Cryptic Lineages of the Genus Escherichia (original) (raw)
Abstract
Extended multilocus sequence typing (MLST) analysis of atypical Escherichia isolates was used to identify five novel phylogenetic clades (CI to CV) among isolates from environmental, human, and animal sources. Analysis of individual housekeeping loci showed that E. coli and its sister clade, CI, remain largely indistinguishable and represent nascent evolutionary lineages. Conversely, clades of similar age (CIII and CIV) were found to be phylogenetically distinct. When all Escherichia lineages (named and unnamed) were evaluated, we found evidence that E scherichia fergusonii has evolved at an accelerated rate compared to E. coli, CI, CIII, CIV, and CV, suggesting that this species is younger than estimated by the molecular clock method. Although the five novel clades were phylogenetically distinct, we were unable to identify a discriminating biochemical marker for all but one of them (CIII) with traditional phenotypic profiling. CIII had a statistically different phenotype from E. coli that resulted from the loss of sucrose and sorbitol fermentation and lysine utilization. The lack of phenotypic distinction has likely hindered the ability to differentiate these clades from typical E. coli, and so their ecological significance and importance for applied and clinical microbiology are yet to be determined. However, our sampling suggests that CIII, CIV, and CV represent environmentally adapted Escherichia lineages that may be more abundant outside the host gastrointestinal tract.
It almost goes without saying that E scherichia coli is an important model organism and has been for more than 120 years. E. coli is often used to study bacterial adaptation (49), experimental evolution (9, 36), and speciation (11, 28, 33, 43). Comparisons between representative genomes continue to provide valuable information about these processes (58), and as genetic data accumulate, our understanding of how bacteria adapt to change and evolve is clarified. For example, E. coli lineages do not easily fit a biological species concept (43), as was previously suggested (11), so it may be more practical to identify lineages that share similar ecological niches rather than phylogenetic similarity alone (27).
E. coli adaptation to the gastrointestinal (GI) tract of warm-blooded animals is largely considered to be the most important source of natural selection during its evolution from a common ancestor with Salmonella. E. coli was originally discovered in and has long been associated with the indigenous microbiota of the GI tract of humans and animals. The vast majority of E. coli organisms transiently pass through the GI tract with little or no effect on the host (6, 46). A small number are able to establish and persist within a single host for months and years, presumably because of symbiotic relationships that develop with other constituents of the microbiota. Some isolates that reach the GI tract are pathogenic and are responsible for a variety of symptoms involving both intestinal and extraintestinal infections of humans and animals. Every year there are approximately 1 billion episodes of diarrhea among children under the age of 5 in developing countries, and 2 million of these cases result in death (35). Pathogenic E. coli is the most common bacterial cause of acute diarrhea in this pediatric population (35), making it a leading cause of diarrhea-associated deaths worldwide. The primary route of transmission for some E. coli, especially pathogenic lineages like Shigella spp., is thought to be the fecal-oral route. This observation underlies its global use as an indicator of water quality as well as its use in microbial source tracking studies whose aim is to determine the source of fecal contamination.
What is known about E. coli ecology (i.e., its abundance and distribution and the factors that influence them) is based on data from studies of representative isolates. For two main reasons, historical collections of isolates hold the potential for misleading conclusions. First, when isolates are assembled for analyses, most studies begin by confirming a phenetic species definition (50) (i.e., lactose fermentation, indole production, or lack of citrate utilization). All isolates that fit this definition are included even though they may not fit an alternative species definition, such as one based on phylogenetics (52). Second, the way that samples have been collected has not been uniform across habitats where E. coli is commonly found. A disproportionately large amount of data has been collected from E. coli isolated from humans and domesticated animals. Therefore, there is potential for under-sampled phenotypic and genotypic diversity from non-host-associated habitats, like the environment (5, 15, 59).
Characterizations of isolates from the environment are changing the way we view E. coli life history. A number of studies have found stable genotypes in the environment, suggesting that certain populations readily circulate and persist outside the host GI tract (5, 15, 24, 40, 59, 61, 62). These observations support the hypothesis that a significant proportion of the global E. coli population is not transmitted directly between hosts via the fecal-oral route but instead flow between the environment and hosts. In this study, we present the identification and genetic characterization of five novel Escherichia clades. Using extended multilocus sequence typing (MLST) analysis, we show that representative isolates formed highly supported and distinct genetic clusters. On the basis of biochemical profiling, we found that the clades were highly similar in phenotype and could not be easily distinguished from E. coli. We used PCR screening of genes in the E. coli pan-genome to show that there has been recent gene flow between E. coli and all other Escherichia species and clades and that the percentages of shared loci among clades were not well correlated with the sequence divergence between them. We also show that Escherichia species and clades differ in their rates of evolution and that the species Escherichia fergusonii appears to be evolving under nonneutral conditions. Lastly, since some of the clades appear to be overrepresented in habitats outside of the host GI tract, we discuss their potential importance and how they may add to our biological understanding of the model organism, E. coli.
MATERIALS AND METHODS
MLST and phylogenetic analyses.
Sequencing methodology used for MLST was carried out as part of a system described in detail elsewhere (www.shigatox.net/cgi-bin/mlst7/index). In total, we analyzed the internal fragments of 22 housekeeping genes (Table 1) at locations around the E. coli chromosome. Primers used to amplify these loci were obtained from a publicly available database at Michigan State University (MSU; www.shigatox.net [42]), a publicly available database at the University College Cork (UCC; mlst.ucc.ie/mlst/dbs/Ecoli) or a previously published E. coli MLST study (60), or they were designed de novo to target new MLST loci based on a recent analysis of E. coli MLST gene candidates (29). The PrimerSelect program of the DNASTAR Lasergene 7 package (DNASTAR, Inc., Madison, WI) was used for primer design from previously sequenced E. coli and S almonella enterica genomes. With the exception of three UCC primer pairs (for amplification of adk, fumC, and purA) that have an optimal annealing temperature of 54°C, all primers were designed to span the entire genetic diversity of Escherichia (i.e., all clades and species), to amplify at a single annealing temperature (58°C), and to use the same PCR conditions as the MSU MLST protocol (www.shigatox.net/stec/mlst-new/mlst_pcr.html).
TABLE 1.
Primers used in extended MLST analysis
| Locus | Size (bp) | Primera | Nucleotide position from start of locusb | Size of product (bp) | Annealing temp (°C) | Primer reference or sourcec | |
|---|---|---|---|---|---|---|---|
| Direction or namee | Sequence (5′-3′) | ||||||
| adk | 645 | F | ATTCTGCTTGGCGCTCCGGG | 10 | 584 | 54 | UCC |
| R | CCGTCAACTTTCGCGTATTT | 594 | UCC | ||||
| arcA | 717 | F | GACAGATGGCGCGGAAATGC | 99 | 552 | 58 | MSU |
| R | TCCGGCGTAGATTCGAAATG | 651 | MSU | ||||
| aroE | 819 | F | GGGGGCGTTTAAATCCTTCA | −69 | 759 | 58 | This study |
| R | GCCTCGCTGCTCACACCA | 690 | This study | ||||
| aspC | 1,191 | F | GTTTCGTGCCGATGAACGTC | 57 | 594 | 58 | MSU |
| R | AAACCCTGGTAAGCGAAGTC | 651 | MSU | ||||
| clpX | 1,275 | F | CTGGCGGTCGCGGTATACAA | 262 | 672 | 58 | MSU |
| R | GACAACCGGCAGACGACCAA | 934 | MSU | ||||
| cyaA | 2,547 | F | CTCGTCCGTAGGGCAAAGTT | 312 | 571 | 58 | MSU |
| R | AATCTCGCCGTCGTGCAAAC | 883 | MSU | ||||
| dnaG | 1,746 | F | CGCTGAACCCAATCGTCT | 765 | 696 | 58 | This study |
| R | TCTCTGAATAAGCCAAGTCCA | 1461 | This study | ||||
| fadD | 1,686 | F | GCTGCCGCTGTATCACATTT | 768 | 580 | 58 | MSU |
| R | GCGCAGGAATCCTTCTTCAT | 1348 | MSU | ||||
| fumC | 1,404 | F | TCACAGGTCGCCAGCGCTTCd | 10 | 806 | 54 | UCC |
| R | GTACGCAGCGAAAAAGATTCd | 816 | UCC | ||||
| grpE | 594 | F | CCCGGAAGAAATTATCATGG | 39 | 488 | 58 | MSU |
| R | TCTGCATAATGCCCAGTACG | 527 | MSU | ||||
| gyrB | 2,415 | F | GACGGGCGCGGCATTCC | 220 | 689 | 58 | This study |
| R | CTGTAGCCTTCTTTGTCCA | 909 | This study | ||||
| icdA | 1,251 | F1 | GCAACGTGGTGGCAGAC | −49 | 541 | 58 | This study |
| R1 | TTCGATACCCGCATAAAT | 492 | This study | ||||
| F2 | CTGCGCCAGGAACTGGATCT | 371 | 669 | 58 | MSU | ||
| R2 | ACCGTGGGTGGCTTCAAACA | 1020 | MSU | ||||
| kdsA | 855 | F | AAAAAGTGGTTAGCATTGG | 8 | 502 | 58 | This study |
| R | GCACCGCGATCGCAAAGAAT | 510 | This study | ||||
| lysP | 1,470 | F | CTTACGCCGTGAATTAAAGG | 36 | 628 | 58 | MSU |
| R | GGTTCCCTGGAAAGAGAAGC | 664 | MSU | ||||
| mdh | 939 | F | GTCGATCTGAGCCATATCCCTAC | 130 | 650 | 58 | MSU |
| R | TACTGACCGTCGCCTTCAAC | 780 | MSU | ||||
| metG | 2,034 | F | CACATCCAGGCTGATGTCTG | 85 | 573 | 58 | Johnson |
| R | CATTTTATTTGCCACCTGCTC | 658 | This study | ||||
| mtlD | 1,149 | F | GCAGGTAATATCGGTCGTGG | 22 | 658 | 58 | MSU |
| R | CGAGGTACGCGGTTATAGCAT | 680 | MSU | ||||
| mutS | 2,562 | F | GGCCTATACCCTGAACTACA | 1683 | 596 | 58 | MSU |
| R | GCATAAAGGCAATGGTGTC | 2279 | MSU | ||||
| purA | 1,299 | F | CGCGCTGATGAAAGAGATGA | 234 | 817 | 54 | UCC |
| R | CATACGGTAAGCCACGCAGA | 1051 | UCC | ||||
| recA | 1,062 | F | CGCATTCGCTTTACCCTGACCd | 185 | 734 | 58 | UCC |
| R | TCGTCGAAATCTACGGACCGGAd | 919 | UCC | ||||
| rpoS | 993 | F | CGCCGGATGATCGAGAGTAA | 274 | 618 | 58 | MSU |
| R | GAGGCCAATTTCACGACCTA | 892 | MSU | ||||
| torC | 1,173 | F | TGAATGGGCGCGAATGAAAGA | 375 | 630 | 58 | This study |
| R | GCGCCGTGGCACTGGTTACA | 1005 | This study |
Sequences were obtained from cultured isolates using standard Sanger-style sequencing at double coverage per locus or from the xBASE, version 2.0, sequence database (7) (Table 2). The SeqMan II program of the DNASTAR Lasergene 7 package (DNASTAR, Inc., Madison, WI) was used to align, edit, and trim sequences for analysis. Single-locus sequences were concatenated and loaded into the MEGA4 program (56) for descriptive statistics (the mean proportion of nucleotide differences [_p_-distance], Tajima's Test, and estimates of evolutionary divergence times) as well as for the generation of unweighted-pair group method using averages (UPGMA), maximum composite likelihood, minimum evolution, and neighbor-joining dendrograms. A split network analysis was generated from total concatenated sequences using the SplitsTree, version 4, program (21). The numerical taxonomy system, NTSYSpc, version 2.20e (Exeter Software, Inc., Setauket, NY), was used to perform a principal coordinate analysis of the Tajima's chi-square test statistics and an analysis of variance (ANOVA) on the first principal coordinates was generated using SAS statistical software (SAS Institute, Cary, NC).
TABLE 2.
List of representative isolates and sequences included in the MLST analysis
| Taxonomic group | Isolate no. | Location of isolation | Habitat or isolate origin | Sequence source | Isolate reference or source |
|---|---|---|---|---|---|
| E. coli | EDL933 | Michigan | Human | xBASE | 7 |
| 042 | Lima, Peru | Human | xBASE | 7 | |
| e2348/69 | England | Human | xBASE | 7 | |
| MG1655 | United States | Human | xBASE | 7 | |
| CFT073 | Maryland | Human | xBASE | 7 | |
| B692 | Australia | Bird | MLST | This study | |
| E677 | Australia | Environment | MLST | This study | |
| S. flexneri | 2457T | Japan | Human | xBASE | 7 |
| E. fergusonii | ATCC 35469T | MLST | |||
| B253 | Australia | Bird | MLST | This study | |
| B372 | Australia | Bird | MLST | This study | |
| B691 | Australia | Bird | MLST | This study | |
| E. albertii | 9194 | Bangladesh | Human | MLST | 23 |
| 19982 | Bangladesh | Human | MLST | 23 | |
| B090 | Australia | Bird | MLST | This study | |
| B101 | Australia | Bird | MLST | This study | |
| B156 | Australia | Bird | MLST | This study | |
| B198 | Australia | Bird | MLST | This study | |
| B249 | Australia | Bird | MLST | This study | |
| B992 | Australia | Bird | MLST | This study | |
| B1086 | Australia | Bird | MLST | This study | |
| K-1 | Bangladesh | Human | MLST | 23 | |
| C-425 | Bangladesh | Human | MLST | 23 | |
| CI | B827 | Australia | Bird | MLST | This study |
| E1492 | Australia | Environment | MLST | This study | |
| E807 | Australia | Environment | MLST | This study | |
| H442 | Australia | Human | MLST | This study | |
| M863 | Australia | Mammal | MLST | This study | |
| TW10509 | Guinea Bissau | Human | MLST | This study | |
| TW11930 | Guinea Bissau | Human | MLST | This study | |
| TW11966 | Guinea Bissau | Human | MLST | This study | |
| CII | B1147 | Australia | Bird | MLST | This study |
| CIII | TW09231 | Michigan | Freshwater beach | MLST | This study |
| TW09276 | Michigan | Freshwater beach | MLST | This study | |
| TW09266 | Michigan | Freshwater beach | MLST | This study | |
| TW09254 | Michigan | Freshwater beach | MLST | This study | |
| B685 | Australia | Bird | MLST | This study | |
| TA04 | Australia | Mammal | MLST | This study | |
| CIV | TW14182 | Michigan | Freshwater beach | MLST | This study |
| TW11588 | Puerto Rico | Soil | MLST | This study | |
| B49 | Australia | Bird | MLST | This study | |
| H605 | Australia | Human | MLST | This study | |
| CV | TW09308 | Michigan | Freshwater beach | MLST | This study |
| B1225 | Australia | Bird | MLST | This study | |
| B646 | Australia | Bird | MLST | This study | |
| E1118 | Australia | Environment | MLST | This study | |
| E1195 | Australia | Environment | MLST | This study | |
| E1196 | Australia | Environment | MLST | This study | |
| E471 | Australia | Environment | MLST | This study | |
| E472 | Australia | Environment | MLST | This study | |
| E620 | Australia | Environment | MLST | This study | |
| M1108 | Australia | Mammal | MLST | This study | |
| TA290 | Australia | Mammal | MLST | This study | |
| TW14263 | Michigan | Racoon | MLST | This study | |
| TW14264 | Michigan | Surface water | MLST | This study | |
| TW14265 | Michigan | Surface water | MLST | This study | |
| TW14266 | Michigan | Surface water | MLST | This study | |
| TW14267 | Michigan | Surface water | MLST | This study | |
| RL325/96 | Dog | MLST | 63 | ||
| Z205 | Parrot | MLST | 63 | ||
| S. enterica | Typhi TY2 | United States | Human | xBASE | 7 |
| Typhimurium LT2 | United States | Human | xBASE | 7 | |
| Enteritidis PT4 | xBASE | 7 | |||
| Gallinarum 287/91 | xBASE | 7 | |||
| S. bongori | 12149 | United Kingdom | Human | xBASE | 7 |
Phenotypic analysis.
Biochemical profiles were generated for isolates per the manufacturer's instructions using the BBL Crystal Identification System (Becton, Dickinson and Co., Sparks, MD). Nonmetric multidimensional scaling analysis was performed using the numerical taxonomy system NTSYSpc, version 2.20e (Exeter Software, Inc., Setauket, NY). This statistical technique is often used to both visualize the data and make judgments based on similarity when many variables are considered. It was used to assign a location to each isolate in three-dimensional (3D) space based on its ability to utilize 31 biochemical substrates. The more similar two isolates are, the closer they will be in 3D space. Isolates with identical biochemical abilities will have identical positions.
PCR screening.
The presence/absence of 27 loci previously found in E. coli was determined for all strains of E scherichia albertii, E. fergusonii, and clades I to V. Previously published primers and PCR conditions were used for chuA, TSPE4.C2, and yjaA (8). Likewise, the primers and conditions of Gordon et al. were used to amplify astA, eaa G, eaeA, fimH, fyuA, hlyD, ibeA, iha, iroN, iutA, _kpsMT_II, ompT, papAH, and _sfa_-foc (17). The presence of a generic marker of uropathogenic pathogenicity islands (PAI) was determined using primers and conditions of Johnson et al. (26). The primer sequences and reaction conditions of Tarr et al. were used to screen for stx1 and terC (57). Also, the primers and conditions from Stacy-Phipps et al. (51) were used to screen for the heat-labile toxin gene, elt. Two loci, gadAB and uidA, were included because they are typically present on the chromosome of most E. coli isolates (32). Methods for amplifying the gadAB locus were followed as published by McDaniels et al. (32). Primer sequences for the uidA locus were designed for this study to span the phylogenetic diversity of all isolates in the EcMLST database (www.shigatox.net) as well as the available online sequences of the National Center for Biotechnology Information database. The uidA primer sequences were 5′-CATTACGGCAAAGTGTGGGTCAAT-3′ (forward) and 5′-TCAGCGTAAGGGTAATGCGAGGTA-3′ (reverse), and the reaction conditions used were identical to those published on the EcMLST website (Table 1). Primers for cdtA, espC, pic, and sat as well as their appropriate reaction conditions were developed as part of a recent study of E. coli autotransporter genes (A. M. Nelson et al., unpublished data). The sequences for these loci were as follows: cdtA, 5′-TGCCGCTCTGACAGGTGGACTTA-3′ (forward) and 5′-GCCTTTAAAAACGGGGTGATACA-3′ (reverse); espC, 5′-GTTGGGGCTCGGACGACTTAT-3′ (forward) and 5′-CCGGCACCCTTGAATGTTAATT-3′ (reverse); pic, 5′-CGATGCCCCCGTAGACTTTGTTTC-3′ (forward) and 5′-TACCGTCTCCCCTTTTCAGTCCTC-3′ (reverse); and sat, 5′-TGGTAGCGGTGGTATTATCTTTGA-3′ (forward) and 5′-CGGCTTCTTTCGTTGTATCTGAGT-3′ (reverse). Reaction conditions for these four loci (cdtA, espC, pic, and sat) were as follows: 94°C for 10 min, followed by 40 cycles of 92°C for 30 s, 50°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 7 min.
RESULTS
Isolate collection.
The isolates included in this study were collected over a number of years as part of other research studies and were isolated from a range of habitats and hosts (Table 2). Isolates resembled “typical” E. coli but had noticeably divergent nucleotide sequences. Usually, they were identified during routine MLST analysis, but because they were noticeably divergent, they were previously excluded from published results. The collection included representative isolates from closely related Escherichia species (E. albertii, E. coli, and E. fergusonii) for comparison. Shigella flexneri was included as another representative E. coli strain. We should note for clarity that the genus and species designations of the four named Shigella spp. (Shigella boydii, Shigella dysenteriae, S. flexneri, and Shigella soneii) are clinically rather than phylogenetically defined (see references 12, 23, 30, 41, and 64) and should otherwise be thought of as pathogenic lineages of E. coli.
Extended MLST analysis.
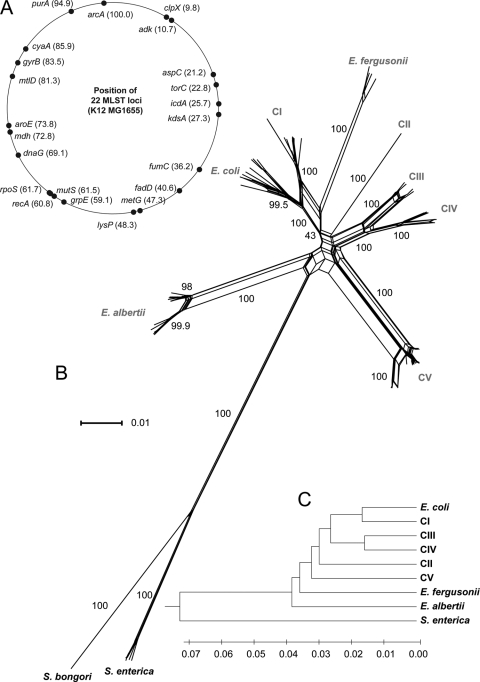
For phylogenetic comparison and evolutionary analyses, we developed an extended set of MLST primer pairs (Table 1) and used them to amplify and sequence 22 conserved loci around the genomes of representative isolates (Fig. 1A). This system yielded an average of 507 (138 variable and 119 phylogenetically informative) nucleotide sites per locus and 11,161 total base pairs of sequence. Concatenated sequences were aligned and used to generate a split network to represent the overall genetic diversity among isolates as well as any phylogenetic incompatibilities within and among taxa. This analysis resulted in nine highly supported clades (Fig. 1B). Isolates of the named species (E. coli, E. fergusonii, E. albertii, and S. enterica) clustered monophyletically with one another, and we named the remaining monophyletic clusters CI to CV.
FIG. 1.
The phylogenetic relationship of novel Escherichia lineages. (A) Position of MLST housekeeping genes relative to the E. coli K-12 MG1655 genome. (B) Split network of isolates showing the phylogenetic position of the five novel clades relative to previously named Escherichia species. (C) UPGMA dendrogram based on the proportion of nucleotide polymorphism differences (_p_-distance).
We next calculated the _p_-distance between clades and represented all pairwise comparisons in a UPGMA tree (Fig. 1C). The overall difference between clades ranged from 3.2% (± 0.13%) between CIII and CIV to 8.8% (± 0.24%) between E. albertii and E. fergusonii. Each of the five novel clades was more closely related to E. coli than to S. enterica (on average, 6.0% different from E. coli compared to 14.6% different from S. enterica).
To assess the extent to which each locus yielded congruent phylogenies, we generated single-locus neighbor-joining trees using the maximum composite likelihood model (for bootstrap values of trees, see Table S1 in the supplemental material). Only three trees (for lysP, rpoS, and fumC) showed a monophyletic relationship (i.e., isolates of a clade clustered together) for all eight clades. (A relationship for the single isolate of CII was not determined because, by definition, it is monophyletic at every locus. In other words, at least two isolates are needed per clade to compare single gene phylogenies). E. coli and CI isolates were rarely monophyletic (only 6 and 7 of 22 trees, respectively) and often clustered together. There were roughly the same number of phylogenetically informative sites among E. fergusonii, CIII, and CIV, yet these clades were monophyletic for 12, 14, and 15 loci, respectively. The groups with the greatest number of congruent trees were CV and E. albertii (18 and 21 trees, respectively). The torC phylogeny was the most incongruent with other single-locus phylogenies (none of the clades were monophyletic at this locus). We found eight examples where isolates from two different clades shared an allele of a locus. Four of the eight examples occurred between E. fergusonii and CI isolates (aroE, grpE, mutS, and purA), and the other four occurred between CI and CV (fadD), CIV and CV (dnaG), CIII and CIV (kdsA), and CI and E. coli (adk).
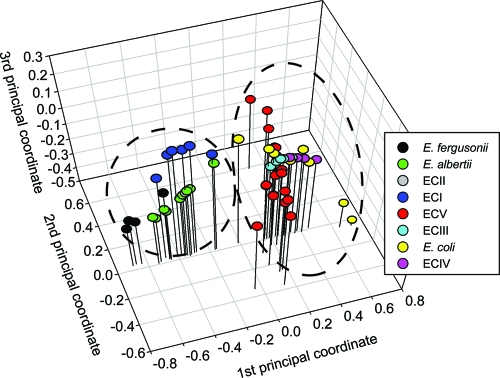
We tested for differences in the relative evolutionary rates among the clades and restricted our analysis to sites in the third codon position so as to maximize the signal from synonymous (neutral) polymorphisms and minimize the effect of selection. A pairwise matrix of chi-square test statistics and probabilities was generated (Tajima's test) for all isolates using S. enterica subsp. enterica serovar Typhimurium LT2 as the outgroup. From this matrix, we calculated the mean probabilities and standard error for each possible clade-wise (i.e., between clade) comparison. The evolutionary rate of E. fergusonii was statistically different from that of E. coli, CIII, CIV, and CV (see Fig. S1 in the supplemental material). There also appeared to be two or possibly three discreet groups of probabilities (i.e., different rates) in the plot. To look for patterns among the relative rates, we conducted a principal coordinate analysis on the pairwise matrix of chi-square test statistics. This analysis was used to visualize any differences among evolutionary rates based on the chi-square test statistic of Tajima. If two isolates have similar evolutionary rates (i.e., similar test statistics), they will be positioned similarly in coordinate space so that any grouping would indicate similar rates. Two main groups were observed. One included isolates of E. coli, CIII, CIV, and CV, and the other included isolates of E. fergusonii, E. albertii, CI, and CII (Fig. 2). ANOVA of the first principal coordinate positions showed that clades could be statistically differentiated [F(7, 59) = 52.11; P < 0.0001] into three groups (_E. fergusonii_-_E. albertii_-CII, _E. albertii_-CII-CI, and CV-CIII-_E. coli_-CIV) although the separation of E. fergusonii from CII and CI is likely to be biased by low sample size (i.e., one of the four E. fergusonii isolates is not statistically different from E. albertii, CII, and CI).
FIG. 2.
Principal coordinates analysis of all pairwise (strain by strain) chi-square test statistics for Tajima's test of relative evolutionary rates. The analysis ordered isolates into two groups, suggesting that there are two distinct evolutionary rates among the phylogenetic groups of Escherichia species in this study. ECI to ECV, Escherichia clades I to V.
We estimated the divergence times of lineages that gave rise to each clade using a minimum evolution tree, which corrects for multiple hits (i.e., polymorphisms) per nucleotide site. Assuming that E. coli split from S. enterica between 100 to 160 million years ago (mya), we estimated that these Escherichia lineages shared a common ancestor between 48 and 75 mya (Table 3). Four lineages (E. albertii, E. fergusonii, CII, and CV) split between 38 and 75 mya, and the youngest four lineages (E. coli, CI, CIII, and CIV) split between 19 and 31 mya. The _E. coli_-CI and CIII-CIV splits occurred more recently and at approximately the same point in time (19 to 31 mya), yet CIII and CIV are already more phylogenetically distinct than E. coli and CI.
TABLE 3.
Estimated divergence times based on minimum evolutionary analysis
| Lineages split | Divergence time (mya)a | ||
|---|---|---|---|
| Early | Middle | Late | |
| Escherichia from S. enterica | 160 | 120 | 100 |
| E. albertii from E. fergusonii, E. coli, and CI to CV | 75 | 58 | 48 |
| CIII, CIV, and CV from E. fergusonii, E. coli, CI, and CII | 64 | 49 | 41 |
| CII from E. fergusonii, E. coli, and CI | 60 | 46 | 38 |
| E. fergusonii from E. coli and CI | 59 | 46 | 38 |
| CV from CIII and CIV | 59 | 45 | 38 |
| CI from E. coli | 31 | 24 | 20 |
| CIII from CIV | 30 | 23 | 19 |
PCR screen for variably present/absent loci.
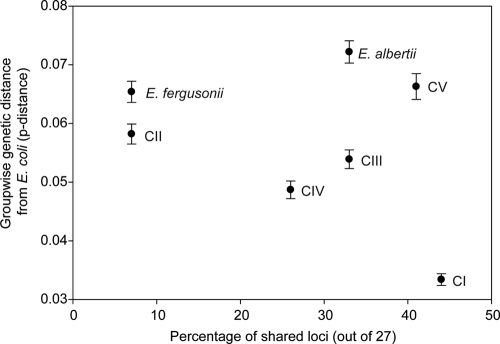
We conducted a PCR-based screen of non-E. coli isolates (E. albertii, E. fergusonii, and CI to CV) for the presence of 27 loci of the E. coli pan-genome. We were unable to detect the presence of nine loci (malX, eaaG, _sfa_-focDE, sat, stx1, hlyD, papA, pic, or espC) in any non-E. coli isolate. For the remaining 18 loci, the number of isolates that produced an amplicon of the correct size ranged from 1 to 40 (see Table S2 in the supplemental material). The loci fyuA, iha, and terC were present in only one isolate from CV, E. albertii, and CIII, respectively. The most abundant loci, uidA (40 isolates), gadAB (39 isolates), and fimH (38 isolates), were also the most cosmopolitan among the Escherichia clades. For example, the uidA locus, which is often used to differentiate E. coli from other species (44), was present in all isolates except E. albertii, E. fergusonii, and 2 of 18 CV isolates. One might expect that closely related isolates (i.e., separated by a short genetic distance) would be more likely to share loci. However, the number of E. coli pan-genome loci shared among the clades was not well correlated with genetic distance. For example, CV and E. albertii share more than or as many of these loci as the more closely related clades CIII and CIV (Fig. 3).
FIG. 3.
Relationship between genetic distance (proportion of nucleotide polymorphisms) and the presence of 27 E. coli pan-genome genes among Escherichia phylogenetic groups. No clear association was found between genetic distance and the percentage of shared loci.
Phenotypic analysis.
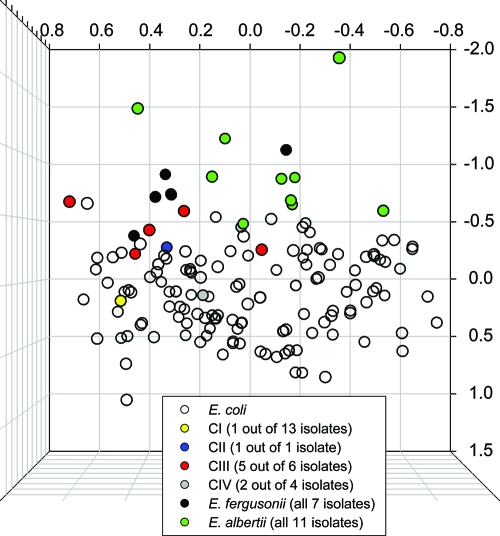
We generated profiles of biochemical and enzymatic reaction patterns for isolates of E. albertii, E. fergusonii, and CI to CV for 31 substrates that are typically used for species level identification of bacteria within the Enterobacteriaceae. For comparison, we included profiles from 692 E. coli isolates that were originally sampled from humans, mammals, and the environment. To visualize the phenotypic variation among and between isolates and to look for patterns, we conducted a numerical taxonomy analysis using nonmetric multidimensional scaling (NMDS). We used the NMDS algorithm to assign each isolate a position in 3D space and plotted isolates with the 50 highest and lowest positions on all three axes (Fig. 4). This analysis showed that isolates of E. coli, E. albertii, and E. fergusonii occupy different areas of 3D space (only unique profiles are represented in the figure). In contrast, 1 of the 6 CIII isolates, 2 of 4 CIV isolates, 12 of 13 CI isolates, and all 18 CV isolates were not represented because they had profiles that could not be distinguished from typical E. coli profiles.
FIG. 4.
NMDS of biochemical profiles for named Escherichia species (green and black circles) and novel Escherichia clades (gray, red, blue, and yellow circles) relative to 692 E. coli isolates (unfilled circles). Only the isolates that represent the 50 most positive and negative positions for each axis are shown. The number of isolates from each clade is given in parentheses. Isolates of the novel clades are highly similar to E. coli (CI to CIV) or completely indistinguishable (CV).
DISCUSSION
The taxonomy of Escherichia.
Since E. coli was made the type species of the genus Escherichia in 1919, five sister species have been added and remain nominally Escherichia. In 1973, Burgess et al. described the isolation of two distinct biotypes from the hindgut of cockroaches (Blatta orientalis) and proposed that these isolates represent a new species called Escherichia blattae (4). This designation was based on its phenotypic similarity to E. coli and, despite the presence of at least four incongruent biochemical reactions, E. blattae remains the second oldest Escherichia species behind E. coli. In 1982, DNA-DNA hybridization was used as a secondary criterion in the description of Escherichia hermannii (2) and Escherichia vulneris (3). Based on a small number of representative isolates, both of these species share minimal genetic similarity with E. coli (∼39% similarity) compared to other members of the Enterobacteriaceae. Furthermore, phylogenetic information at conserved loci demonstrates that they are likely misclassified members of the genus (19).
For the two newest Escherichia species, E. fergusonii (14) and E. albertii (22), phenotypic, DNA hybridization, and molecular phylogenetic data support an Escherichia classification. Hybridizations show a high degree of genetic similarity to E. coli (∼60% similarity), and phylogenetic data cluster E. fergusonii with E. coli (31, 37-39). Analysis of the same loci has not been done for E. albertii, but Hyma et al. recently used MLST to show that representative isolates were monophyletic and shared a common ancestor with E. coli after the _Salmonella_-Escherichia split (23).
Our MLST analysis shows that there are at least five unnamed clades of Escherichia (CI to CV). E. coli, CI, CIII, and CIV are young lineages (i.e., evolutionary children), having diverged in the past 19 to 31 million years. All of the named Escherichia species and unnamed clades were monophyletic at most of the 22 loci examined, except for E. coli and CI, which were indistinguishable at most loci. As is common for bacteria, when polyphyletic phylogenies are observed (18, 47), the cause of the phylogenetic incongruence is often unclear because it is difficult to differentiate between the effects of mutation, recombination, natural selection, and drift. A recent study comparing the single gene phylogenies of 1,878 core genes in 20 E. coli genomes found that over half of the trees (55%) were incongruent (58). Furthermore, the authors suggested that differences in tree topologies were most likely due to a lack of phylogenetic signal (i.e., low number of shared polymorphisms) and not conflicting phylogenies (i.e., recombination). The _E. coli_-CI data seem to support this hypothesis. However, we did not find a “lack of phylogenetic signal” between the youngest two clades (CIII and CIV). This observation suggests that there has been ample time for E. coli and CI to become distinct, yet some evolutionary process, like recombination, may be acting to maintain their similarity. Regardless, our data suggest that different evolutionary processes may be responsible for these nascent lineages.
Recently, an evolutionary model has been proposed to explain the presence of shared polymorphisms between E. coli and S. enterica. In the temporal fragmentation model of speciation (43), recombination between diverging lineages may continue for some time at loci that are unlinked to mutations underlying ecological distinctiveness (up to ∼70 million years for some orthologs of E. coli and S. enterica). This is different from the biological species concept, where there is no interspecific recombination (11). Recombination in the temporal fragmentation model allows for locus-specific rates of recombination as a function of the genomic location of beneficial mutations that arise during speciation. In other words, the phylogenetic distinctiveness of two diverging lineages should be proportional to the number of ecologically important mutations in their genomes.
Since CIII and CIV are more phylogenetically distinct than E. coli and CI, the model predicts that there should be more ecologically important mutations between the genomes of CIII and CIV than between the genomes of E. coli and CI. Also, ecologically important mutations should be more likely to happen near loci that show a monophyletic relationship (i.e., clade specific). Since three loci gave monophyletic relationships for all clades (fumC, lysP, and rpoS), we can predict that ecologically important mutations occur more often in areas around these genes. Future studies comparing gene content differences among the clades are needed to test this hypothesis against the possibility that clade-specific polymorphisms arose through random drift, as was suggested for S. enterica lineages (13).
The phylogenetic distinctiveness of E. coli and CI will continue to be an issue until perhaps more isolates are collected and more rigorous analyses are done. The situation appears to be quite different for CII, for which we have a single representative, and CV. These two clades are much more diverse at the nucleotide level, and CV is more phylogenetically distinct from the other groups than E. fergusonii and almost as distinct as E. albertii. Two isolates of CV (Z205 and RL325/96) were originally discovered by routine MLST analysis of seven housekeeping loci (63). After observing how divergent they were from typical E. coli, the authors concluded that such isolates represented rare, “living fossils” of E. coli. Accordingly, they suggested that the genetic diversity of present-day E. coli is the result of a massive population bottleneck that occurred between 10 to 30 mya. Our data provide little evidence of an E. coli bottleneck, but they clearly show that CV is phylogenetically distinct and one of the oldest Escherichia lineages (38 to 59 mya).
Gene flow among Escherichia clades.
In the analysis of 20 E. coli genomes presented by Touchon et al., pan-genome genes (i.e., those that were variably present or absent in genomes) outnumbered core genome genes (i.e., those that were shared among all genomes) by approximately nine to one (58). Such results illustrate that only ∼42% of the coding capacity of any one E. coli genome is conserved among other members of the species, suggesting that loci are gained and lost rapidly as isolates colonize new habitats. To assess the potential for gene flow between E. coli and related clades, we looked at the distribution of certain loci that have been previously screened for in large isolate collections. Many of these loci are, or have been, implicated in enhancing an isolate's ability to cause intestinal or extraintestinal disease (17, 26, 57) while some are cosmopolitan and found in almost all E. coli isolates (e.g., uidA, gadAB, and fimH). We should note that it was not our intention to correlate the presence of these loci with virulence potential. They were simply selected to assess the presence/absence of E. coli pan-genome genes in distantly related lineages. Since the acquisition of horizontally acquired genetic material is dependent on sequence divergence (13, 45), we hypothesized that gene flow among the Escherichia clades would be negatively correlated with genetic distance from E. coli. Our results provide little support for this hypothesis. We were surprised to find that two of the most distantly related clades (E. albertii and CV) and the most closely related clade (CI) shared similar percentages of loci (33% and 41% compared to 44%). This suggests that sequence divergence, based on shared housekeeping loci, is not a good predictor of the presence or absence of pan-genome genes.
Evidence for different evolutionary rates.
According to the molecular clock hypothesis, the rate of neutral nucleotide substitution between two diverging lineages should be relatively equal through time (54). The method we chose to test for rate differences (Tajima's relative rates test) has the expectation of equality between lineages, regardless of the model of substitution or variation in rates among the sites (54). Our results suggest that there are at least two distinct evolutionary rates among Escherichia lineages and that E. fergusonii has statistically more changes than would be expected under neutral conditions. These results are consistent with the hypothesis that E. fergusonii is under selection to change or has experienced multiple bottleneck events during its evolution. It was recently shown by Touchon and colleagues that the genome sequence of the E. fergusonii type strain (ATCC 35469T) has undergone multiple rearrangements compared to 20 E. coli genomes (58). It is tempting to hypothesize that such mutations are driving the accelerated rate we observed because such rearrangements can occur faster than single nucleotide changes.
Results from the relative rates test suggest that the estimated divergence time for the E. fergusonii split from E. coli and CI (38 to 59 mya) is likely to be too long (i.e., E. fergusonii is younger than estimated). Touchon et al. also show that ATCC 35469T carries more E. coli core genes than S. dysenteriae (58), which is actually a pathogenic lineage of E. coli (30, 41). Shigella species have universally undergone accelerated gene deletion, presumably as a result of reduced selection, and S. dysenteriae maintains the fewest genes of strain K-12 compared to 11 other pathogenic strains (20). However, whether or not E. fergusonii and other lineages with similar rates (E. albertii, CII, and CI) are truly under selection remains to be tested. Tests of neutrality often examine the relationship between the number of segregating nucleotide sites and nucleotide diversity (e.g., Tajima's D) within evolving populations (48, 55). We calculated this test statistic between isolates in this study, and the data are consistent with the effects of natural selection on E. fergusonii, E. albertii, and CV (data not shown). However, it may be inappropriate to assume that isolates from different clades are members of the same evolving population, which is a main assumption for this statistic, so a more rigorous test for selection in this context is required.
Evidence for ecological differences.
There are remarkably few collections of any Escherichia species where the isolates have been sampled in such a manner that allows an adequate assessment of their abundance and distribution in different habitats. E. fergusonii and E. albertii were both discovered because certain strains caused disease in humans (14, 22). Upon further analysis of samples from Australian animals and the environment, we found that both of these species were rare, compared to E. coli, but they were detected only in birds (seven E. albertii and four E. fergusonii isolates detected in 634 birds and none detected in any of the other 1,754 vertebrate hosts examined) (D. M. Gordon, unpublished data). Assuming that birds represent the primary habitat for these clades, this observation suggests that their effective population size is considerably smaller than that of E. coli, which is quite prevalent in birds and mammals (16).
Our phenotypic analysis showed that the novel clades (CI to CV) were often indistinguishable from E. coli. It stands to reason that these clades have been sampled before but were misidentified. Another potential reason that they have not been characterized until now is that few studies resolve the genetic relatedness of isolates using MLST. For example, to our knowledge, there has been only one MLST analysis of randomly sampled E. coli from the environment (59). During this study, CIII, CIV, and CV isolates were identified in a single sample of 205 isolates from freshwater beaches along Lake Huron, Michigan, in the United States (59), suggesting that such clades represent environmentally adapted bacteria that persist better outside of a warm-blooded host than typical E. coli isolates. While assembling isolates for the present study, we found that other CV isolates were sampled from the environment (Australia) and surface water (United States), as well as a raccoon (United States), birds (Australia), mammals (Australia), and the two isolates mentioned above from a dog and a parrot (locations of hosts were not reported). CIII and CIV had similar habitat ranges and were isolated from soil (Puerto Rico), water (United States), a human (Australia), birds, and mammals. These data suggest that the novel clades have a wide habitat range, but since they have not been reported in host-associated E. coli studies in the current literature, they appear to be overrepresented in habitats other than the GI tract. Furthermore, the presence of clades III, IV, and V in the United States, Puerto Rico, and Australia suggests that the isolates have a worldwide distribution.
There are currently three online E. coli databases with epidemiologic and MLST data (www.mlst.ucc.ie/mlst/dbs/Ecoli, www.shigatox.net, and www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html), all of which are heavily biased toward pathogenic isolates and isolates originally sampled from humans. The main conclusion that can be drawn by querying these databases is that the novel clades are rarely represented. For example, Wirth and colleagues (from the www.mlst.ucc.ie/mlst/dbs/Ecoli database) attempted to “cover a large portion of the known bacterial diversity within” E. coli in an analysis of 462 isolates primarily from diseased and healthy hosts (63). In all, the authors found two isolates of CV. For another database (www.shigatox.net), there has been no effort to include nonpathogenic isolates, so it may be more accurate to refer to it as a strictly pathogenic database. Querying this database revealed that only a few CI isolates are represented among the more than 3,000 different sequence types. The virtual absence of representatives of CI to CV in any database suggests that members of these clades have been, for one reason or another, excluded from analyses and publication or that they rarely cause disease. Future characterizations of isolates by MLST from humans and animals will certainly clarify this issue.
The panel of 31 biochemical reactions we used was unable to distinguish CI, CII, CIV, or CV from E. coli. The only clade that was phenotypically distinct was CIII (five of six isolates). Upon inspection, however, this difference did not result from a gain of biochemical function but from the loss of sucrose and sorbitol fermentation and lysine utilization. An obvious explanation for the lack of phenotypic differentiation is that the appropriate biochemical markers for these taxa were not considered. Considering that we used reactions that were developed to identify only the known members of the Enterobacteriacea family, this is a likely explanation for our results. An alternative hypothesis, and one that requires more testing, is that there are no measurable differences in phenotype between these clades. Regardless, more biochemical profiling of more representative isolates may lead to a better understanding of the ecological potential of these clades.
Importance of novel Escherichia clades.
The fact that these novel clades are highly similar to E. coli on the basis of traditional biochemical assays and yet have very different evolutionary histories and genotypes is perplexing from a taxonomic standpoint. Furthermore, the reliance on phenotypically and not phylogenetically defined taxa has undoubtedly hindered their previous identification, and so little can be said about their ecologic, genetic, or functional potential. When describing E. hermannii in 1982, Brenner et al. (2) asked, “Of what use is a species that cannot be identified phenotypically?” The practical merit of this question is obvious, yet these clades provide the means for an answer.
E. coli is recognized as an indicator of fecal pollution by the United States, Australia, the European Union, and the World Health Organization (53). Recent studies challenge its utility as an indicator with the identification of “naturalized” E. coli strains that persist and grow autochthonously under natural environmental conditions (5, 24, 25). These observations help to explain the occurrence of E. coli bloom events in freshwater lakes (40), population structure differences between host and host-associated secondary habitats (15), and temporally stable population structures in the environment (59). We sampled numerous isolates of these novel clades from water and the environment, and there is no way of knowing how abundant they may be in such habitats. On the other hand, the lack of systematic, random samples from habitats other than the human GI tract makes it impossible to argue against the possibility that these clades “may be the most abundant Escherichia in the gut of elephants” (T. S. Whittam, personal communication). Their importance, therefore, remains to be shown, but now that they have been identified, information about their ecology and genetic potential promises to impact our understanding of the Escherichia genus as a whole.
Effects of potential sampling bias.
These are the first data supporting the existence of cryptic Escherichia species (i.e., two or more distinct species classified as a single species [1]). Because this delineation is based on genetic polymorphisms and not phenotypic profiling, the differentiation of CI to CV from the named Escherichia spp. is unlikely to change with increased analyses. However, these data say little about the abundance and distribution of genetic diversity within these clades, and there is no way of knowing how many additional Escherichia lineages exist in habitats yet to be explored. Sequence-based characterizations of large, randomly sampled isolates from multiple habitats are needed to assess the ecology of these organisms.
Supplementary Material
[Supplemental material]
Footnotes
▿
Published ahead of print on 21 August 2009.
‡
This work is dedicated to Thomas Whittam, now deceased, for his inspiring support and guidance throughout this project.
REFERENCES
- 1.Bickford, D., D. J. Lohman, N. S. Sodhi, P. K. Ng, R. Meier, K. Winker, K. K. Ingram, and I. Das. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22**:**148-155. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, D. J., B. R. Davis, A. G. Steigerwalt, C. F. Riddle, A. C. McWhorter, S. D. Allen, J. J. Farmer III, Y. Saitoh, and G. R. Fanning. 1982. Atypical biogroups of Escherichia coli found in clinical specimens and description of Escherichia hermannii sp. nov. J. Clin. Microbiol. 15**:**703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner, D. J., A. C. McWhorter, J. K. Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15**:**1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess, N. R., S. N. McDermott, and J. Whiting. 1973. Aerobic bacteria occurring in the hind-gut of the cockroach, Blatta orientalis. J. Hyg. 71**:**1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8**:**504-513. [DOI] [PubMed] [Google Scholar]
- 6.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98**:**467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri, R. R., and M. J. Pallen. 2006. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acids Res. 34**:**D335-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66**:**4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, T. F., S. K. Remold, R. E. Lenski, and D. Schneider. 2008. Expression profiles reveal parallel evolution of epistatic interactions involving the CRP regulon in Escherichia coli. PLoS Genet. 4**:**e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doolittle, R. F., D. F. Feng, S. Tsang, G. Cho, and E. Little. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271**:**470-477. [DOI] [PubMed] [Google Scholar]
- 11.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173**:**7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escobar-Paramo, P., C. Giudicelli, C. Parsot, and E. Denamur. 2003. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J. Mol. Evol. 57**:**140-148. [DOI] [PubMed] [Google Scholar]
- 13.Falush, D., M. Torpdahl, X. Didelot, D. F. Conrad, D. J. Wilson, and M. Achtman. 2006. Mismatch induced speciation in Salmonella: model and data. Philos. Trans. R Soc. Lond. B 361**:**2045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farmer, J. J., III, G. R. Fanning, B. R. Davis, C. M. O'Hara, C. Riddle, F. W. Hickman-Brenner, M. A. Asbury, V. A. Lowery III, and D. J. Brenner. 1985. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21**:**77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148**:**1513-1522. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, D. M., and A. Cowling. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149**:**3575-3586. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D. M., S. E. Stern, and P. J. Collignon. 2005. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151**:**15-23. [DOI] [PubMed] [Google Scholar]
- 18.Hanage, W. P., C. Fraser, and B. G. Spratt. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3**:**6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartl, D. L. 1992. Population genetics of microbial organisms. Curr. Opin. Genet. Dev. 2**:**937-942. [DOI] [PubMed] [Google Scholar]
- 20.Hershberg, R., H. Tang, and D. A. Petrov. 2007. Reduced selection leads to accelerated gene loss in Shigella. Genome Biol. 8**:**R164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23**:**254-267. [DOI] [PubMed] [Google Scholar]
- 22.Huys, G., M. Cnockaert, J. M. Janda, and J. Swings. 2003. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 53**:**807-810. [DOI] [PubMed] [Google Scholar]
- 23.Hyma, K. E., D. W. Lacher, A. M. Nelson, A. C. Bumbaugh, J. M. Janda, N. A. Strockbine, V. B. Young, and T. S. Whittam. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187**:**619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72**:**612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii, S., and M. J. Sadowsky. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23**:**101-108. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181**:**261-272. [DOI] [PubMed] [Google Scholar]
- 27.Koeppel, A., E. B. Perry, J. Sikorski, D. Krizanc, A. Warner, D. M. Ward, A. P. Rooney, E. Brambilla, N. Connor, R. M. Ratcliff, E. Nevo, and F. M. Cohan. 2008. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl. Acad. Sci. USA 105**:**2504-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konstantinidis, K. T., A. Ramette, and J. M. Tiedje. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B 361**:**1929-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konstantinidis, K. T., A. Ramette, and J. M. Tiedje. 2006. Toward a more robust assessment of intraspecies diversity, using fewer genetic markers. Appl. Environ. Microbiol. 72**:**7286-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4**:**1125-1132. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence, J. G., H. Ochman, and D. L. Hartl. 1991. Molecular and evolutionary relationships among enteric bacteria. J. Gen. Microbiol. 137**:**1911-1921. [DOI] [PubMed] [Google Scholar]
- 32.McDaniels, A. E., E. W. Rice, A. L. Reyes, C. H. Johnson, R. A. Haugland, and G. N. Stelma, Jr. 1996. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β-d-glucuronidase. Appl. Environ. Microbiol. 62**:**3350-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman, H., and E. A. Groisman. 1994. The origin and evolution of species differences in Escherichia coli and Salmonella typhimurium. EXS 69**:**479-493. [DOI] [PubMed] [Google Scholar]
- 34.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26**:**74-86. [DOI] [PubMed] [Google Scholar]
- 35.O'Ryan, M., V. Prado, and L. K. Pickering. 2005. A millennium update on pediatric diarrheal illness in the developing world. Semin. Pediatr. Infect. Dis. 16**:**125-136. [DOI] [PubMed] [Google Scholar]
- 36.Ostrowski, E. A., R. J. Woods, and R. E. Lenski. 2008. The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proc. Biol. Sci. 275**:**277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paradis, S., M. Boissinot, N. Paquette, S. D. Belanger, E. A. Martel, D. K. Boudreau, F. J. Picard, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2005. Phylogeny of the Enterobacteriaceae based on genes encoding elongation factor Tu and F-ATPase beta-subunit. Int. J. Syst. Evol. Microbiol. 55**:**2013-2025. [DOI] [PubMed] [Google Scholar]
- 38.Perez Luz, S., F. Rodriguez-Valera, R. Lan, and P. R. Reeves. 1998. Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J. Bacteriol. 180**:**2144-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pham, H. N., K. Ohkusu, N. Mishima, M. Noda, M. Monir Shah, X. Sun, M. Hayashi, and T. Ezaki. 2007. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn. Microbiol. Infect. Dis. 58**:**153-161. [DOI] [PubMed] [Google Scholar]
- 40.Power, M. L., J. Littlefield-Wyer, D. M. Gordon, D. A. Veal, and M. B. Slade. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7**:**631-640. [DOI] [PubMed] [Google Scholar]
- 41.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97**:**10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi, W., D. W. Lacher, A. C. Bumbaugh, K. E. Hyma, L. M. Ouellette, T. M. Large, C. L. Tarr, and T. S. Whittam. 2004. EcMLST: an online database for multi locus sequence typing of pathogenic Escherichia coli, p. 520-521. In Proceedings of the 2004 IEEE Computational Systems Bioinformatics Conference, Los Alamitos, CA. Institute of Electrical and Electronics Engineers, Washington, DC.
- 43.Retchless, A. C., and J. G. Lawrence. 2007. Temporal fragmentation of speciation in bacteria. Science 317**:**1093-1096. [DOI] [PubMed] [Google Scholar]
- 44.Rice, E. W., M. J. Allen, D. J. Brenner, and S. C. Edberg. 1991. Assay for β-glucuronidase in species of the genus Escherichia and its applications for drinking-water analysis. Appl. Environ. Microbiol. 57**:**592-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, M. S., and F. M. Cohan. 1993. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics 134**:**401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savageau, M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122**:**732-744. [Google Scholar]
- 47.Sheppard, S. K., N. D. McCarthy, D. Falush, and M. C. Maiden. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320**:**237-239. [DOI] [PubMed] [Google Scholar]
- 48.Simonsen, K. L., G. A. Churchill, and C. F. Aquadro. 1995. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141**:**413-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sleight, S. C., C. Orlic, D. Schneider, and R. E. Lenski. 2008. Genetic basis of evolutionary adaptation by Escherichia coli to stressful cycles of freezing, thawing and growth. Genetics 180**:**431-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sneath, P. H., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, CA.
- 51.Stacy-Phipps, S., J. J. Mecca, and J. B. Weiss. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33**:**1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staley, J. T. 2006. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos Trans. R Soc. Lond. B 361**:**1899-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Standridge, J. 2008. E. coli as a public health indicator of drinking water quality. J. Am. Water Works Assoc. 100**:**65-75. [Google Scholar]
- 54.Tajima, F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135**:**599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123**:**585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24**:**1596-1599. [DOI] [PubMed] [Google Scholar]
- 57.Tarr, C. L., T. M. Large, C. L. Moeller, D. W. Lacher, P. I. Tarr, D. W. Acheson, and T. S. Whittam. 2002. Molecular characterization of a serotype O121:H19 clone, a distinct Shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70**:**6853-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Touchon, M., C. Hoede, O. Tenaillon, V. Barbe, S. Baeriswyl, P. Bidet, E. Bingen, S. Bonacorsi, C. Bouchier, O. Bouvet, A. Calteau, H. Chiapello, O. Clermont, S. Cruveiller, A. Danchin, M. Diard, C. Dossat, M. E. Karoui, E. Frapy, L. Garry, J. M. Ghigo, A. M. Gilles, J. Johnson, C. Le Bouguenec, M. Lescat, S. Mangenot, V. Martinez-Jehanne, I. Matic, X. Nassif, S. Oztas, M. A. Petit, C. Pichon, Z. Rouy, C. S. Ruf, D. Schneider, J. Tourret, B. Vacherie, D. Vallenet, C. Medigue, E. P. Rocha, and E. Denamur. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5**:**e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walk, S. T., E. W. Alm, L. M. Calhoun, J. M. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9**:**2274-2288. [DOI] [PubMed] [Google Scholar]
- 60.Weissman, S. J., S. Chattopadhyay, P. Aprikian, M. Obata-Yasuoka, Y. Yarova-Yarovaya, A. Stapleton, W. Ba-Thein, D. Dykhuizen, J. R. Johnson, and E. V. Sokurenko. 2006. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 59**:**975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler Alm, E., J. Burke, and A. Spain. 2003. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 37**:**3978-3982. [DOI] [PubMed] [Google Scholar]
- 62.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie van Leeuwenhoek 55**:**23-32. [DOI] [PubMed] [Google Scholar]
- 63.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60**:**1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, J., H. Nie, L. Chen, X. Zhang, F. Yang, X. Xu, Y. Zhu, J. Yu, and Q. Jin. 2007. Revisiting the molecular evolutionary history of Shigella spp. J. Mol. Evol. 64**:**71-79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]