The Drosophila HP1 homologue Rhino is required for transposon silencing and piRNA production by dual strand clusters (original) (raw)
. Author manuscript; available in PMC: 2010 Mar 18.
Summary
piRNAs silence transposons and maintain genome integrity during germ-line development. In Drosophila, transposon-rich heterochromatic clusters encode piRNAs either on both genomic strands (dual-strand clusters) or predominantly one genomic strand (uni-strand clusters). Primary piRNAs derived from these clusters are proposed to drive a ping-pong amplification cycle catalyzed by proteins that localize to the perinuclear nuage. We show that the HP1 homologue Rhino is required for nuage organization, transposon silencing, and ping-pong amplification of piRNAs. rhi mutations virtually eliminate piRNAs from the dual-strand clusters and block production of putative precursor RNAs from both strands of the major 42AB dual-strand cluster, but do not block production of transcripts or piRNAs from the uni-strand clusters. Furthermore, Rhino protein associates with the 42AB dual-strand cluster, but does not bind to uni-strand cluster 2 or flamenco. Rhino thus appears to promote transcription of dual-strand clusters, leading to production of piRNAs that drive the ping-pong amplification cycle.
Introduction
Mutations in the Drosophila piRNA pathway disrupt transposon silencing, cause DNA break accumulation during female germline development, and lead to defects in posterior and dorsoventral axis specification (Brennecke et al., 2007; Chambeyron et al., 2008; Klattenhoff et al., 2007; Vagin et al., 2006). The axis specification defects associated with piRNA pathway mutations are dramatically suppressed by mutations in mnk and mei-41, which encode Chk2 and ATR kinase homologues that function in DNA damage signaling (Chen et al., 2007; Klattenhoff et al., 2007; Pane et al., 2007). The developmental defects linked to piRNA pathway mutations thus appear to be secondary to DNA damage, which may result from transposon mobilization. PIWI proteins bind piRNAs, and mutations in genes encoding mouse and Zebrafish piwi homologues lead to transposon over-expression and germline-specific apoptosis (Carmell et al., 2007; Houwing et al., 2007), which could be triggered by DNA damage. The piRNA pathway may therefore have a conserved function in transposon silencing and maintenance of germline genome integrity.
The majority of Drosophila piRNAs appear to be derived from transposon rich clusters, most of which are localized in pericentromeric and sub-telomeric heterochromatin (Brennecke et al., 2007). The majority of clusters produce piRNAs from both genomic strands (dual-strand clusters). However, two major clusters on the X-chromosome produce piRNAs predominantly from one genomic strand (uni-strand clusters) (Brennecke et al., 2007; Brennecke et al., 2008). One of these uni-strand clusters maps to flamenco, a locus required for transposon silencing in the somatic follicle cells (Brennecke et al., 2007; Mevel-Ninio et al., 2007; Pelisson et al., 2007; Pelisson et al., 1994; Prud'homme et al., 1995; Sarot et al., 2004). The flamenco cluster contains fragments of a number of transposons, including Zam, idefix, and gypsy, and flamenco mutations disrupt silencing of these transposons (Desset et al., 2008; Mevel-Ninio et al., 2007; Prud'homme et al., 1995). In addition, transgenes carrying fragments of transposons in this cluster show _flamenco_-dependent silencing (Sarot et al., 2004). These findings suggest that piRNAs encoded by flamenco trans-silence complementary transposons located outside this cluster (Brennecke et al., 2007).
The mechanism of trans-silencing by piRNA is not well understood. piRNA-PIWI protein complexes catalyze homology-dependent target cleavage, suggesting that target transposon mRNAs are co-transcriptionally or post-transcriptionally degraded (Gunawardane et al., 2007; Saito et al., 2006). However, several Drosophila piRNA pathway mutations have been reported to modify position effect variegation (PEV) (Brower-Toland et al., 2007; Pal-Bhadra et al., 2002; Pal-Bhadra et al., 2004), which is linked to spreading of transcriptionally silent heterochromatin from pericentric and telomeric regions (Girton and Johansen, 2008). Piwi protein also binds to heterochromatin in somatic cells, and interacts with Heterochromatin protein-1 (HP1) in yeast two-hybrid and immunoprecipitation assays (Brower-Toland et al., 2007). piRNA-Piwi protein complexes could therefore silence target transposons by directing assembly of heterochromatin-like domains. In fission yeast, which do not have piRNAs, siRNAs and Argonaute 1 (Ago1) appear to recognize nascent transcripts at the centromere, triggering both transcript destruction and HP1 recruitment and assembly of centromeric heterochromatin (Buhler et al., 2006; Verdel and Moazed, 2005). A similar combination of homology dependent cleavage and heterochromatin assembly could drive piRNA based silencing in the Drosophila germline.
The mechanism of piRNAs biogenesis also remains to be fully elucidated. Dicer endonucleases cleave double-stranded precursors to produce miRNAs and siRNAs (reviewed in Ghildiyal and Zamore, 2009), but piRNA production is Dicer independent (Houwing et al., 2007; Vagin et al., 2006). A subset of sense and antisense piRNAs overlap by 10 base pairs and show a strong bias toward an A at position 10 of the sense strand and a complementary U at the 5′ end of the antisense strand, suggesting that positions 1 and 10 base pair (Brennecke et al., 2007; Gunawardane et al., 2007). As Argonautes cleave their targets between positions 10 and 11 of the guide strand (Gunawardane et al., 2007; Saito et al., 2006), these finding suggest that piRNAs are produced by a “ping-pong” amplification cycle in which antisense strand piRNAs bound to Argonaute proteins cleave complementary RNAs to produce the 5′ end of sense piRNAs, which in turn direct a reciprocal reaction that generates the 5 end of antisense strand piRNAs (Brennecke et al., 2007; Gunawardane et al., 2007). However, most piRNAs cannot be assigned to ping-pong pairs, some clusters produce piRNAs from only one strand (Brennecke et al., 2007), and the mechanism of 3′ end generation has not been determined. It is also unclear how ping-pong amplification is initiated, since the cycle depends on pre-existing primary piRNAs.
Here, we show that Rhino (Rhi), a member of the Heterochromatin Protein 1 (HP1) subfamily of chromo box proteins (Volpe et al., 2001), is required for transposon silencing, production of piRNAs by dual-strand heterochromatic clusters, and efficient ping-pong amplification. Significantly, Rhi protein associates with the 42AB dual-strand cluster, and is required for production of longer RNAs from both strands of this cluster. Rhi thus appears to promote expression of trigger RNAs that are processed to from primary piRNAs that drive ping-pong amplification and transposon silencing. We also show that protein coding genes carrying transposons and transposon fragments within introns escape silencing, suggesting that piRNA silencing is imposed after RNA processing. Furthermore, rhi mutations disrupt nuage, a perinuclear structure that is enriched in piRNA pathway components. We therefore speculate that the nuage functions as a perinuclear surveillance machine that scans RNAs exiting the nucleus and destroys transcripts with piRNA complementarity.
Results
Drosophila piRNA pathway mutations lead to germ-line DNA damage and disrupt axis specification through activation of Chk2 and ATR kinases, which function in DNA damage signaling (Chen et al., 2007; Cook et al., 2004; Pane et al., 2007). Mutations in the rhi locus lead to very similar patterning defects. The mei-41 and mnk genes encode ATR and Chk2, respectively (Brodsky et al., 2004; Hari et al., 1995). To determine if the axis specification defects associated with rhi result from damage signaling, we generated double mutants with mnk and mei-41 and quantified axis specification by scoring for assembly of dorsal appendages, which are egg shell structures that form in response to dorsal signaling during oocyte development (Supplementary Table 1). Only 17% (n=700) of embryos from rhiKG/rhi2 females had two wild-type appendages. However, 80% (n=689) of embryos from mnk;rhiKG/rhi2 double mutant females had two appendages (Supplementary Table 1). In addition, 33 % (n=732) of embryos from mei-41;rhiKG/rhi2 double mutant females had two appendages (Supplementary Table 1). Consistent with these observations, rhi mutations disrupt dorsal localization of Gurken and posterior localization of Vasa in the oocyte, and localization of both proteins is restored in mnk; rhiKG/rhi2 double mutants (Figure 1).
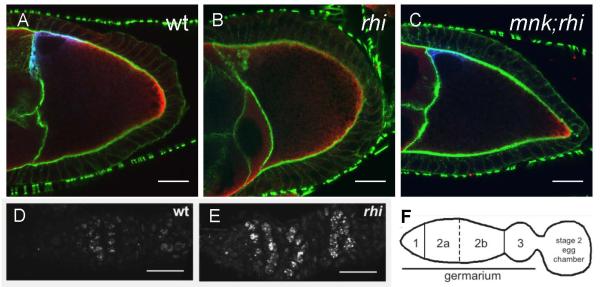
Figure 1. DNA damage signaling in rhi mutants.
A-C. Mutations in mnk, which encodes the DNA damage signaling kinase Chk2, suppress the Gurken and Vasa protein localization defects in rhi mutants. (A) In a stage 9 wild type oocyte, Grk (blue) is localized at the dorsal anterior cortex near the oocyte nucleus and Vas (red) is localized at the posterior cortex. Actin filaments (green) mark the cell boundaries. (B) In rhi egg chambers, this localization pattern is lost, with Grk and Vas dispersed throughout the oocyte. (C) mnk suppresses the rhi phenotype, and rescues Grk and Vas localization during late oogenesis. Images were acquired under identical conditions. Projections of 2 serial 0.6 mm optical sections are shown. Scale bars are 20 mm. D-F. rhi mutants have increased DNA damage in the germline. (D) Foci of γ̃H2Av are observed in wild type ovaries in region 2a and 2b of the germarium and correspond to the DSBs induced during meiotic recombination. (E) In rhi mutants, much larger foci also appear in region 2a of the germarium but persist in region 3 and the developing egg chambers. Samples were labeled and images were acquired under identical conditions. Projections of 5 serial 1mm optical sections are shown. Posterior is oriented to the right. Scale bars are 10 mm. (F) A schematic representation of the regions of the germarium and a developing egg chamber.
Both ATM and ATR kinases have been reported to activate Chk2 (Wang et al., 2006). Mutations in the Drosophila atm gene are lethal, but caffeine inhibits ATM and to a lesser extent ATR (Sarkaria et al., 1999). Strikingly, 88% (n=473) of embryos from rhi mutant mothers fed caffeine had wild-type dorsal appendages (Supplementary Table 1). Similarly, only 2% (n=277) of embryos from armi mutant females had two dorsal appendages, compared with 11% (n=477) following caffeine treatment (Supplementary Table 1). In addition, 56% (n=575) of embryos from mei41D3/mei41D3; armi72.1/armi1 females had wild-type appendages, but 83% (n=226) of embryos from mei41D3/mei41D3; armi72.1/armi1 double mutants fed with caffeine had two appendages (Supplementary Table 1). Caffeine combined with mei-41 mutations thus lead to levels of suppression that are similar to mnk single mutations, suggesting that ATM and ATR redundantly activate Chk2 in armi and rhi mutants.
The mei-W68 locus encodes the Drosophila Spo11 homologue, which is required for meiotic double-strand break formation (McKim and Hayashi-Hagihara, 1998). However, mei-W68 mutations fail to suppress the dorsal appendage defects associated with rhi (Supplementary Table 1), indicating that DNA damage signaling in rhi mutants is not due to defects in meiotic break repair.
The phosphorylated form of the Drosophila histone H2AX (γ–H2Av) accumulates near DNA double strand break sites (Gong et al., 2005; Modesti and Kanaar, 2001; Redon et al., 2002). In wild-type ovaries, γ–H2Av foci are generally restricted to region 2 of the germarium, where meiotic double strand breaks are formed (Figure 1D-F) (Jang et al., 2003). As the cysts mature and pass through region 3 of the germarium, γ–H2Av labeling is reduced. Stage 2 egg chambers, which bud from the germarium, show only low levels of γ–H2Av labeling. In rhi mutants, prominent γ–H2Av foci are present in germ-line cells of the germarium, and these foci persist and increase in intensity as cysts mature and bud to form stage 2 egg chambers (Figure 1D and E). rhi mutations thus appear to trigger germline-specific DNA breaks and damage signaling through ATM, ATR, and Chk2.
Transposon silencing and gene expression
The piRNA pathway is required for transposon silencing in the Drosophila female germ line (Vagin et al., 2006), but has also been implicated in heterochromatic gene silencing in somatic cells (Brower-Toland et al., 2007; Pal-Bhadra et al., 2002; Pal-Bhadra et al., 2004). We therefore assayed both transposon and protein-coding gene expression using whole genome tiling arrays (Figure 2). In both rhi and armi mutants, most transposon families show a relatively modest 1.5- to 2-fold increase in expression, which is not statistically significant (FDR >0.02). However, a subset of transposon families are dramatically over-expressed in both rhi and armi mutants (Figure 2 B and C; blue points indicate FDR<0.02). For example, HeT-A expression increased 70-fold in rhino and 117-fold in armi (Supplementary Table 2). In total, 15 of 17 transposon families that are significantly over-expressed in rhi are also over-expressed in armi (Supplementary Figure 1). 11 families are over-expressed with an FDR <0.02 in armi mutants, but not in rhi (Supplementary Figure 1). Rhino thus appears to silence a subset of the transposons silenced by Armi. This could reflect a role for Armi in transposon silencing in both somatic follicle cells and the germ line (Klattenhoff et al., 2007), while Rhi appears to be restricted to the germline (see below).
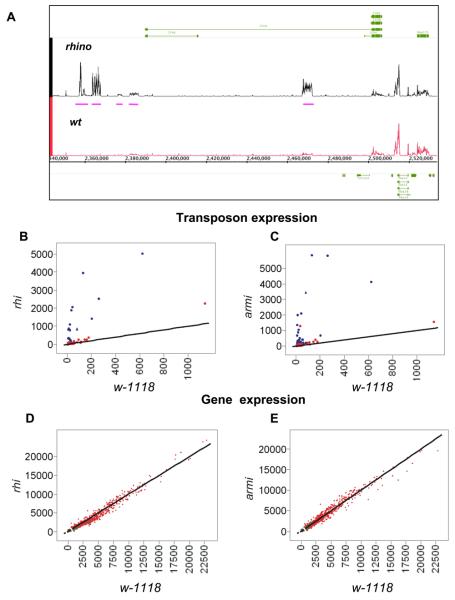
Figure 2. Gene and transposon expression in rhi mutant ovaries.
A. Genome Browser view of tiling array data in near jing, a protein-coding gene in pericentromeric heterochromatin on chromosome 2R. Expression of jing exons (green bars) is unaltered by rhi mutants. However, several intronic and extragenic transposons are significantly over-expressed (pink bars). B and C. Genome wide analysis of transposon family expression in rhi and armi mutants. Tiling arrays were used to quantify expression of 95 transposon families in rhi, armi, and w-1118 controls. Graphs show expression in rhi and armi plotted against expression in w-1118. The lines intercept the origin and have a slope of 1, and thus indicate equal expression in both genotypes. Significantly over-expressed transposon families are indicated by blue data points. D and E. Genome wide comparison of protein coding gene expression in rhi and armi mutants, plotted against expression in w1118. Heterochromatic genes are indicated by green data points and euchromatic genes are indicated by red data point. Both classes cluster around the diagonal, indicating similar expression levels in mutant and controls.
Both rhi and armi mutations increased expression of LTR elements, non-LTR retrotransposons, and IR-elements (Supplementary Figure 2) (Vagin et al., 2006), Similar patterns of transposon over-expression are observed in aub and ago3 mutants, which disrupt piRNA biogenesis (Li et al., 2009). Mutations in established piRNA pathway genes and in the rhino locus thus disrupt transposon silencing, independent of transposition mechanism.
piRNAs from the suppressor of stellate locus silence the Stellate gene during male germline development, and Stellate protein over-expression leads to Stellate crystal formation during spermatogenesis (Aravin et al., 2001; Bozzetti et al., 1995; Livak, 1984; Livak, 1990; Palumbo et al., 1994). However, rhi mutations do not lead to Stellate crystal formation or compromise male fertility (Supplementary Figure 3 and data not shown).
HP1 and several genes in the piRNA pathway have been implicated in position effect variegation, which is linked to spreading of heterochromatin from centromeric and telomeric regions (Elgin and Grewal, 2003; Pal-Bhadra et al., 2004). However, neither rhino nor armi led to statistically significant changes in the expression of any protein coding genes, including the 613 annotated heterochromatic genes (Smith et al., 2007) (Figure 2D and E, green points indicate heterchromatic genes). piRNA pathway and rhi mutations thus do not produce changes in heterochromatin organization sufficient to alter protein coding gene expression during oogenesis.
Figure 2A shows a Genome Browser view of the region containing the heterochromatic gene jing. Expression of exons that comprise the mature jing transcript are essentially identical in w1118 and rhi, but expression of a flea transposon located in a major intron increases 7 fold (FDR<0.02), and several transposons in the intergenic regions near jing are also over-expressed (Figure 2A, rhino, pink bars). The repeated nature of natural transposons and the design of the arrays makes it impossible to determine which specific transposon copy or copies are over-expressed, but we can conclude that at least one member of the transposon family is over-expressed. Over 1300 protein coding genes carry transposon insertions within introns, and thus have primary transcripts that could base pair with piRNAs. This includes ago3, which encodes an Arognaute protein that is expressed in the female germline and is required for ping-pong amplification of piRNAs (Li et al., 2009). Our array studies show that expression of ago3, and the other protein coding genes carrying intronic transposon insertions, does not significantly change in rhi or armi mutants (Figure 2D and E and data not shown). These observations suggest that piRNA dependent silencing may be imposed after splicing, which removed transposon homology from protein coding genes.
Rhi localization
To define the subcellular distribution of Rhi, we generated a GFP-rhi transgene and raised anti-Rhi antibodies, which were used to localize the protein in vivo and immunolabel whole mount egg chambers. Both methods revealed germ-line-specific nuclear foci that are present throughout oogenesis (Figure 3A-D). In addition, germ-line specific expression of the GFP-Rhi fusion protein rescued fertility and axial patterning in rhi mutations (Table 1). Rhi thus appears to function specifically within the germ-line cells of the ovary.
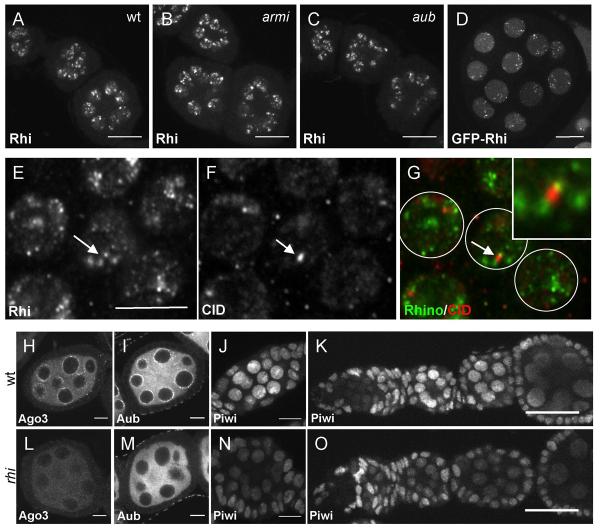
Figure 3. Rhi localization is independent of the piRNA pathway, but localization of the PIWI proteins Ago3 and Aub requires Rhi.
A-C. Rhi localization appears similar in (A) wild type, (B) armi and (C) aub stage 2 to stage 4 egg chambers. Projections of 5 serial 1mm optical sections are shown. Scale bar is 20 mm. D. GFP-Rhi transgene shows localization pattern similar to endogenous Rhi detected with anti-Rhi antiserum in the germline nuclei of stage 4-5 egg chambers. Scale bar is 10 mm. E-G. Wild type ovaries immunostained with (E and G) anti-Rhi antiserum and (F and G) anti-CID antibody show that some Rhi foci localize adjacent to CID foci (arrows) consistent with binding to peri-centromeric heterochromatin in some but not all chromosomes. Scale bar is 5 mm. H-O. rhi mutation disrupts localization of PIWI class Argonautes. Stage 4-5 (H, I, L and M), stage 2-3 (J and N), and germarium to stage 3-4 egg chambers (K and O) of wild type and rhi mutant ovaries were immunostained with corresponding antibodies. Projection of 3 serial 1mm optical sections. Scale bars (H, I, J, L, M and N) 10 mm (J,K and O) 20 mm. Wild type localization of Ago3 and Aub proteins to peri-nuclear nuage is disrupted in rhi mutants. Piwi protein localizes to the nuclei of both germline and somatic cells in wild type egg chambers. Only germline nuclear localization of Piwi in early stages is disrupted by mutations in rhi.
To determine if Rhi foci are associated with centromeres, we labeled for Rhi and CID, the Drosophila homologue of the centromere-specific, histone H3-like CENP-A (Blower and Karpen, 2001). Rhi accumulated in regions adjacent to most CID foci in germ-line nuclei, consistent with localization to pericentromeric heterochromatin (Figure 3E-G). However, many Rhi foci were not obviously linked to CID. Some of these foci could be linked to telomeres or other chromatin domains. Resolving this question will require higher resolution molecular approaches.
To determine if Rhi localization depends on the piRNA pathway, we immunolabeled egg chambers mutant for aub and armi. Rhino localization to nuclear foci was not disrupted by either mutation (Figure 3A-C). In striking contrast, rhi mutations disrupt localization of Aub and Ago3 to nuage, a perinuclear structure implicated in RNA processing (Figure 3H, I, L and M). Vasa is a core component of nuage, and perinuclear localization of Vasa was also lost in rhi mutants (Supplementary Figure 4). Piwi localizes to nuclei in both germ-line cells and the somatic follicle cells. In wild-type ovaries, Piwi is most abundant in germ-line nuclei during early stages of oogenesis (Figure 3K). In rhi mutants, nuclear localization of Piwi is reduced during these early stages (Figure 3N and O). However, in later stage egg chambers, which make up the bulk of the ovary, Piwi localization in rhi is similar to wild type controls (Figure 3J, K, N and O). These findings suggest rhi functions upstream of Ago3 and Aub, but may have a less critical role in Piwi-dependent processes.
piRNA expression is ablated for most transposon families in rhino mutants
To determine if Rhi is required for piRNA expression, we sequenced small RNAs from control and rhi mutant ovaries. Unlike miRNAs, piRNAs carry 2′ methoxy, 3′ hydroxy termini that render them resistant to oxidation and stabilize these RNAs in vivo (Vagin et al., 2006). To enrich for piRNAs and increase effective sequencing depth, we oxidized RNA samples prior to library construction and sequencing, and normalized the data to surviving non-coding RNA fragments (Ghildiyal et al., 2008; Seitz et al., 2008) (see Supplementary Table 3 for sequencing statistics). These studies indicate that rhi mutations reduce total piRNA abundance by approximately 80% (Figure 4A and B). Northern blotting for specific piRNAs and miRNAs support these findings (Supplementary Figure 5). Defects in 3′ modification destabilize piRNAs and would lead to preferential loss of piRNAs in oxidized samples. We therefore deep sequenced un-oxidized RNAs and normalized piRNA abundance to miRNAs. These studies confirn that rhi mutations reduce piRNA abundance by 80%, and indicate that this reduction does not result from a defect in end modification (data not shown).
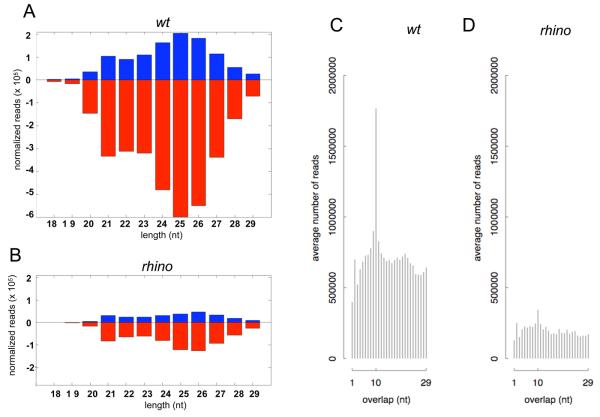
Figure 4. piRNA production in rhi mutants.
A. Length histogram of piRNAs expressed in wild type ovaries. B. Length histogram of piRNAs produced in rhi mutants. Sense and antisense piRNAs are reduced by approximately 80%, and peak length shifts from 25 nt to 26 nt. C. Histogram of overlapping sense and antisense piRNA in wild type ovaries, showing a pronounced peak at 10 nt, characteristic of ping-pong amplification. D. Histogram of overlapping sense and antisense piRNA in rhi mutant ovaries. The 10 nt peak is nearly eliminated, suggesting a breakdown in the ping-pong amplification cycle.
The majority of Drosophila piRNAs are derived from transposons and other repeated elements (Aravin et al., 2003; Brennecke et al., 2007). We analyzed the impact of rhi mutations on piRNA expression from 95 families with at least 500 matching reads in control samples (Supplementary Table 3; Li et al., 2009)). rhi mutations lead to a 50% or greater reduction in antisense piRNA abundance for 83% of these transposon families, and a 98% reduction in antisense piRNAs for approximately 30% of these elements (Supplementary Figure 7). For 66 of 95 families, both sense and antisense piRNAs are reduced. For example, rhi mutations nearly eliminate sense and anti-sense piRNAs from the telomeric transposon HeT-A (Figure 5A). Eight transposon families continue to express at least 50% of wild-type sense strand piRNAs, but show an 80% or greater reduction in antisense piRNAs. The jockey element falls into this class. Mutations in rhi reduce sense strand piRNAs linked to jockey by only 10%, but antisense strand piRNAs are reduced by 95% (Figure 4B, jockey). For all of the transposon families that show reduced antisense piRNAs, including those that retain sense strand piRNAs, there is a clear reduction in opposite strand piRNAs that overlap by 10 nt, consistent with defects in ping-pong amplification (Figure 5Ac and Bc). A comparison of the P-values for the 10nt overlap bias across all transposon families confirms that the loss of ping-pong pairs in rhi is very highly significant (Supplementary Figure 6, P= 3e-10). The loss of species that overlap by 10nt is also clear from an analysis of total piRNAs (Figure 4 C and D). The rhi mutations thus leads to a near collapse of the ping-pong cycle amplification cycle.
Figure 5. Transposon-specific changes in piRNA abundance.
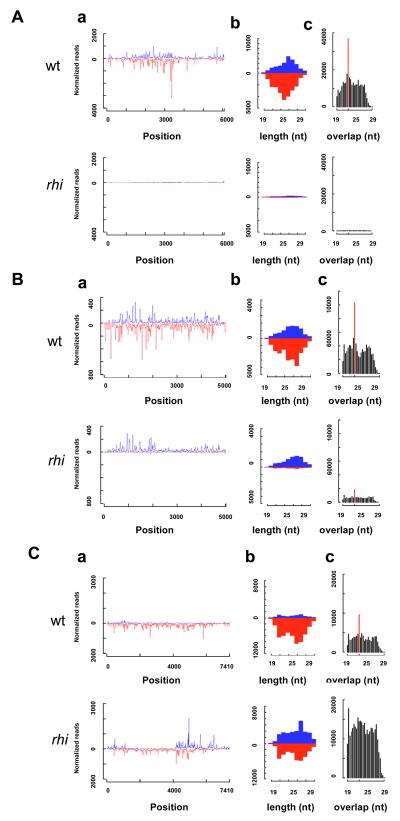
Aa. Sense and antisense piRNA reads mapping to the consensus Het-A sequence. Ab. Length histograms for all HetA piRNAs. Ac. Frequency distribution of overlapping HetA piRNA. A statistically significant 10 nt bias (red bar) is characteristic of ping-pong amplification. Ba. Sense and antisense piRNAs reads mapping to the consensus jockey sequence. Bb. Length histograms for all jockey piRNAs. Bc. Frequency distribution of overlapping jockey piRNAs. Ca. Sense and antisense piRNA reads mapping to the consensus blood sequence. Cb. Length histograms for all blood piRNAs. Cc. Frequency distribution of overlapping blood piRNA. The for the majority of transposons, including HetA, rhi mutations dramatically reduce sense and anti-sense piRNAs and nearly elinimate piRNA that overlap by 10nt. For a subset of elements, represented by jockey,rhi leads to a loss of anti-sense piRNAs but no significant reduction in sense strand piRNAs. A very limited number of transposons, including blood, show no change or an increase in sense strand piRNAs in rhi mutants. Mutations in rhi reduce piRNAs with a 10 nt overlap, even for elements that show an increase in piRNAs from opposite strands (Cc, blood).
Only 10 of 95 transposon families continue to express antisense piRNAs at or above 75% of wild-type levels in rhi mutants (blood, mdg-1, Tabor, Stalker, Stalker 2, Stalker3, Stalker4, 412, 297, gypsy 5; Supplementary Table 5). Eight of these families (blood, mdg-1, Tabor, Stalker, Stalker 2, Stalker3, Stalker4, 412) also show an increase in sense strand piRNAs (Figure 5Ca; Supplementary figure 8). The sense strand piRNAs generally map to the same regions as peaks of antisense piRNAs (Figure 5Ca, blood; Supplementary figure 8). This pattern could indicate that antisense strand piRNA direct production of the sense strand piRNAs. Alternatively, specific regions within full-length elements or fragments of elements that lie within specific clusters may be preferentially utilized during piRNA production. The available data cannot distinguish between these alternatives.
An analysis of piRNAs encoded by the 10 transposon families that show Rhi-independent piRNA production revealed three patterns with respect to overlapping sense and antisense species. The overlapping piRNAs encoded by Stalker3 did not show a statistically significant (P>0.001) 10 nt overlap bias in either wild-type or rhi mutants, indicating that their production is independent of ping-pong amplification. However, six families showed a statistically significant 10nt overlap peak in both wt and rhi mutants, indicating that at least some of the piRNAs are produced by a ping-pong cycle that is independent of Rhi (Tabor, Stalker, Stalker 2, Stalker4, 412, 297; Supplementary Figure 8). The final class of elements includes blood, mdg1 and gypsy5, which show a statistically significant ping-pong peak in wild type, but loose the 10 nt overlap bias in rhi mutants (Figure 5Cc, blood, Supplementary Figure 8). For this class, Rhi thus appears to promote production of only a subset of piRNAs through ping-pong amplification. Intriguingly, rhi leads to a 10-fold increase in blood expression, suggesting the minor ping-pong pool of piRNAs may be critical to transposon silencing (Supplementary Figure 9).
Overlapping ping-pong pairs show transposon family-specific nucleotide biases at positions 1 and 10 that appear to reflect the specific PIWI proteins that participate in the amplification cycle (Brennecke et al., 2007; Gunawardane et al., 2007). For example, elements for which sense strand piRNAs are primarily bound by Ago3 and antisense strand piRNAs are primarily bound by Aub, show an A bias at position 10 of the sense strand and a U bias at position 1 of the antisense strand (Brennecke et al., 2007; Gunawardane et al., 2007). Families that retain a statistically significant ping-pong peak generally retain the pattern of nucleotide bias observed in wild type (Supplementary Figure 8), suggesting that rhi reduces the efficiency of the ping-pong amplification, but does not alter the specific PIWI proteins that participate in the cycle.
Antisense piRNAs can base pair with target RNAs and guide cleavage by PIWI proteins, and are therefore presumed to be the effectors of transposon silencing. To determine if loss of antisense piRNAs in rhi mutants correlates with loss of silencing, we plotted the fold-change in transposon expression (_rhi/_wt) against the fold-change in antisense piRNAs (Supplementary Figure 9). All of the transposon families that increased in expression by 20 fold or greater in rhi mutants also showed a 75% or greater reduction in antisense piRNA abundance. In addition, none of the families that retained antisense piRNA expression at 80% or higher levels were significantly over-expressed (FRD<0.02; Supplementary Figure 9). However, many transposon families that show a reduction in antisense piRNAs abundance of over 10 fold did not show a statistically significant increase in expression (Supplementary Figure 9). These elements may be silenced by a piRNA independent mechanism. Alternatively, piRNAs linked to these elements could silence these elements, perhaps by inhibiting translation, without altering target transcript stability.
piRNA clusters
The majority of piRNAs match transposons that are present in multiple copies in the genome and cannot be uniquely mapped. However, piRNAs encoded by polymorphic transposons, divergent transposon fragments, or other unique sequences can be mapped. Chromosome profiles of these “unique mappers” reveal dispersed piRNA peaks in the euchromatic chromosome arms and a limited number of prominent peri-centromeric and sub-telomeric clusters, which appear to be the source of the majority of piRNAs (Brennecke et al., 2007). rhi mutations essentially eliminate piRNAs mapping to pericentromeric heterochromatin on all of the autosomes (Supplementary Figure 10 and Figure 6). By contrast, piRNAs mapping to the pericentromeric region on the X are retained (Supplementary Figure 10 and Figure 6).
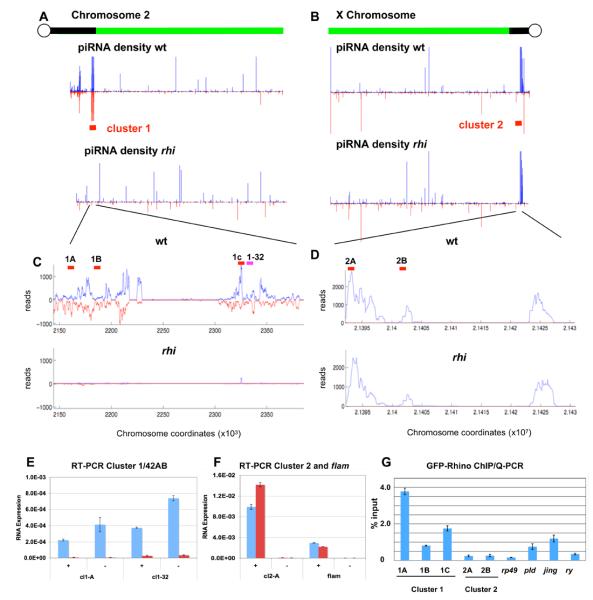
Figure 6. Rhino is required for piRNA production by dual-strand heterochromatic clusters.
A and B. Chromosome 2R and X density profiles of uniquely mapping plus (blue) and minus (red) strand piRNAs in wild type and rhi mutants. piRNAs map to dispersed loci on the chromosome arms and prominent heterochromatic clusters. Pericentromeric piRNAs from chromosome 2R (A) and all other autosomes (Supplementary Figure 10) are dramatically reduced in rhi mutants. Pericentromeric piRNAs on the X chromosome show relatively little change. C and D. Higher resolution maps of clusters 1 and 2, which map to the indicated regions on 2R and X, respectively. Mutaitons in rhi nearly eliminated piRNAs encoded by cluster 1, which is the major dual-strand cluster, but have little impact on piRNAs from cluster 2, which is the major uni-strand cluster. Quantitative strand-specific RT-PCR for RNA derived from dual strand cluster 1 (E) and uni-strand clusters 2 and flam (F). In wild type ovaries, RNA is detected from both the plus (+) and minus (−) strands of cluster 1, at two independent locations (F cl1-A and cl1-32, blue bars). RNAs from both strands of cluster 1 are dramatically reduced in rhi mutants (red bars). Significant levels of RNA are only detected from the plus strand of cluster 2 and flam (F, blue bars), and rhi does not block expression of these RNAs (F, red bars). G. Chromatin immunprecipitation/quantitive PCR analysis of Rhino binding to cluster 1/42AB, the euchromatic genes rp49 and ry, and the herterochromatic genes pld and jing, which flank cluser 1. Rhino protein is highly enriched at cluster 1 relative to cluster 2 and the euchromatic genes. The protein coding genes flanking cluster 1 show intermediate levels of binding, suggesting that Rhino may spread to regions flanking the dual strand clusters. Anti-GFP antibodies were used to precipitate Rhino-GFP from cross-linked ovary chromatin fractions. The approximate positions of the Q-PCR primer pairs used in ChIP and RT-PCR reactions are indicated by the red bars in panels C and D. The pink bar in C indicates the approximate position of an additional primer pair used in RT-PCR.
Most heterochromatic clusters produce piRNAs from both the plus and minus genomic strands (dual-strand clusters), but two major pericentromeric clusters on the X chromosome produce piRNA almost exclusively from one strand (uni-strand clusters) (Brennecke et al., 2007). We find that rhi reduces by 30 to 50 fold piRNAs from both strands of the top 11 dual-strand clusters (Supplementary Figure 11, blue bars). For example, piRNA production from cluster 1/42AB, which is estimated to produce up to 30% of all piRNAs(Brennecke et al., 2007), is reduced by over 97% (Figure 6A-C). In striking contrast, piRNAs encoded by the uni-strand clusters are only minimally impacted by rhi (Supplementary Figure 11, red bars). As shown in Figure 6 B and D, piRNAs from cluster 2 are derived almost exclusively from one strand, and production of these piRNAs is nearly unchanged in rhi mutants (Figure 6B and D). This does not appear to reflect expression of cluster 2 piRNAs exclusively in the somatic follicle cells, since Ago3 is germline specific and ago3 mutations reduce piRNAs linked to this locus by close to 20 fold (Supplementary Figure 12). In addition, unique piRNAs mapping to this cluster immunoprecipitate with the germline specific PIWI proteins Aub and Ago3 (Supplementary Figure 12). Both dual-strand and uni-strand clusters thus appear to be expressed in the germline, but rhi mutations only disrupt piRNA production by the dual-strand clusters. Consistent with these findings, 9 of the 10 transposon families that continue to express high levels of antisense piRNAs in rhi (75% of wild type or greater) have insertions in one or both of the major uni-strand clusters (Supplementary Table 4).
To determine if Rhino protein associates with clusters, we performed chromatin immunoprecipitation (ChIP) using anti-GFP antibodies and flies expressing a functional Rhino-GFP transgene specifically in the germline. To control for non-specific binding, precipitation was performed using non-immune IgG on chromatin from ovaries expressing the GFP-rhino transgene. Additionally, anti-GFP antibodies were used on chromatin isolated from wild-type flies that do not express the GFP fusion. Quantitiative-PCR (qPCR) assays showed only background signal in both of these control reactions (not shown). The anti-GFP fractions were assayed for three regions of cluster 1/42AB, two regions of cluster 2, two regions in the heterochromatic protein coding genes jing and pld that flank the 42A/B cluster, and the euchromatic protein coding genes rp49 and ry (Figure 6G). GFP-Rhi binding, measured as a fraction of input chromatin, was enriched at all three sites in the dual-strand cluster relative to the euchromatic protein coding genes (Figure 6F). By contrast, the two sites in uni-strand cluster 2 showed no enrichment relative to rp49 or ry controls (Figure 6F). Regions in the two heterchromatic genes immediately flanking the 42A/B cluster showed binding that was 3 to 4 fold lower than the peak region in the cluster (1A), and appoximately 3 fold higher than binding to euchromatic genes. Rhino thus appears to be enriched at dual-strand heterochromatic clusters, and may spread somewhat beyond the computationally defined limits of these clusters.
Cluster transcription
The piRNA clusters are proposed to produce long precursor RNAs that are processed to form primary piRNAs, which in turn trigger the ping-pong amplification cycle by targeting sense strand transposon transcripts. To determine if Rhino is required for RNA production by clusters, we used quantitative reverse transcriptase (RT) PCR to assay RNAs derived from both strands of cluster 1/42AB, cluster 2, and flam. Reactions without RT produced no significant signal, and the low level of signal obtained in the absence of the strand specific RT primers was subtracted from the signal obtained with the strand specific primers. Consistent with production of piRNAs from both genomic strands, we detected longer RNAs from both strands at two independent locations in cluster 1/42AB (Figure 6E, blue bars). Significantly, RNAs from both strands were nearly eliminated in rhi homozygous mutants (Figure 6E, red bars). At cluster 1 and flam, which produce piRNA almost exclusively form the plus strand (Figure 6D, Brennecke et al), RT-PCR detected RNA from only the plus strands (Figure 6F, blue bars). In striking contrast to cluster 1/42AB, rhi mutations led to a slight increase in plus strand transcript from cluster 2 and only a modest decrease in plus strand RNA from flam. These observations suggest that Rhino promotes production of precursor RNAs from dual-strand cluster 1/42AB, and possibly all dual-strand clusters.
Brennecke et al. (2007) proposed that anti-sense piRNAs derived from the clusters initiate ping-pong amplification by cleaving sense strand transcripts from target transposons. However, unique piRNAs derived from opposite strands of cluster1/42AB show a strong 10nt overlap bias (Supplementary figure 13), indicating that they are produced by ping-pong processing of precursor RNAs derived from the cluster. Anti-sense piRNAs derived through cluster based ping-pong amplification thus appear to target sense strand RNAs derived from functional transposons located throughout the genome.
Discussion
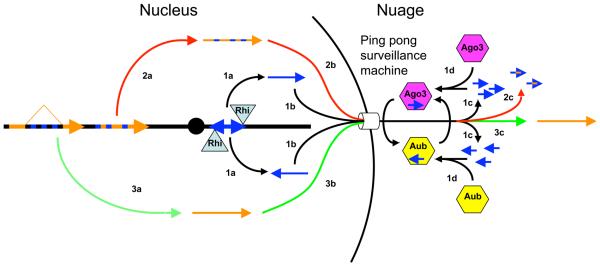
piRNAs encoded by transposon-rich heterochromatic clusters have been proposed to initiate a ping-pong cycle that amplifies the piRNA pool and mediates transposon silencing (Brennecke et al., 2007; Lin, 2007; O'Donnell and Boeke, 2007). However, the mechanisms of piRNA biogenesis and silencing are not well understood, and it is unclear how the piRNA clusters are differentiated from other chromatin domains. We show that the HP1 homologue Rhino is required for production of piRNAs from dual strand clusters and associates with the major 42AB cluster by ChIP. Significantly, we also identify putative piRNA precursor RNAs from both strands of the 42AB cluster, and show that Rhino is required for production of these RNAs. These findings lead us to propose that Rhi binding promotes transcription of dual strand clusters, and that the resulting RNAs are processed to form primary piRNAs that drive the ping-pong amplification cycle and transposon silencing (Figure 7, black pathway).
Figure 7. Model for Rhino-dependent transposon silencing.
1 (black arrows). Rhino binds to dual-strand clusters and promotes production of RNAs from both genomic strands (1a), which are exported from the nucleus (1b) and processed into piRNAs by a ping-pong cycle driven by Ago3 (pink hexagon) and Aub (yellow hexagon), localized to the perinuclear nuage (1c). 2 (red arrows). Transposons carrying piRNA homology are transcribed (2a), exported from the nucleus (2b), and degraded as they encounter the “ping-pong surveillance machine” within the nuage. 3 (green arrows). Protein coding genes with intronic transposon insertions are spliced, which removes piRNA homology (3a). These transcripts are exported from the nucleus (3b) escape recognition by the surveillance system, and are translated (3c). Sequences matching piRNAs are indicated by blue. Other transcribed regions are in orange.
While Rhino protein appears to be restricted to germline nuclei, rhi mutations disrupt perinuclear localization of Ago3 and Aub (Figure 3), which catalyze the ping-pong amplification cycle (Li et al., 2009). Mutations in krimper, which encodes a component of the perinuclear nuage, also disrupt transposon silencing and piRNA production (Lim and Kai, 2007). piRNA silencing and nuage assembly thus appear to be co-dependent processes. These observations, with our finding that protein coding genes carrying piRNA homology within introns escape silencing by the piRNA pathway (Figure 2A), suggest that transcripts are scanned for piRNA homology within the nuage, after splicing and nuclear export. Mature protein coding mRNAs thus pass through the nuage and are translated because piRNA homology has been removed by splicing. By contrast, mature transposon transcripts carry piRNA complementarity are recognized by the perinuclear ping-pong machine, leading to destruction. Interestingly, mutations in the mouse maelstrom gene disrupt nuage and lead to male sterility and significant over-expression of LINE-1 elements (Soper et al., 2008). Nuage may therefore have a conserved function in transposon RNA surveillance and silencing.
In S. pombe, siRNAs bound to Ago1 appear to recruit HP1 to centromeres through interactions with nascent transcripts, thus triggering heterochromatin assembly and transcriptional silencing. Our data indicate that the HP1 homologue Rhino is required for transposon silencing, but this process appears to be mechanistically distinct from centromeric heterochromatin silencing in yeast. For example, localization of the Rhino HP1 homologue to nuclear foci is independent of piRNA production, and Rhino binding appears to promote transcription of heterchromatic clusters. This in turn generates piRNAs that may direct silencing through post-transcriptional target cleavage. However, piRNAs bound to PIWI proteins have been implicated in heterochromatin assembly in somatic cells, and this process could be related evolutionarily to heterochromatin assembly in fission yeast.
Intriguingly, rhi is a rapidly evolving gene, and all three Rhi protein domains (chromo, chromo shadow and hinge) show evidence of strong positive selection (Vermaak et al., 2005). Based on these observations, Vermaak et al. (2005) proposed that rhino is involved in a genetic conflict within the germline. The observations reported here suggest that the conflict between transposon propagation and maintenance of germ line DNA integrity drives rhi evolution, and that the heterochromatic dual-strand clusters have a key role in this battle. Rhino appears to define heterochromatic domains that produce transposon silencing piRNAs. Rhino could therefore have evolved to bind transposon integration proteins, which would promote transposition into clusters and production of trans-silencing piRNAs. In this model, the transposon integration machinery would evolve to escape Rhino binding and silencing. The rapid pace of rhino evolution makes identification of homologues in other species difficult (Vermaak et al., 2005), but the conserved role for piRNAs in germline development suggests that HP1 variants may have critical roles in the conflict between selfish elements and genome integrity in other species, including humans.
Experimental Procedures
Drosophila stocks
All animals were raised at 25°C Oregon R, w1118 and cn1; ry506 were used as controls, as noted. The following alleles were used: mnkP6 (Brodsky et al., 2004; Takada et al., 2003); rhiKG00910 (rhiKG) and rhi02086 (_rhi_2) (Volpe et al., 2001); armi72.1 and armi1 (Cook et al., 2004); mei41D3, (Hari et al., 1995; Hawley and Tartof, 1983); P[lacW]mei-W68K05603, mei-W681 (McKim and Hayashi-Hagihara, 1998). The mnkP6 allele was kindly provided by M. Brodsky (Brodsky et al., 2004). All other stocks were obtained from the Bloomington Drosophila Stock Center (Consortium, 2003; http://flybase.org/). Standard genetic procedures were used to generate double mutant combinations.
Immunohistochemistry
Antibody production is described in Supplementary Experimental Procedures. Egg chamber fixation and whole-mount antibody labeling were performed as previously described (Theurkauf, 1994). Vas protein was labeled with rabbit polyclonal anti-Vas antibody (Liang et al., 1994) at 1:1000. Gurken protein was labeled with mouse monoclonal anti-Gurken antibody (obtained from the Developmental Studies Hybridoma Bank, University of Iowa) at 1:10. Rhi protein was labeled with a guinea pig polyclonal anti-Rhi antiserum developed by our group (see above) at 1:2000. Piwi, Aub and Ago3 were labeled with rabbit polyclonal anti-Piwi, anti-Aub and anti-Ago3 antibodies developed for this study (see above) at 1:1000. Antibody against γ-H2Av was kindly provided by Kim McKim (Gong et al., 2005) and egg chambers were labeled as described previously (Belmont et al., 1989). CID was labeled with an affinity purified chicken anti-CID antibody provided by Gary Karpen at 1:100 (Blower and Karpen, 2001). HOAP was labeled with a polyclonal rabbit anti-Hoap antibody generated by our group (see above) at 1:1000. Rhodamine-conjugated phalloidin (Molecular Probes) was used at 1:100 to stain F-Actin, and TOTO3 (Molecular Probes) was used at 1:500 (0.2 mM final concentration) to visualize DNA.
Labeled tissue was mounted and analyzed using a Leica TCS-SP inverted laser-scanning microscope as described previously (Cha et al).
GFP-Rhino transgene
The GFP-Rhi transgene was generated by recombining the Rhi-DONR (see above) construct with a modified pCasper vector containing the GFP sequence and Gateway cloning cassette B (Invitrogen). The resulting vector contained GFP fused in frame to the N-terminus of Rhino under the control of the Gal4 promoter. Transgenic animals were generated using standard embryo microinjection techniques at Genetic Services, Inc.
RNA isolation and tiling array hybridization
Total RNA from was isolated from manually dissected ovaries from 2-4 day old flies using RNeasy (Qiagen) according to the manufacturer's instructions. The RNA was quantified by absorbance at 260 nm. Three independent RNA isolates from each genotype was then assayed as follows: Double-stranded cDNA was prepared using GeneChip® WT Amplified Double-Stranded cDNA Synthesis Kit (Affymetrix). DNA was labeled using GeneChip® WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix). Labeled DNA was hybridized to GeneChip® Drosophila Tiling 2.0R Arrays (Affymetrix) using GeneChip® Hybridization, Wash, and Stain Kit (Affymetrix) at the University of Massachusetts Medical School genomic core facility.
To determine if genetic background or DNA damage significantly alters gene or transposon expression, we assayed ovarian RNA isolated from two common laboratory strains (w-1118 and cn,bw), the meiotic repair mutant okra, the DNA damage signaling mutant mnk, and mnk;okra double mutants. Pair-wise comparisons show little difference in genome wide patterns of gene or transposon expression in any of these five strains (Supplementary Figure 12). The background for the rhi heteroallelic combination used here is cn/+; ry/+, which is genetically wild type. Since the armi allelic combination used here is in a homozygous w-1118 background, this genotype was used a control in our array studies.
The tiling array data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE14370 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14370).
Small RNA isolation, oxidation, and sequencing were performed as described elsewhere (Li et al., 2009). Bioinformatics methods, chromatin immunoprecipitation and strand specific RT-PCR procedures are described in Supplementary Experimental Procedures.
Supplementary Material
01
02
03
04
Acknowledgements
We thank Celeste Berg for rhi mutant stocks, Maria Zapp and Ellie Kittler in the UMass Deep Sequencing Core and Phyllis Spartick in the UMass Genomics core for expert assistance with small RNA sequencing and tiling array analyses, and members of the Zamore, Weng and Theurkauf labs for critical discussions during the course of these studies. The Genomics Core was supported by the Diabetes and Endocrinology research Center grant DK032502. This work was supported in part by grants from the National Institutes of Health to W.E.T. (HD049116) and P.D.Z. (GM62862 and GM65236). H.X. was supported in part by Pfizer, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzetti MP, Massari S, Finelli P, Meggio F, Pinna LA, Boldyreff B, Issinger OG, Palumbo G, Ciriaco C, Bonaccorsi S, et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc Natl Acad Sci U S A. 1995;92:6067–6071. doi: 10.1073/pnas.92.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone C, Aravin A, Sachidanandam R, Stark A, Hannon G. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS ONE. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895–898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore P. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton J, Johansen K. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv Genet. 2008;61:1–43. doi: 10.1016/S0065-2660(07)00001-6. [DOI] [PubMed] [Google Scholar]
- Gong WJ, McKim KS, Hawley RS. All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 2005;1:e67. doi: 10.1371/journal.pgen.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 1999. Purifying Antibodies; pp. 70–80. [Google Scholar]
- Hawley R, Tartof K. The effect of mei-41 on rDNA redundancy in Drosophila melanogaster. Genetics. 1983;104:63–80. doi: 10.1093/genetics/104.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Jang JK, Sherizen DE, Bhagat R, Manheim EA, McKim KS. Relationship of DNA double-strand breaks to synapsis in Drosophila. J Cell Sci. 2003;116:3069–3077. doi: 10.1242/jcs.00614. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics. 1990;124:303–316. doi: 10.1093/genetics/124.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Hayashi-Hagihara A. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 1998;12:2932–2942. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M, Kanaar R. DNA repair: spot(light)s on chromatin. Curr Biol. 2001;11:R229–232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Palumbo G, Bonaccorsi S, Robbins LG, Pimpinelli S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics. 1994;138:1181–1197. doi: 10.1093/genetics/138.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Payen-Groschene G, Terzian C, Bucheton A. Restrictive flamenco alleles are maintained in Drosophila melanogaster population cages, despite the absence of their endogenous gypsy retroviral targets. Mol Biol Evol. 2007;24:498–504. doi: 10.1093/molbev/msl176. [DOI] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. Embo J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Shu S, Mungall C, Karpen G. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper S, van der Heijden G, Hardiman T, Goodheart M, Martin S, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Kelkar A, Theurkauf WE. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003;113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics. 2001;159:1117–1134. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Redpath JL, Fan ST, Stanbridge EJ. ATR dependent activation of Chk2. J Cell Physiol. 2006;208:613–619. doi: 10.1002/jcp.20700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04