Without Argonaute3, Aubergine-bound piRNAs collapse but Piwi-bound piRNAs persist (original) (raw)
. Author manuscript; available in PMC: 2009 Nov 1.
Summary
Piwi-interacting RNAs (piRNAs) silence transposons in the germ line of animals. They are thought to derive from long primary transcripts spanning transposon-rich genomic loci, “piRNA clusters.” piRNAs are proposed to direct an auto-amplification loop in which an antisense piRNA, bound to Aubergine or Piwi protein, directs the cleavage of sense RNA, triggering production of a sense piRNA bound to the PIWI protein Argonaute3 (Ago3). In turn, the new piRNA is envisioned to direct cleavage of a cluster transcript, initiating production of a second antisense piRNA. Here, we describe strong loss-of-function mutations in ago3, allowing a direct genetic test of this model. We find that Ago3 acts to amplify piRNA pools and to enforce on them an antisense bias, increasing the number of piRNAs that can act to silence transposons. We also detect a second piRNA pathway centered on Piwi and functioning without benefit of Ago3-catalyzed amplification. Transposons targeted by this second pathway often reside in the flamenco locus, which is expressed in somatic ovarian follicle cells, suggesting a role for piRNAs beyond the germ line.
Introduction
The ability to tame transposons while retaining them in the genome is a particular specialty of eukaryotes. Transposons, repetitive sequences, and other forms of “selfish” DNA comprise as much as 42% of the human genome and nearly 30% of the genome of Drosophila melanogaster. In metazoa, transposons are silenced by the piRNA pathway, which is guided by 23–30 nt RNAs (Vagin et al., 2006; Brennecke et al., 2007).
The piRNA pathway is distinct from other RNA silencing pathways in that its small RNA guides are not produced by dicing. In contrast, both small interfering RNAs (siRNAs) and microRNAs (miRNAs) are cleaved by double-stranded RNA-specific endonucleases, Dicers, to yield double-stranded intermediates–siRNA or miRNA/miRNA* duplexes–that are loaded into members of the Argonaute family of proteins (reviewed in Ghildiyal and Zamore, 2009). piRNAs, too, act as guides for Argonaute proteins, but they appear not to exist as stable double-stranded intermediates at any point in their biogenesis (Vagin et al., 2006; Houwing et al., 2007). piRNAs bind PIWI proteins, a sub-family of Argonaute proteins that are expressed in germ-line cells (Aravin et al., 2006; Girard et al., 2006; Lau et al., 2006; Vagin et al., 2006; Brennecke et al., 2007; Gunawardane et al., 2007; Batista et al., 2008; Das et al., 2008; Wang and Reinke, 2008). PIWI proteins were first identified by their roles in maintaining (Cox et al., 1998; Cox et al., 2000) and patterning (Wilson et al., 1996; Harris and Macdonald, 2001) Drosophila germ cells. The defects in the organization of embryonic pattern in piRNA pathway mutations are likely an indirect consequence of their larger role in maintaining genomic stability (Klattenhoff et al., 2007). The three Drosophila PIWI proteins, Piwi, Aubergine (Aub), and Argonaute3 (Ago3), are expressed in the male and female germ line cells (Wilson et al., 1996; Cox et al., 1998; Cox et al., 2000; Williams and Rubin, 2002; Brennecke et al., 2007; Gunawardane et al., 2007; Nishida et al., 2007).
The prevailing model for piRNA biogenesis–the “ping-pong” model–reflects the discovery that the first 10 nt of piRNAs bound to Aub or Piwi, which are largely antisense and typically begin with uridine, are often complementary to the first 10 nt of piRNAs bound to Ago3, which are largely sense and typically bear an adenosine at position 10 (Brennecke et al., 2007; Gunawardane et al., 2007). Many Argonaute proteins can act as RNA-guided RNA endonucleases, and all such Argonautes cut their target RNAs 5′ to the base that pairs with the tenth nucleotide of the small RNA guide; all three fly PIWI proteins retain their endonuclease activity (Saito et al., 2006; Gunawardane et al., 2007). Thus, the observed 10 nt 5′ complementarity between piRNAs suggests that the 5′ ends of piRNAs bound to Aub or Piwi are defined by Ago3-catalyzed cleavage, and, reciprocally, that the 5′ ends of piRNAs bound to Ago3 are defined by piRNAs bound to Aub or Piwi. The ping-pong model seeks to explain these observations, as well as the role of piRNA cluster transcripts in piRNA biogenesis, the function of piRNAs in silencing transposons, and the extraordinary antisense bias of piRNAs generally. At its core, the model proposes that piRNAs participate in an amplification loop in which transposon sense transcripts (e.g., transposon mRNAs) trigger the production of new, antisense piRNAs. Ago3, guided by sense piRNAs, lies at the heart of the amplification loop.
To test for the ping-pong model, we isolated strong loss-of-function mutations in ago3. Here, we report the detailed analysis of ago3 and aub mutant flies. Our data provides strong support for an amplification cycle in which Ago3 amplifies piRNA pools and enforces on them a strong antisense bias, increasing the number of piRNAs that can act to destroy transposon mRNAs. Moreover, we detect a second, perhaps somatic, piRNA pathway centered on Piwi and functioning without benefit of Ago3-catalyzed amplification. Most of the transposons targeted by this second pathway reside in the flamenco piRNA cluster, which was first identified as a repressor of transposon expression in somatic follicle cells (Pelisson et al., 1994).
Results
Loss-of-Function ago3 Alleles
The Drosophila ago3 gene resides in pericentromeric heterochromatin on the left arm of chromosome 3 (Figure 1A). Heterochromatin is refractory to _P_-element or piggyBac transposon insertion and to targeted recombination, complicating isolation of ago3 mutants. We used TILLING (Cooper et al., 2008) to identify three mutant alleles (t1, t2, and t3) that create premature stop codons in ago3 (Figure 1B) and obtained one chromosomal deficiency (Df(3L)TTT) that deletes at least six genes, including ago3 (Figure S1A). One homozygous (ago3t3/ago3t3) and three trans-heterozygous (ago3t1/ago3t2, ago3t2/ago3t3, and ago3t1/ago3t3) combinations, as well as each mutant allele in trans to Df(3L)TTT, were viable. Full-length Ago3 protein was essentially undetectable in all seven allelic combinations (Figure 1C and Supplementary Figure S1B), and ago3t2/TM6B or ago3t3/TM6B heterozygous ovaries contained half as much Ago3 protein as Oregon R wild-type ovaries (Figure 1D), suggesting that the ago3 alleles correspond to strong loss-of-function mutations. We refer to ago3t2/ago3t3 trans-heterozygotes as ago3 for brevity.
Figure 1.
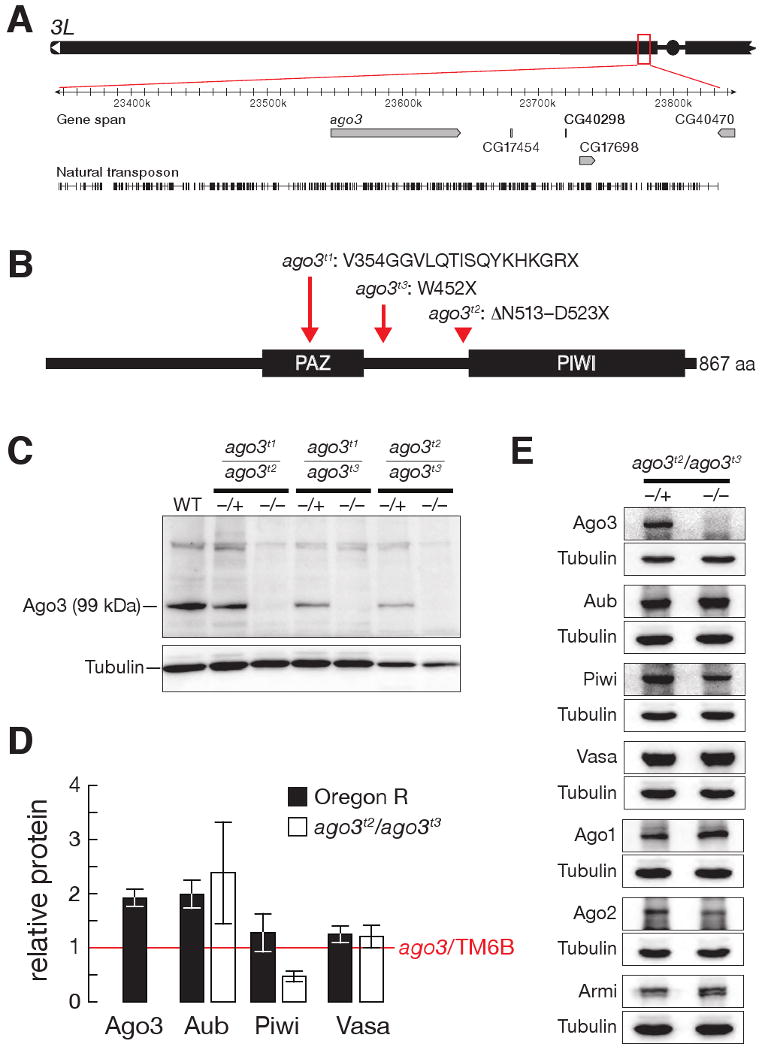
ago3 mutants. (A) The ago3 gene resides in pericentromeric heterochromatin on the left arm of chromosome 3. (B) ago3 mutant alleles were identified by TILLING. (C) Full-length Ago3 protein was not detected in trans-heterozygous ago3 ovaries, but was readily detected in heterozygotes and wild-type. (D) Protein levels in wild-type and ago3 ovaries, relative to ago3/TM6B (red line). The average ± standard deviation for at least three independent biological samples is shown. (E) Representative data for Aub, Piwi, Vasa, Argonaute1 (Ago1), Argonaute2 (Ago2), and Armitage (Armi) in ago3 ovaries. Tubulin served as a loading control.
Mutually Interdependent Localization of PIWI Proteins
The intracellular localization of each PIWI protein appears to require other PIWI proteins. In the Drosophila female germ line, Piwi is largely in the nucleus, whereas Aub and Ago3 are cytoplasmic and concentrated in “nuage,” a ring around the cytoplasmic face of the nuclei of the transcriptionally active germ-line nurse cells (Findley et al., 2003; Brennecke et al., 2007; Pane et al., 2007). The putative RNA helicase Vasa, which marks the germ-line in most animals, is also in nuage (Liang et al., 1994). Mutations that disrupt piRNA biogenesis, but not those that block siRNA or miRNA production, disrupt localization of Aub to nuage (Lim and Kai, 2007).
Ago3 is also required for Aub to localize to nuage (Figure 2A). However, Ago3 is not required for Aub expression or stability: the concentration of Aub protein and the amount of Aub in the cytoplasm increased in ago3 ovaries relative to their heterozygous siblings (Figures 1D and 1E). Reciprocally, Aub plays a role in the localization of Ago3 to nuage, although some Ago3 persisted in nuage in an aub hypomorphic mutant allelic combination (aubHN2/aubQC42). The function of nuage is unknown, but it may have a complex substructure, because the localization of Vasa within nuage only partially coincides with that of Ago3 (Figure 2B) and Aub (Figure 2C).
Figure 2.
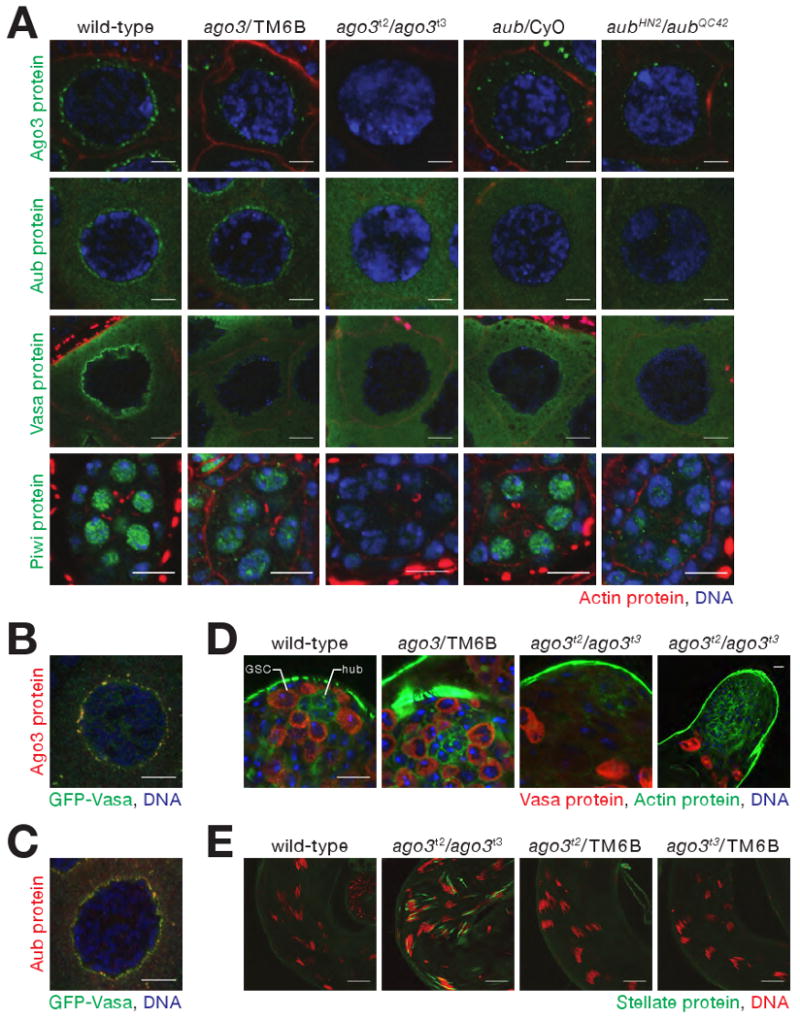
Consequences of loss of Ago3 in the Drosophila germ line. (A) Mutually interdependent incorporation of Ago3 and Aub, into nuage in ovaries. Images correspond to single confocal sections (63× magnification); scale bar: 5 μm (Ago3, Aub, and Vasa panels) or 10 μm (Piwi). (B) and (C) Vasa localization within nuage is only partially congruent with that of Ago3 and Aub. Images correspond to single confocal sections of stage 4/5 egg chambers (63× lens, 4× zoom). Scale bars: 10 μm. (D) In 5−7 day old male adults, Vasa-expressing germ-line stem cells (GSC), which normally surround the somatic hub cells (hub), were not detected in ago3 testes. Since Vasa expression increases in ago3, the two images from ago3 mutants were acquired using reduced gain relative to wild-type and heterozygous testes. Scale bar: 10 μm (first three images) or 20 μm (fourth). (E) Stellate silencing in testes requires Ago3. Scale bars: 20 μm.
The interdependence of Ago3 and Aub for their localization to nuage likely reflects an underlying requirement for these two proteins for nuage assembly: the peri-nuclear localization of Vasa, a marker for nuage, was absent in both ago3 and aub mutants, although the abundance of Vasa was unaltered in ago3 (Figure 1D and 1E). The abundance of Armitage, Argonaute1, and Argonaute2, proteins required for small silencing RNA biogenesis or function, were similarly unaltered in ago3 mutants (Figure 1E). In ago3/TM6B heterozygotes, which produce half as much Ago3 protein as wild-type, and in aub/CyO heterozygotes, less Aub, Ago3, or Vasa was present in nuage than in wild-type, suggesting that assembly of nuage is exquisitely sensitive to the amount of Ago3 and Aub (Figure 2A).
The peak of nuclear-localized Piwi protein expression occurs earlier in oogenesis than when Ago3 and Aub are maximally perinuclear (Cox et al., 2000). Less Piwi is found in the nucleoplasm of ago3/TM6B or aub/CyO heterozygotes than in wild-type, and little or no Piwi is present in the nuclei of the ago3 or aub homozygotes (Figure 2A); ago3 ovaries contain about half as much Piwi as their heterozygous siblings (Figure 1E). (The localization of Aub and Ago3 cannot be studied in piwi, because piwi mutants arrest early in oogenesis.) We conclude that the correct intracellular localization of Ago3, Aub, and Piwi is inter-dependent.
As reported previously for aub testes, Vasa protein expression increased in male germ cells in ago3 testes, relative to wild-type or ago3/TM6B (data not shown), consistent with the proposal that the AT-chX-1 and AT-chX-2 piRNAs down-regulate vasa expression (Nishida et al., 2007); AT-chX-1 piRNA levels were dramatically reduced in ago3 testes (Figure 3A).
Figure 3.
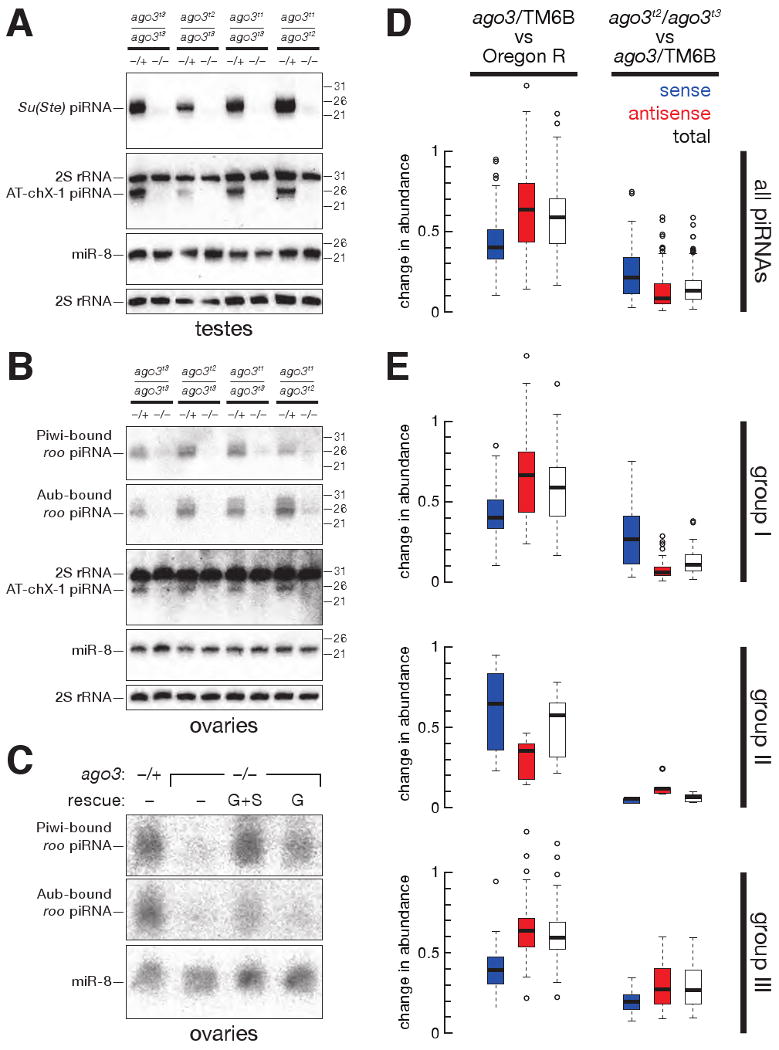
Aub- and Piwi-bound piRNAs disappear without Ago3. (A) Accumulation of Su(Ste) and AT-chX-1 piRNAs in testes required Ago3. (B) Accumulation of roo antisense piRNAs, as well as the AT-chX-1 piRNA, in ovaries required Ago3. (C) Accumulation of Piwi- and Aub-bound roo antisense piRNAs in the ovary was rescued in ago3 mutants by a single-copy transgene expressing Ago3 in the germ line (G) or in both the germ line and the soma (G+S). (D) Box plots illustrating the change in abundance of piRNAs, analyzed by transposon family, in ago3/TM6B versus Oregon R (left panel) and ago3 versus ago3/TM6B (right panel) ovaries. (E) Box plots illustrating the change in abundance for piRNAs for each of 95 transposon families, separated by group.
ago3 Mutations Affect Fecundity
ago3 females are sterile. ago3 females initially laid far fewer eggs than their heterozygous siblings and by day 10 stopped laying altogether (Figure S2A). The egg-laying deficit was rescued by a single-copy transgene expressing Ago3 in the germ line or in both the germ line and the soma. Of those eggs laid, none of the embryos hatched, compared with ∼90% for wild-type (Table S1). For 29%-67% of the embryos produced by ago3 mothers, the dorsal appendages were fused or absent (Table S1), indicating a maternally inherited patterning defect, as has been reported for other piRNA pathway mutations (Schupbach and Wieschaus, 1991; Wilson et al., 1996; Cook et al., 2004; Klattenhoff et al., 2007; Pane et al., 2007) Loss of Ago3 in the germ line, rather than a secondary mutation present in the ago3 mutants, caused the patterning defects, as a single-copy transgene expressing Ago3 from the germ line-specific nanos promoter rescued the dorsal appendage defects to essentially wild-type rates (Table S1).
In males, Ago3 is required to maintain germ-line stem cells (Figure 2D). We examined testes from 5–7 day old wild-type, ago3/TM6B, and ago3 males, using Vasa protein expression to identify germ-line cells. In wild-type and ago3/TM6B, Vasa-expressing cells were present at the tip of the testes, in a ring around the hub cells, a group of somatic cells that support the adjacent germ-line stem cells (Fuller, 1993). In contrast, the corresponding region in ago3 was devoid of Vasa-expressing cells, suggesting that germ-line stem cells are not properly maintained in the absence of Ago3. Consistent with a failure to maintain germ-line stem cells, ago3 males are semi-fertile, siring fewer offspring than males with the same genetic background (Figure S2B).
Silencing Selfish Genetic Elements in Germ Line Requires Ago3
In males, antisense piRNAs derived from the Suppressor of Stellate (Su(Ste)) locus on the Y chromosome silence the X-linked Stellate locus during spermatogenesis (Balakireva et al., 1992; Palumbo et al., 1994; Bozzetti et al., 1995; Aravin et al., 2001; Aravin et al., 2003; Vagin et al., 2006). Su(Ste) piRNAs were the first piRNAs to be identified (Aravin et al., 2001), and, in Su(Ste) mutants, piRNAs targeting Stellate are lost causing Stellate protein crystals to form in primary spermatocytes (Livak, 1984; Pimpinelli et al., 1985; Livak, 1990). Stellate silencing requires the piRNA pathway genes, aub, spindle E, armitage, squash, and zucchini (Schmidt et al., 1999; Stapleton et al., 2001; Tomari et al., 2004; Pane et al., 2007). Stellate protein crystals form in primary spermatocytes in ago3 testes (Figure 2E).
Virtually all Su(Ste) piRNAs are antisense (Vagin et al., 2006) and bound to Aub, not Piwi (Nishida et al., 2007). Su(Ste) piRNAs fail to accumulate in aub mutants, but accumulate to higher than normal levels in piwi testes (Vagin et al., 2006). We used Northern hybridization to examine Su(Ste) piRNA production in the testes (Figure 3A). Su(Ste) piRNAs, as well as AT-chX-1 piRNA, were strongly reduced in ago3 males, relative to their heterozygous siblings, for all seven allelic combinations examined (Figure 3A and data not shown). Thus, both Aub and Ago3 are required to silence Stellate, and both proteins are required to produce or stabilize those Su(Ste) piRNAs normally bound to Aub. We speculate that _Stellate_-derived, Ago3-bound sense piRNAs, while rare, amplify Aub-bound, antisense Su(Ste) piRNAs in testes.
piRNAs have been best characterized in the Drosophila ovary. piRNAs derived from roo LTR retrotransposons are among the most abundant ovarian piRNAs and are disproportionately antisense to roo coding sequences; these antisense piRNAs are bound to Aub and Piwi (Vagin et al., 2006; Gunawardane et al., 2007), but not to Ago3 (Brennecke et al., 2007). roo antisense piRNAs failed to accumulate in ago3 ovaries, but were readily detectable in ago3/TM6B ovaries (Figure 3B). Production of _roo_-derived piRNAs was rescued by a single-copy transgene expressing Ago3 from the actin5c promoter and more weakly rescued by a single-copy transgene expressing Ago3 from the nanos promoter (Figure 3C).
Genome-wide piRNA Analysis
To obtain a broader view of the function of Ago3 in piRNA biogenesis, we sequenced piRNAs from wild-type (Oregon R), ago3/TM6B, ago3t2/ago3t3, aub/CyO, and aubHN2/aubQC42 ovaries. Libraries were prepared from oxidized small RNAs to permit greater effective sequencing depth. In all, 3,282,391 genome-matching, non-ncRNA, non-miRNA, 23–29 nt long small RNAs were sequenced (Table S2). Despite the large number of sequences obtained from each genotype, most piRNA species were sequenced only once. This remained true even when all data sets were pooled, suggesting that piRNAs comprise the most diverse class of regulatory molecules in the fly. Together, our piRNA sequences from wild-type, ago3/TM6B and ago3 ovaries cover 10% of the fly genome.
In parallel, we immunoprecipitated Ago3, Aub, and Piwi from ago3/TM6B ovaries and Aub and Piwi from ago3 ovaries, and then constructed and sequenced libraries of the small RNAs bound to each protein (Table S2). We do not know the extent to which PIWI proteins co-purify, e.g., because they are bound to a common RNA or present in a common complex. To avoid potential mis-assignment of piRNAs to a specific PIWI protein, we analyzed only those piRNAs that associated uniquely with Ago3, Aub, or Piwi.
We calibrated the abundance of the piRNAs uniquely associated with Ago3, Aub, or Piwi such that the aggregate abundance of a subset of piRNAs in an immunoprecipitation dataset equaled that in the total piRNA data set from the corresponding genotype. This subset of piRNAs was defined as those that mapped only once to the fly genome and were sequenced at least once in both of the two data sets. This strategy allows direct comparison of the relative abundance of piRNAs uniquely bound to one PIWI protein with those bound to another, as well as comparison of the uniquely bound piRNAs between ago3 heterozygotes and mutants.
Ago3 Limits Sense piRNA Accumulation and Amplifies Antisense piRNAs
Both ago3 and ago3/TM6B ovaries had fewer piRNAs than wild-type (Figure 3D). Ago3 was previously found to bind mainly piRNAs corresponding to the sense, mRNA strand of transposons. Our data suggest that the intracellular concentration of Ago3 limits the accumulation of sense piRNAs: the median abundance sense piRNAs, analyzed by transposon family, in ago3/TM6B heterozygotes was ∼40% of wild-type. Antisense piRNAs were less affected by halving the amount of Ago3: the median abundance of antisense, transposon-mapping piRNAs in ago3/TM6B ovaries was ∼64% of wild-type. While complete loss of Ago3 further depressed the abundance of transposon-mapping, sense piRNAs, antisense piRNA accumulation in ago3 ovaries collapsed. The median abundance by transposon family for antisense piRNAs in ago3 ovaries was less than a tenth of the median abundance in the heterozygotes, and less than one-twentieth the median abundance in wild-type. The data support the view that for most selfish genetic element families, Ago3, presumably guided by sense piRNAs, acts to amplify antisense piRNAs bound to Aub.
Immunoprecipitation data confirmed this idea. In ago3/TM6B ovaries, 71% of small RNAs uniquely bound to Aub were antisense, but in ago3 mutants, only 41% of the remaining Aub-bound piRNAs were antisense (Table S3). Thus, the characteristic antisense bias of Aub-bound piRNAs is enforced by Ago3. The strand bias of Piwi-bound piRNAs was less affected by loss of Ago3: in the ago3 heterozygotes, 73% of the piRNAs uniquely associated with Piwi were antisense; in the mutants, 62% of the remaining piRNAs uniquely bound to Piwi were antisense. That the strandedness of Piwi-bound piRNAs changes at all suggests that Ago3 plays a role in the production of at least some antisense, Piwi-bound piRNAs.
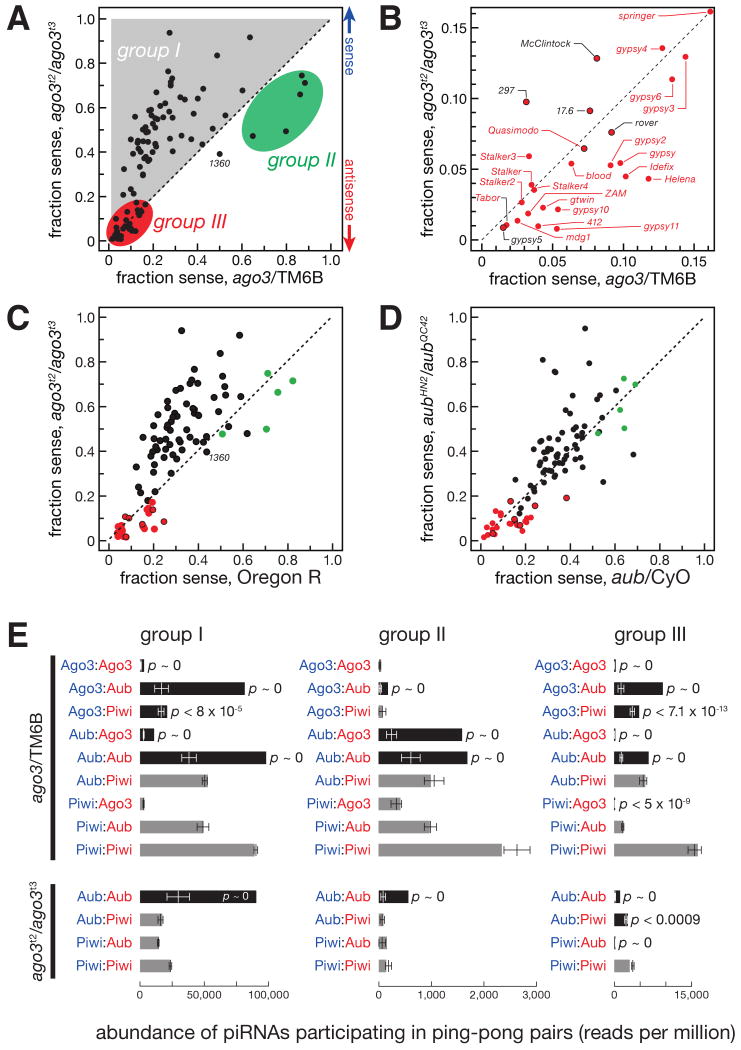
Three piRNA Groups
While antisense piRNAs are generally more abundant than sense piRNAs, individual transposon families have distinct ratios of sense and antisense piRNAs. To determine the role of Ago3 in establishing these ratios, we analyzed the fraction of sense piRNAs (sense/(sense + antisense)) for each of the 95 transposon families for which we obtained ≥ 500 reads in ago3/TM6B ovaries. We compared the fraction of sense piRNAs in the heterozygotes to the fraction of sense piRNAs in ago3 mutants (Figures 4A and 4B). We also compared the fraction of sense piRNAs in the heterozygotes to both the fraction of sense piRNAs in ago3 mutants and the fold-change in antisense piRNAs between the two genotypes (Figure S3).
Figure 4.
Three distinct groups of piRNAs. (A) The sense fraction of the piRNAs was compared for each transposon family between ago3 and ago3/TM6B ovaries. (B) An enlargement of the lower left corner of (A). Transposons labeled in red are present in the flamenco locus. Here and in (C) and (D), group III transposon families not present in flamenco are denoted with a red-filled black circle. (C) The sense fraction of piRNAs was compared between ago3 and Oregon R ovaries. (D) The sense fraction of the piRNAs was compared between aub and aub/CyO ovaries. (E) The normalized abundance of ping-pong pairs detected for all nine (ago3/TM6B) or four (ago3) possible PIWI protein pairings for the piRNA species uniquely associated with a single PIWI protein. Black bars, Bonferroni-corrected _p_-value < 0.005; gray bars, _p_-value > 0.005. _p_-values < 10-30 are reported as p ∼ 0. Values are average ± two standard deviations.
We detected three groups of transposons. For 63 transposon families, the abundance of antisense piRNAs declined dramatically, causing an increase in the fraction of sense piRNAs (Figure S3). Among these 63 group I transposon families, 61 also had a decreased antisense bias comparing ago3 to Oregon R (Figure 4C). (For convenience, the enigmatic transposon family 1360 was included in group I in our subsequent analyses.)
Five transposon families compose group II. These all had more sense than antisense piRNAs in ago3 heterozygotes, and four of the five had more sense than antisense piRNAs in wild-type ovaries, as previously noted (Brennecke et al., 2007). In ago3 ovaries, both the fraction (Figures 4A, 4C, and S3) and the absolute amount (Figure 3E) of sense piRNAs declined for group II transposons, relative to heterozygous or wild-type ovaries.
piRNAs mapping to the 26 group III transposons were disproportionately antisense, ranging from 80 to nearly 100 percent antisense in ago3 heterozygotes (Figures 4A and 4B). In ago3 ovaries, group III transposons generally retained their antisense piRNAs to a greater extent than those in groups I and II (Figures 3E and S3).
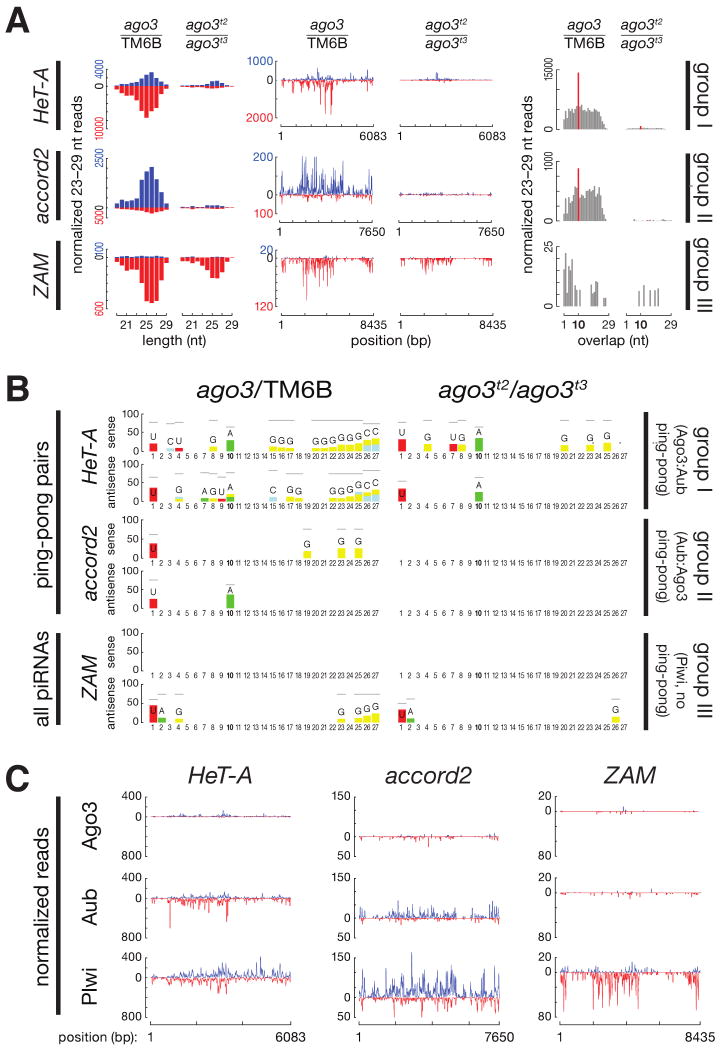
Paradigmatic examples of each group–HeT-A for group I, accord2 for group II, and ZAM for group III–are analyzed in greater detail in Figure 5. Figures S4, S5, and S6 present the corresponding data for each of the 95 transposons, as well as the tandem repeat mst40.
Figure 5.
Paradigmatic examples of each transposon group. (A) piRNA length distribution, abundance relative to consensus position, and ping-pong pair abundance. Blue, sense piRNAs; red, antisense. (B) Sequence-excess logos generated by subtracting the background from the relative frequency of each nucleotide at each position of the 23–29 nt RNAs in each sample. Only the nucleotide positions where foreground was significantly higher than background are shown (Bonferroni-corrected _p_-values < 0.001). Gray horizontal bars indicate the maximum possible value for each position. (C) Normalized and calibrated abundance of piRNAs uniquely associated with Ago3, Aub, or Piwi in ago3/TM6B or ago3 ovaries.
Group I Transposons Require Ago3 for Antisense piRNA Amplification
HeT-A is the quintessential group I transposon (Figure 5). The piRNAs that map to group I transposons show a strong “ping-pong” signature that derives from the 10 nt overlap between antisense piRNAs bound to Aub and sense piRNAs bound to Ago3 and Aub (Figures 4E). For group I transposons, although Ago3 associates almost exclusively with sense piRNAs, it is the Aub-bound antisense piRNAs that disappear in its absence (Figure S6). A central postulate of the ping-pong model is that the action of Ago3, rather than Aub, drives the production of more antisense than sense piRNAs (see Supplemental Discussion). Consistent with this proposal, the fraction of sense piRNAs for most group I transposons shows little overall change in aub ovaries (Figure 4D).
Group II Transposons act “backwards”
For group II transposons, antisense piRNA levels appear to reflect the abundance of Ago3: the median abundance by transposon family of group II antisense piRNAs in ago3 heterozygotes was ∼35% of wild-type (Figure 3E). Sense piRNAs declined less when the abundance of Ago3 was halved. However, in the absence of Ago3, group II sense piRNAs declined ∼17-fold from their levels in heterozygotes and ∼27-fold from their levels in wild type. Group II antisense piRNAs also decreased, but less dramatically. The “backwards” behavior of group II piRNAs, relative to group I, suggests that the production or accumulation of Aub-bound piRNAs generally requires Ago3, irrespective of their sense or antisense identity.
piRNAs from group I and group II transposons normally partition among PIWI proteins according to their orientation and sequence bias (Figures S4, S5, and S6). Group I antisense (or group II sense) piRNAs typically begin with U (U1) and bind Aub, whereas group I sense (or group II antisense) piRNAs show less 5′ nucleotide bias but typically bear an A at position 10 (A10) and bind Ago3 (e.g., accord2 in Figure 5). In the absence of Ago3, both sense and antisense piRNAs remained associated with Aub (Figure S6). Some of these showed ping-pong pairing, but without respect to orientation, so that the U1 that normally characterizes antisense piRNAs and the A10 that normally characterizes sense piRNAs became conflated (Figure 5B and Figure S4). In the absence of Ago3, those Aub-bound piRNAs with an A at position 10 began with U more often than expected by chance (Figure S7) and began with U about as often as did those Aub-bound piRNAs without an A at position 10. These observations suggest that a 5′ U favors binding of a piRNA to Aub. Moreover, the data suggest that the A10 signature of Ago3-bound piRNAs is a consequence of the preponderance of Aub-bound piRNAs that start with U, consistent with the idea that Aub directs the production of Ago3-bound piRNAs.
piRNAs for Group III Transposons Are Produced by Both Aub- and Ago3-Dependent and Aub- and Ago3-Independent Pathways
In the absence of Ago3, the abundance of group III antisense piRNAs decreased far less than for group I (group I median = 0.06, group III median = 0.27; Figure 3E and S3). Antisense piRNAs from group III transposon families were mostly bound to Piwi: the median ratio between the amount of antisense piRNAs bound by Piwi and the amount bound by Aub in ago3 ovaries was 31.3 for group III transposon families, a five-fold increase from the median Piwi/Aub ratio for group III transposons in ago3/TM6B ovaries. This contrasts with the median Piwi/Aub ratios for groups I (2.00) or II (2.94) transposon families (Figure S8A). Furthermore, loss of Ago3 caused group III antisense piRNAs bound to Piwi to decrease far less than antisense piRNAs bound to Aub; for group I, piRNAs bound to Piwi and Aub declined to a comparable extent (Figure S8B). Group III antisense piRNAs were also less affected by loss of Aub than the other two groups (Figures S9 and S10). Collectively, these observations suggest that group III transposons are predominantly silenced by a Piwi-dependent, Ago3-independent pathway.
While Piwi-bound piRNAs persisted in the absence of Ago3 to a greater extent than Aub-bound piRNAs, the absence of Ago3 clearly reduced the abundance of Piwi-bound piRNAs for all groups (Figure S8B). Perhaps some but not all Piwi-bound piRNAs are generated in the germ line by an Ago3-directed amplification cycle. Supporting this view, we detected a statistically significant (p < 7.1 × 10-13) 10 nt overlap between sense Ago3-bound piRNAs and antisense Piwi-bound group III piRNAs (Figure 4E). Such group III Ago3 (sense):Piwi (antisense) piRNA ping-pong pairs were about half as abundant as Ago3:Aub pairs.
Most group III antisense piRNAs bound to Aub were eliminated in ago3 mutants (Figure S8), suggesting that our group III piRNA data conflates at least two distinct pathways: a Piwi-dependent, Ago3- and Aub-independent pathway in which piRNA-directed piRNA amplification plays a little if any role and an Aub-dependent pathway in which sense piRNAs bound to Ago3 act catalytically to amplify the antisense piRNAs associated with Aub. piRNAs acting in the Piwi-dependent pathway were disproportionately antisense, and this antisense bias required neither Ago3 nor Aub: production of antisense piRNAs for group III transposons remained essentially unchanged in aubHN2/aubQC42 mutants (Figure 4D).
Group III Transposons Often Reside in the flamenco piRNA Cluster
Remarkably, of the 26 transposon families comprising group III, 20 are present in the flamenco locus (Pelisson et al., 1994; Brennecke et al., 2007). flamenco was originally identified as required for the silencing of gypsy (Prud'homme et al., 1995) and later shown to be required to silence ZAM and Idefix (Mevel-Ninio et al., 2007; Desset et al., 2008). gypsy, ZAM, and idefix are all group III transposons. Reporter experiments suggest that Piwi is required to silence these transposons (Sarot et al., 2004; Desset et al., 2008). Given that Piwi, but not Aub or Ago3, is readily detected in ovarian somatic follicle cells (Cox et al., 2000; Saito et al., 2006; Gunawardane et al., 2007; Nishida et al., 2007); that gypsy silencing requires piwi but not aub (Pelisson et al., 2007); that the gypsy promoter drives expression of gypsy in the somatic follicle cells, which produce retrovirus-like particles that then infect the oocyte (Pelisson et al., 1994); that ZAM and Idefix are silenced in the somatic follicle cells by Piwi (Desset et al., 2008); and that follicle cell clones mutant for piwi or flamenco have remarkably similar defects(Cox et al., 1998; Mevel-Ninio et al., 2007), it is tempting to speculate that group III transposons in general are repressed by a Piwi-dependent, Ago3- and Aub-independent pathway that operates in follicle cells. In this view, antisense, Piwi-bound piRNAs would provide the primary somatic cell defense against group III transposon expression, whereas an Aub- and Ago3-dependent pathway provides a secondary defense in the germ line, the ultimate target of these transposons.
For groups I and II, loss of Ago3 switches the bias of both Aub and Piwi-bound piRNAs from strongly antisense-biased (or strongly sense-biased for group II) to slightly sense-biased (or antisense-biased for group II). That is, in wild-type flies, Ago3 acts to skew the strandedness of piRNA pools for group I and group II transposons. For group III transposons, however, loss of Ago3 slightly increased the antisense bias of piRNAs uniquely bound to Piwi: in ago3/TM6B ovaries, 88% of such piRNAs were antisense; in ago3, 90% were antisense (Table S3). We conclude that most Piwi-bound group III piRNAs are made directly from antisense transcripts such as the hypothesized precursor transcript that spans the flamenco locus. This explanation is consistent with the previous proposal that flamenco triggers silencing of transposons such as gypsy, ZAM, and idefix in somatic follicle cells–a cell type that expresses little if any Ago3 or Aub.
Ago3 amplifies piRNAs
Overall, piRNAs associated with Piwi were ∼15-fold and piRNAs bound to Aub were ∼6.4-fold more abundant than those bound to Ago3. The greater abundance of Piwi- and Aub-bound piRNAs is consistent both with the idea that Ago3 is less abundant than the other two PIWI proteins and with the proposal that Ago3 acts catalytically to amplify Piwi- and Aub-bound piRNAs. Consistent with the idea that group III piRNAs are largely Ago3-independent, the likelihood of a group III piRNA being associated with Piwi rather than Aub more than doubled in ago3 ovaries: the Piwi-bound to Aub-bound piRNA ratio was 5.0 in ago3/TM6B, but 11.7 in ago3. This trend was not observed for group I piRNAs, where Piwi-bound piRNAs were twice as abundant as Aub-bound for ago3/TM6B, but only 1.6 times more abundant in ago3 ovaries. Thus, for group III piRNAs, the absence of Ago3-catalyzed amplification shifts the piRNA pool towards the Piwi-dependent pathway.
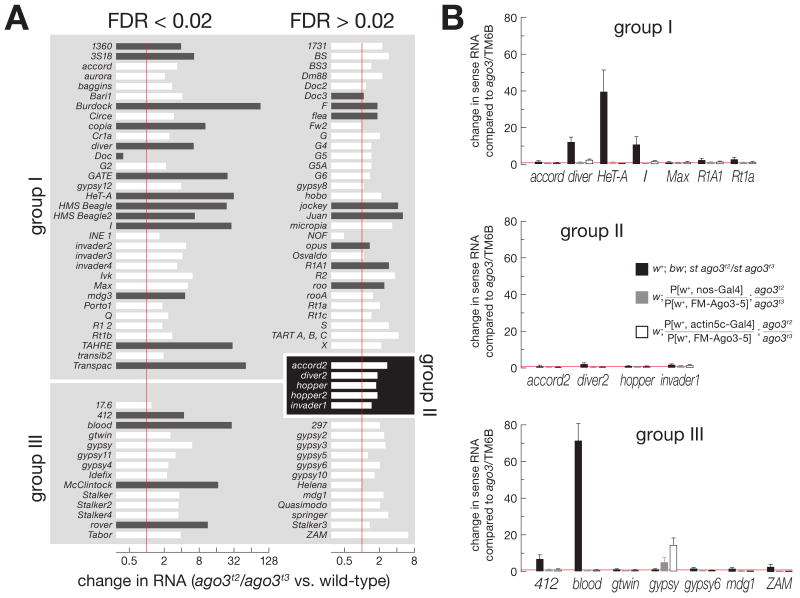
Loss of Ago3 Increases Group I Transposon Expression
What is the molecular consequence of piRNA loss? We used whole-genome tiling microarrays to measure the effect of loss of Ago3 on gene and transposon expression (Figure 6A and S11) and quantitative RT-PCR of selected transposons to corroborate the microarray data (Figure 6B). The abundance of genic mRNA levels were generally unchanged in both ago3 (Figure S11A) and aub ovaries (Figure S12A), compared with wild-type controls (w1118). In contrast, using a false-discovery rate (FDR) < 0.02, the expression of 33 of the 64 group I transposons and 14 of the 26 group III transposons increased in the absence of Ago3 (Figure 6A and S11B). However, the level of expression for many of these in both wild-type and ago3 ovaries was less than the 50th percentile of expression for mRNA in wild-type, making accurate quantification of their change in expression challenging. Using this threshold for expression, 14 of the 64 group I and 4 of the 26 group III transposon families showed increased expression in the absence of Ago3 (Figure 6A).
Figure 6.
Loss of Ago3 increases expression of some group I and group III, but not group II, transposons. (A) Expression of group I, group II, and group III transposon families in ago3 ovaries, relative to wild-type (w1118) ovaries, was assayed using whole-genome tiling microarrays. White bars: expression of the transposon family in both ago3/TM6B and ago3 ovaries was less than that of the 50th percentile for expression of all mRNAs in wild-type, suggesting that expression change cannot be reliably quantified. Black bars: transposon families with expression greater than this threshold in one or both genotypes. Significant (false-discovery rate (FDR) < 0.02) and non-significant data (FDR > 0.02) are separated. (B) Quantitative RT-PCR was used to assess transposon expression, relative to Actin, for ago3 ovaries (black bars) and ovaries expressing one copy of a UAS-Ago3 transgene driven by _nanos_-Gal4 (gray bars) or _actin5c_-Gal4 (white bars), relative to ago3/TM6B ovaries.
Despite the loss of group II piRNAs in ago3 mutants, expression of the five group II transposons was not significantly altered at a FDR < 0.02 (Figure 6A). Perhaps the production of Aub-bound sense piRNAs for group II transposons is futile and does not silence these elements, since they were likewise not activated in an aub mutant (Figures S12B and S13).
Compared with wild-type, expression of 18 of the 64 group I transposon families increased in aub ovaries (8 of these 18 had expression in both w1118 and ago3 greater than the 50th percentile for mRNA expression in wild-type), including 14 that also increased in ago3 ovaries (Figures S12B and S13). At a FDR < 0.02, the expression of only 2 group III transposons–rover and _McClintock_– was increased; both were also desilenced in ago3 ovaries. These data support the idea that group I transposons rely on Ago3 and Aub for silencing, whereas group III transposons are silenced by Piwi, with help from Ago3, likely in the germ line only, for some transposons.
Germ Line Expression of Ago3 Rescues Group I Transposon Silencing
Our data suggest that Ago3 is required for piRNA production and silencing of group I transposons in the female Drosophila germ line. As a final test of this hypothesis, we expressed Ago3 in ago3 ovaries using the germ-line restricted nanos promoter. We used quantitative RT-PCR to measure the levels of mRNA for seven group I transposons, including five whose expression was increased in the absence of Ago3: HeT-A, Diver, I-element, R1A1, and Rt1a. For each, germ-line expression of Ago3 using the nanos promoter rescued silencing (Figure 6B).
Mis-expression of Ago3 in the Soma Interferes with Group III Transposon Silencing
Argonaute proteins can compete for binding small RNAs. For example, decreasing the concentration of Ago1 in cultured S2 cells increases the loading of miRNAs into Ago2, whereas decreasing Ago2 increases miRNA loading into Ago1 (Förstemann et al., 2007; Horwich et al., 2007). Similarly, mis-expression of Aub in the soma inhibits Ago2-mediated RNAi (Specchia et al., 2008). To test the role of Ago3 in silencing transposons in somatic ovarian cells, we used the actin5c promoter to express Ago3 in both the germ line and the soma of ago3 ovaries.
Surprisingly, ectopic expression of Ago3 in the soma increased the expression of the group III transposon gypsy, but enhanced silencing of the group I transposon HeT-A (Figure 6B). First, these data confirm our observation that increased Ago3 expression in the female germ line increases silencing of group I transposons, perhaps because the concentration of Ago3 in the germ-line nurse cells limits piRNA amplification. Second, the data suggest that Ago3 cannot silence those transposons normally expressed in the somatic cells of the ovary. Perhaps Ago3 competes with Piwi for piRNAs, but, unlike Piwi, cannot act directly to silence transposons. Further supporting the idea that PIWI proteins compete for piRNAs, gypsy was silenced to a ∼43-fold greater extent in aub ovaries than in aub/CyO (Figure S13). We speculate that when Aub levels are low, more gypsy piRNAs associate with Piwi, leading to enhanced silencing of gypsy.
Discussion
Disentangling Multiple piRNA Pathways
Because ovaries contain both germ-line and somatic cells, our data conflate two distinct cell lineages. Combining our data with extensive genetic studies of gypsy and other transposon families represented in the flamenco locus, we have attempted to disentangle germ-line and somatic piRNA function (Figure 7). We propose that the somatic piRNA pathway is the more straightforward, involving only Piwi and not Ago3 or Aub. Our data suggest that Piwi cannot act alone to amplify piRNAs. We envision that Piwi-bound piRNAs in the soma are produced by a ribonuclease that randomly generates single-stranded guides that are subsequently loaded into Piwi and trimmed to length. Although Piwi-bound piRNAs generally begin with U and Piwi shows in vitro a preference for binding small RNA that begins with U (Yin and Lin, 2007), current evidence cannot distinguish between a putative piRNA-generating ribonuclease cleaving mainly at U and Piwi selecting U1 piRNAs from a set of RNAs with all possible 5′ nucleotides.
Figure 7.
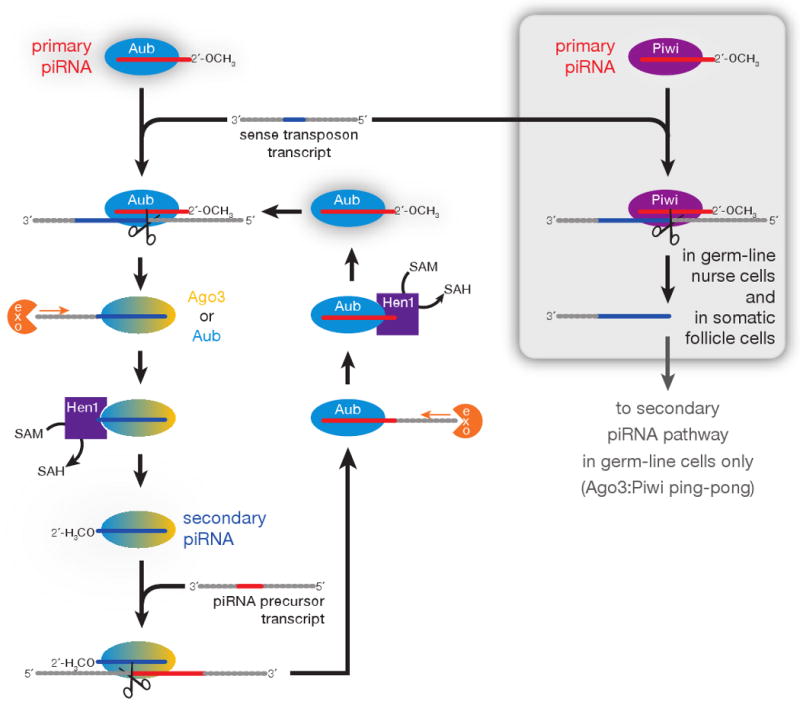
A model for piRNA biogenesis. The Aub- and Ago3-dependent piRNA amplification cycle is envisioned to operate only in the germ line, whereas a Piwi-dependent, Aub- and Ago3-independent pathway is shown for somatic cells. In the germ line, Piwi can also partner with Ago3 to amplify piRNAs.
Without an amplification cycle to ensure an antisense bias, some other mechanism must operate to explain why Piwi-bound piRNAs are overwhelmingly antisense. A plausible but somewhat unsatisfying explanation comes from flamenco itself, whose constituent transposons are nearly all oriented in a single direction, so that the ∼160 kb flamenco transcript is almost entirely antisense to the transposons. How such a non-random array of transposons could arise is unknown. Other non-randomly oriented piRNA clusters may explain the smaller number of transposons in group III that are not present in flamenco.
The transposons in most piRNA clusters do not show such a pronounced non-random orientation. These likely act in the germ line to produce primary piRNAs that load into Aub. The observed antisense bias of Aub-bound piRNAs arises subsequently, when Aub generates Ago3-bound secondary piRNAs and Ago3 acts, in turn, to produce Aub-bound secondary piRNAs. We propose that in the absence of Ago3, the sense/antisense ratio of Aub-bound piRNAs reverts to the inherent sense/antisense bias of the transposable element sequences present in the transcripts of piRNA clusters.
For this cycle to skew the Aub-bound piRNA population toward antisense, the substrate for cleavage by primary piRNA-bound Aub must be largely sense RNA. The best candidate for such sense RNA is mRNA derived from actively transcribed transposon copies. If such sense mRNA were largely found in the cytoplasm, it would be spatially segregated from the cluster transcripts, which we envision to be retained in the nucleus. Supporting this idea, sense transcripts from the group I transposon, I element, normally accumulate only in the nuclei of germ-line nurse cells (Chambeyron et al., 2008), likely because they are destroyed in the cytoplasm by Aub-bound primary piRNAs and Aub-bound piRNAs produced by Ago3-dependent amplification. In the absence of Aub, these sense transposon transcripts accumulate in the cytoplasm instead, consistent with the strong desilencing of I element in aub and ago3 mutants (Figure 6, S12, and S13, and Vagin et al., 2006).
Nuage and the Paradox of piRNA Production
The piRNA ping-pong hypothesis predicts a role for Ago3 in the production of Aub-bound antisense piRNAs, but our finding that loss of Ago3 also reduced the abundance of Piwi-bound antisense piRNAs was unexpected. The majority of Aub and Ago3 is found in nuage and in the cytoplasm, but Piwi is predominantly nuclear. How then can Ago3 direct the production of Piwi-bound piRNAs? Perhaps Piwi transits the nuage en route from its site of synthesis, the cytoplasm, to where it accumulates, the nucleus. In this view, cytoplasmic Piwi is predicted to lack a small RNA guide. Piwi would then acquire its small RNA guide in the nuage, through a process that requires Ago3. Loading a piRNA into Piwi might then license it for entry into the nucleus, where it could act post-transcriptionally or transcriptionally to silence transposon expression. In this view, mutations in genes required for nuage assembly or stability, such as vasa, as well as genes required for Piwi loading would reduce the amount of nuclear Piwi. A similar mechanism may operate in mammals, where the PIWI protein MILI is found in cytoplasmic granules, whereas MIWI2 is nuclear. In the absence of MILI, MIWI2 delocalizes from the nucleus to the cytoplasm, although MIWI2 is not required for the localization of MILI (Aravin et al., 2008).
Such a model cannot explain the loading of Piwi in the somatic follicle cells, which contain little or no Ago3 or Aub and which do not contain nuage. A simple but untested hypothesis for these cells is that in the absence of nuage, empty Piwi readily enters the nucleus, where it obtains its small RNA guide. We might reasonably expect that in germ cells the absence of nuage would impair the loading of Piwi by eliminating the Ago3-dependent, germ-line specific Piwi-loading process, but also facilitate entry of some empty Piwi into the nucleus, where it could obtain small RNA guides. Consistent with this idea, we do detect some Piwi in the nucleus in ago3 ovaries. The simplicity of this hypothesis, of course, belies the complexity of testing it.
Why two distinct piRNA production pathways?
Retrotransposons “reproduce” by producing sense RNA encoding transposases and other proteins that allow them to jump to new locations in the germ cell genome. The conservation of the piRNA ping-pong cycle in animals (Aravin et al., 2001; Grimson et al., 2008) suggests that it is an ancient and conserved germ-line defense against retrotransposition. In flies, the gypsy family of retroelements appears to have moved its reproductive cycle to the somatic follicle cells adjacent to the germ line, which it infects using retrovirus-like particles. gypsy thus appears to avoid germ-line piRNA surveillance by transcribing and packaging its RNA in the soma. Perhaps expression of Piwi in Drosophila follicle cells reflects an adaptive evolutionary counter move to the gypsy reproductive strategy. The simplicity of the direct loading of Piwi with antisense piRNAs derived from flamenco may have made this counter defense more evolutionarily accessible than a strategy requiring expression of all the proteins needed for the Ago3:Aub ping-pong mechanism. In the future, more extensive analysis of the cellular and genetic requirements for ping-pong-independent and ping-pong-dependent piRNA mechanisms in Drosophila melanogaster and in more ancient animal species may provide a test for these ideas.
Experimental Procedures
Detailed experimental and computational procedures are described in Supplemental Materials. Sequence data are available via the NCBI trace archives (http://www.ncbi.nlm.nih.gov/Traces/) using accession number SRA007727. Microarray data are available via the NCBI gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE14370.
Isolation of ago3t1, ago3t2, and ago3t3 alleles
From the 6,000 EMS-mutagenized fly lines (Koundakjian et al., 2004) screened by the Seattle TILLING project (http://tilling.fhcrc.org/), candidate lines that contained mutations that induced premature stop codons in the ago3 coding sequence were characterized by sequencing of genomic PCR amplicons.
Molecular cloning and generation of transgenic flies
Ago3 was cloned from Oregon R ovary cDNA using 3′ RACE. Ovary RNA was reverse transcribed. First strand cDNA was treated with RNase H. Full length Ago3 coding sequence was amplified by PCR, cloned, and recombined into the pPFMW vector. Insert junctions were confirmed by sequencing. Plasmid DNA was injected and transgenic flies identified.
Immunohistochemistry and microscopy
Egg chamber fixation and whole-mount antibody labeling were performed as previously described (Theurkauf, 1994). Samples were analyzed using a Leica TCS-SP inverted laser-scanning microscope, with identical imaging conditions for each set of wild-type and mutant.
Immunoprecipitation
Immunoprecipitation was performed using anti-Ago3, anti-Aub or anti-Piwi antibodies bound to Protein A Sepharose beads. Input, supernatant, and bound samples were subject to western blotting analysis to confirm immunoprecipitation.
Small RNA cloning and sequencing
Total RNA was isolated from manually dissected ovaries from 2–4 day flies. After 2S rRNA depletion, 18–29 nt small RNA was purified and oxidized followed by ethanol precipitation. 3′ ligated product was purified from a denaturing urea-polyacrylamide gel, and then ligated to a 5′ RNA adapter. The small RNA library was amplified and then purified from an agarose gel. Purified libraries were sequenced using a Solexa Genome Analyzer (Illumina, San Diego, CA, USA).
Supplementary Material
01
02
03
04
05
06
Acknowledgments
We thank Mikiko and Haruhiko Siomi and Greg Hannon for antibodies, Erica Selva for flies expressing _nanos_-Gal4 from a transgene on the second chromosome, Paul Lasko for GFP-Vasa flies, Bradley Till for assistance with tilling primer design, Alicia Boucher for assistance with fly husbandry, Gwen Farley for technical assistance, and members of the Zamore and Weng laboratories for advice, suggestions, and critical comments on the text. This work was supported in part by grants from the National Institutes of Health to PDZ (GM62862 and GM65236) and WET (GM050898 and HD049116) and from NSERC Canada and CIHR to BMH. HX was supported in part by Pfizer, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA Pathway Primed by Individual Transposons Is Linked to De Novo DNA Methylation in Mice. Molecular Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva MD, Shevelyov Y, Nurminsky DI, Livak KJ, Gvozdev VA. Structural organization and diversification of Y-linked sequences comprising Su(Ste) genes in Drosophila melanogaster. Nucleic Acids Res. 1992;20:3731–3736. doi: 10.1093/nar/20.14.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte DJ, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzetti MP, Massari S, Finelli P, Meggio F, Pinna LA, Boldyreff B, Issinger OG, Palumbo G, Ciriaco C, Bonaccorsi S, et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc Natl Acad Sci U S A. 1995;92:6067–6071. doi: 10.1073/pnas.92.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Henikoff S. Fly-TILL: Reverse genetics using a living point mutation resource. Fly (Austin) 2008;2:300–302. doi: 10.4161/fly.7366. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Development. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM Locus Directs the Somatic Silencing of Two Retrotransposons through both Piwi-Dependent and -Independent Pathways. PLoS ONE. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee LM, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute protein complexes after their production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, A Martinez Alfonso, editors. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 71–148. [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A Slicer-Mediated Mechanism for Repeat-Associated siRNA 5′ End Formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167:203–206. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics. 1990;124:303–316. doi: 10.1093/genetics/124.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo G, Bonaccorsi S, Robbins LG, Pimpinelli S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics. 1994;138:1181–1197. doi: 10.1093/genetics/138.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S, Sullivan W, Prout M, Sandler L. On biological functions mapping to the heterochromatin of Drosophila melanogaster. Genetics. 1985;109:701–724. doi: 10.1093/genetics/109.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schafer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V, Benna C, Mazzotta GM, Piccin A, Zordan MA, Costa R, Bozzetti MP. aubergine Gene Overexpression in Somatic Tissues of auberginesting Mutants Interferes With the RNAi Pathway of a yellow Hairpin dsRNA in Drosophila melanogaster. Genetics. 2008;178:1271–1282. doi: 10.1534/genetics.107.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton W, Das S, McKee BD. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma. 2001;110:228–240. doi: 10.1007/s004120100136. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci U S A. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06