vi) at 37°C for 20 minutes followed by reconstitution as outlined above.
ATPase assay
The ATPase activity of all MsbA mutants was determined using a colorimetric reaction as described previously12. Briefly, spin labeled MsbA mutants in liposomes were incubated with different concentrations of ATP at 37 °C for 20 minutes. The reaction was stopped by the addition of SDS. Released inorganic phosphate (Pi) which is directly proportional to the ATPase activity was determined by color development using a 1:1 solution of ammonium molybdate (2% in 1 M HCl) and ascorbic acid (12% in 1 M HCl) followed by absorbance measurement at 850 nm wavelength. The amount of phosphate released was determined by comparing with inorganic phosphate standards.
Electron Paramagnetic Resonance (EPR) spectroscopy
CW-EPR spectra of spin-labeled MsbA mutants were collected on a Varian E-109 spectrometer equipped with a loop-gap resonator 31,32. Samples were loaded in glass capillary tubes. The microwave power was 2 mW incident, the modulation amplitude was 1.6 G and the scan width used was 160 G. Power saturation experiments were carried out on a Bruker Elexsys 500 spectrometer at room temperature. Spin-labeled MsbA samples were loaded in gas-permeable TPX capillaries and the measurements were carried out under nitrogen and in the presence of 20 % oxygen or 50 mM NiEDDA. The data were analyzed to obtain the parameter P1/2. The EPR accessibility parameter Π was calculated as previously described33 except that spin labeled T4 lysozyme (135) was used as a standard.
Supplementary Material
01
02
Acknowledgments
This work was supported by National Institutes of Health Grant R01-GM077659 to Hassane S. Mchaourab. We thank Dr. Hanane Koteiche and Derek Claxton for critical reading of the manuscript and Jared Godar for critical reading of the manuscript and assistance with figure presentations. We also thank Dr. Geoffrey Chang (Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA) for sharing with us the coordinates of the apo MsbA structure.
Abbreviations
ABC
ATP-binding cassette
EPR
electron paramagnetic resonance
CW-EPR
continuous wave EPR
WT*
cysteine-less MsbA
MTSSL
1-oxyl-2,2,5,5-tetramethylpyrrolinyl-3-methyl)-methanethiosulfonate spin label
AMPPNP
5′-adenylyl imidodiphosphate
α-ddm
_n_-dodecyl-αD-maltoside
SEC
size exclusion chromatography
NiEDDA
nickel-ethylenediaminediacetic acid
NBD
nucleotide-binding domain
TMD
transmembrane domain
TM
transmembrane
IH
intracellular helix
EL
extracellular loop
Appendix A
Appendix B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annual Review of Biochemistry. 2004;73:241–68. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 2.Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Current Opinion in Structural Biology. 2008;18:726–33. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–70. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 4.Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter.[see comment] Nature. 2006;443:180–5. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 5.Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science. 2008;321:246–50. doi: 10.1126/science.1156213. [DOI] [PubMed] [Google Scholar]
- 6.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Molecular Cell. 2009;33:528–36. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism.[see comment] Science. 2002;296:1091–8. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 8.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–21. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 9.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315:373–7. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 10.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19005–10. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borbat PP, Surendhran K, Bortolus M, Zou P, Freed JH, Mchaourab HS. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. Plos Biology. 2007;5:e271. doi: 10.1371/journal.pbio.0050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J, Yang G, Mchaourab HS. Structural basis of energy transduction in the transport cycle of MsbA.[see comment] Science. 2005;308:1023–8. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- 13.Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12837–42. doi: 10.1073/pnas.0803799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doerrler WT, Raetz CR. ATPase activity of the MsbA lipid flippase of Escherichia coli [erratum appears in J Biol Chem 2002 Nov 15, 277(46):44588] Journal of Biological Chemistry. 2002;277:36697–705. doi: 10.1074/jbc.M205857200. [DOI] [PubMed] [Google Scholar]
- 15.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Molecular Cell. 2002;10:139–49. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–83. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 17.Chang G, Roth CB, Reyes CL, Pornillos O, Chen YJ, Chen AP. Retraction.[see comment][retraction of Chang G, Roth CB. Science. 2001 Sep 7;293(5536):1793–800; PMID: 11546864] Science. 2006;314:1875. [Google Scholar]
- 18.Zou P, Bortolus M, Mchaourab HS. Conformational Cycle of the ABC transporter MsbA in Liposomes. Detailed Analysis using Double Electron-Electron Resonance Spectroscopy. Journal of Molecular Biology. 2009 doi: 10.1016/j.jmb.2009.08.050. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–71. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma S, Davidson AL. Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. Journal of Bacteriology. 2000;182:6570–6. doi: 10.1128/jb.182.23.6570-6576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 22.McHaourab HS, Oh KJ, Fang CJ, Hubbell WL. Conformation of T4 lysozyme in solution. Hinge-bending motion and the substrate-induced conformational transition studied by site-directed spin labeling. Biochemistry. 1997;36:307–16. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- 23.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proceedings of the National Academy of Sciences of the United States of America; 1995. pp. 8239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.[see comment] Science. 2009;323:1718–22. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smriti, Zou P, Mchaourab HS. Mapping Daunorubicin-binding Sites in the ATP-binding Cassette Transporter MsbA Using Site-specific Quenching by Spin Labels. Journal of Biological Chemistry. 2009;284:13904–13. doi: 10.1074/jbc.M900837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural mechanisms of QacR induction and multidrug recognition. Science. 2001;294:2158–63. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- 27.Peters KM, Schuman JT, Skurray RA, Brown MH, Brennan RG, Schumacher MA. QacR-cation recognition is mediated by a redundancy of residues capable of charge neutralization. Biochemistry. 2008;47:8122–9. doi: 10.1021/bi8008246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumacher MA, Miller MC, Brennan RG. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO Journal. 2004;23:2923–30. doi: 10.1038/sj.emboj.7600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nature Structural & Molecular Biology. 2004;11:918–26. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 30.Koteiche HA, Reeves MD, Mchaourab HS. Structure of the substrate binding pocket of the multidrug transporter EmrE: site-directed spin labeling of transmembrane segment 1. Biochemistry. 2003;42:6099–105. doi: 10.1021/bi0342867. [DOI] [PubMed] [Google Scholar]
- 31.Koteiche HA, McHaourab HS. The determinants of the oligomeric structure in Hsp16.5 are encoded in the alpha-crystallin domain. FEBS Letters. 2002;519:16–22. doi: 10.1016/s0014-5793(02)02688-1. [DOI] [PubMed] [Google Scholar]
- 32.Sathish HA, Stein RA, Yang G, Mchaourab HS. Mechanism of chaperone function in small heat-shock proteins. Fluorescence studies of the conformations of T4 lysozyme bound to alphaB-crystallin. J Biol Chem. 2003;278:44214–44221. doi: 10.1074/jbc.M307578200. [DOI] [PubMed] [Google Scholar]
- 33.Altenbach C, Froncisz W, Hemker R, Mchaourab H. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys J. 2005;89:2103–12. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02