Population genomics in a disease targeted primary cell model (original) (raw)
Abstract
The common genetic variants associated with complex traits typically lie in noncoding DNA and may alter gene regulation in a cell type-specific manner. Consequently, the choice of tissue or cell model in the dissection of disease associations is important. We carried out an expression quantitative trait loci (eQTL) study of primary human osteoblasts (HOb) derived from 95 unrelated donors of Swedish origin, each represented by two independently derived primary lines to provide biological replication. We combined our data with publicly available information from a genome-wide association study (GWAS) of bone mineral density (BMD). The top 2000 BMD-associated SNPs (P < ∼10−3) were tested for _cis_-association of gene expression in HObs and in lymphoblastoid cell lines (LCLs) using publicly available data and showed that HObs have a significantly greater enrichment (threefold) of converging _cis_-eQTLs as compared to LCLs. The top 10 BMD loci with SNPs showing strong _cis_-effects on gene expression in HObs (P = 6 × 10−10 − 7 × 10−16) were selected for further validation using a staged design in two cohorts of Caucasian male subjects. All 10 variants were tested in the Swedish MrOS Cohort (n = 3014), providing evidence for two novel BMD loci (SRR and MSH3). These variants were then tested in the Rotterdam Study (n = 2090), yielding converging evidence for BMD association at the 17p13.3 SRR locus (_P_combined = 5.6 × 10−5). The _cis_-regulatory effect was further fine-mapped to the proximal promoter of the SRR gene (rs3744270, _r_2 = 0.5, P = 2.6 × 10−15). Our results suggest that primary cells relevant to disease phenotypes complement traditional approaches for prioritization and validation of GWAS hits for follow-up studies.
Genome-wide association studies (GWAS) of polygenic traits such as bone mineral density (BMD) (Richards et al. 2008; Sigurdsson et al. 2008; Styrkarsdottir et al. 2008) are continuing to generate long lists of genomic regions with potential involvement in the disease in question. However, the effect sizes of common variants linked to disease are sufficiently small that even slight differences in discovery versus follow-up cohorts can have drastic consequences on the power of replication (Moonesinghe et al. 2008). Therefore, even some of the nonreplicating loci among GWAS could include true positive associations, rather than commonly held notion that nonreplication equals false associations arising by chance (Chio et al. 2009). Furthermore, GWAS replication efforts restricted to only top hits may miss many true hits as demonstrated in a recent study showing that single nucleotide polymorphisms (SNPs) that achieved genome-wide significance in the follow-up studies were not ranked in the top 1000 SNPs in the initial scan (Thomas et al. 2008). Consequently, alternative approaches for rational prioritization of GWAS hits are needed.
In an attempt to elucidate the functional role of significant disease loci from GWAS, data from expression quantitative trait loci (eQTL) studies, linking noncoding variants to expression differences at the cellular level, have been integrated with the information from disease GWAS (Emilsson et al. 2008; Schadt et al. 2008; Fraser and Xie 2009). Since genetic variation in gene expression is likely to be an important underlying mechanism in complex disease susceptibility, the use of eQTL data generated from cell models are believed to be a useful tool in the dissection of disease associations. However, the limited access to human primary cells have resulted in many of the human eQTL studies utilizing Epstein-Barr virus transformed lymphoblastoid cell lines (LCLs) (Dixon et al. 2007) such as those analyzed in the HapMap project (Stranger et al. 2007). More recently, complex tissues such as unsorted peripheral blood mononuclear cells (PBMC) (Emilsson et al. 2008; Heinzen et al. 2008), liver (Schadt et al. 2008), adipose (Emilsson et al. 2008), and cortical brain tissues (Heinzen et al. 2008) have been used as RNA sources. Although eQTL studies in tissues may better represent disease physiology, the cell specificity of _cis_-regulation indicates that in heterogeneous tissues the genotype–expression correlations are confounded and large samples may be needed to pinpoint the true causal variants. However, even in cell models, nongenetic confounders can be significant as demonstrated in a recent study using the HapMap LCL samples showing that biological noise and in vitro artifacts can explain much of the population variation in gene expression (Choy et al. 2008).
Here, we use a primary cell model of cultured human osteoblasts (HOb) to aid in the genetic dissection of BMD, the only standardized phenotype currently used for the diagnosis of osteoporosis. Osteoblasts are responsible for the production of bone matrix in the bone remodeling cycle and have been used extensively as a cell model for various metabolic bone diseases (Ducy et al. 2000). The osteoblasts are obtained through explant-outgrowth from donors undergoing hip or knee replacement surgery and, although the cell source cannot be defined unambiguously, they exhibit a full range of osteoblastic features (Jonsson et al. 1999). We recently showed in a comprehensive comparative transcriptome study that a vast number of genes and networks involved in bone metabolism are expressed predominantly in human bone cells (obtained from bone explants) as compared to the most closely related cell types: human fibroblasts and chondrocytes (Grundberg et al. 2008).
The first GWAS of bone phenotypes was recently presented (Styrkarsdottir et al. 2008) highlighting five genomic loci associated with BMD and included well-known susceptibility genes, e.g., TNFSF11, TNFRSF11A, TNFRSF11B, and ESR1. BMD is a highly heritable (Pocock et al. 1987), complex trait and, similar to human height (Gudbjartsson et al. 2008; Lettre et al. 2008; Weedon et al. 2008), tens or possibly hundreds of loci are believed to influence the variation of BMD, indicating that many loci remain to be identified.
We designed a population genomic study of gene expression in human osteoblasts and addressed potential environmental confounding in cell culture by including true biological replicates. Biological replicates increase our power to find true _cis_-eQTLs and we establish that most _trans_-associations are false. We show that population genomic data generated in osteoblasts are more relevant to bone disease than similar data generated in lymphoblastoid cells. Consequently, we apply the _cis_-eQTL data from cultured osteoblasts for prioritization of genomic loci at P < 10−3 identified in the GWAS of BMD (Styrkarsdottir et al. 2008) and identify a novel BMD associated gene (SRR) where promoter variation alters gene expression.
Results
We obtained whole-genome expression profiles from cultured bone cells (HOb) derived from 95 unrelated individuals (n = 42 females, n = 53 males, year of birth 1913–1982; median year of birth 1950) each with two biological replicates, using Illumina HumanRef8 v2 arrays. No effect of age on gene expression was seen as assessed in a principal component analysis (PCA) (Supplemental Fig. 1). The biological replicates represent multiple cell cultures of osteoblasts derived from independent bone chips from the same individual, and were cultured separately throughout the experiment (∼60 d/sample). PCA on normalized expression data did not identify any outliers, and the expression scores from the biological replicates were averaged to obtain a single measure for the 18,144 genes included on the array. We assessed whether technical effects influence gene expression intensities by correlating the expression signal for all probes for the first biological replicate with that of the second replicate across all 94 samples. We found a strong correlation between the biological replicates (median r = 0.99) indicating a minor effect of technical confounders on the expression data.
Of the total number of probes included, ∼6% overlapped at least one SNP (dbSNP build 126; Supplemental Table 1). We note that the majority of the overlapping SNPs are either (1) not validated or (2) rare with minor allele frequency (MAF) <0.1, and thus would probably not bias the results with spurious eQTLs given the sample size of the present study. However, in order to exclude this possibility, all probes overlapping a SNP were removed in the subsequent comparative analyses. We obtained whole-genome genotyping information of all samples using the Illumina Hap550K chips.
We tested all the genotyped SNPs with MAF > 0.1 and which passed our quality control (n = 383,547 SNPs) in association with gene expression (cis or trans) using a linear regression model. One individual with a low genotyping rate was excluded, resulting in a total of 94 samples included in the final association study of SNPs and expression traits. All expression traits were initially adjusted for sex and year of birth but, since it was recently shown (Leek and Storey 2007) that in addition to these measured variables there are sources of signal due to unknown or unmeasured factors that could reduce power or introduce sources of spurious signals in expression studies, we modified the regression analysis accordingly. We performed a surrogate variable analysis (SVA) in order to identify these factors and found one significant surrogate variable with no correlation to the known covariates, date of birth (r = −0.29) and sex (r = 0.09), respectively. We then included the newly identified surrogate variable as a covariate in the regression model. However, adding the “unmodeled factor” that was identified in the SVA as a covariate together with sex and year of birth did not increase the biological accuracy, and only a minor change in ranking of _cis_-eQTLs was seen following SVA adjustments (Supplemental Fig. 2). In fact, 83% of the top 1000 _cis_-eQTLs overlapped the top 1000 _cis_-eQTLs obtained after SVA adjustments. Thus, in all subsequent analysis only sex and year of birth were included as covariates.
Association of SNPs with gene expression in human primary osteoblasts
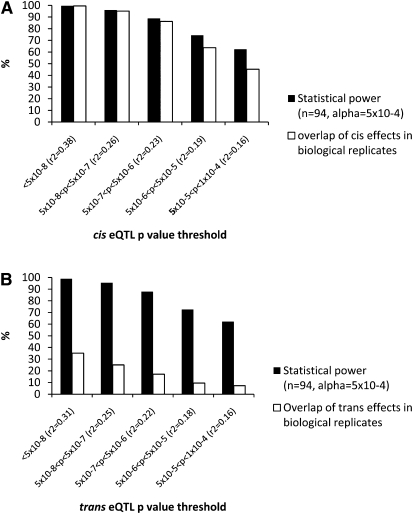
In order to assess replicability of _cis_- versus _trans_-associations, we separated the two replicates and performed regression analysis using each biological replicate separately. A total of 1,404,011 tests were carried out in 17,080 genes, yielding an average of 77 association tests per gene. At P < 3.5 × 10−8 (Bonferroni adjusted _P_ = 0.05/1,404,011) we found 590 and 654 _cis_-eQTLs, in the first and second biological replicate, respectively (testing limited to 250 kb of flanking sequence). Comparing _cis_-eQTLs for the two biological replicates analyzed separately we found a high overlap (>80%) of significant associations with medium to large effect sizes (_r_2 > 0.19, P < 5.0 × 10−5; Fig. 1A). Using this threshold, we found a total of 2484 significant _cis_-eQTLs in the first biological replicate where 2058 of these were also found in the second biological replicate. The observed overlap of shared _cis_-eQTLs between independently derived cell panels closely parallels the proportion predicted by statistical power down to _P_ ∼ 10−5 level in biological replicates. This suggests that most _cis_-eQTLs at _P_ ≤ 10−5 in our data are true. Conversely, _trans_-regulatory SNPs (located on a different chromosome than the measured gene) show much less than expected overlap in biological replicates based on power calculations, suggesting that the majority of them are false. Even _trans_-eQTLs with large effect sizes (_r_2 > 0.30) observed in one replicate show only 36% overlap in the second biological replicate despite ∼100% power in replication (Fig. 1B).
Figure 1.
eQTLs in biological replicates of primary human osteoblasts. All expression traits from two biological replicates (replicate 1 = R1 and replicate 2 = R2) of 94 cultured human osteoblasts were tested in separate analysis for association with SNPs with putative _cis_- or _trans_-regulatory effects. The overlap between the replicates (white bars) was calculated at different _P_-value thresholds of R1 ranging from P < 5 × 10−8 – 1 × 10−4 (_x_-axis) and compared with the average statistical power (black bars) to detect the differences using alpha = 5 × 10−4 (R2) and effect sizes (_r_2) corresponding to each significant eQTL in R1. (A) _cis_-eQTLs are defined as expression traits associated with SNPs located ±250 kb region flanking either side of the gene. (B) _Trans_-eQTLs include SNP–gene associations located on different chromosomes.
Driven by these observations, we focused solely on eQTLs that have putative _cis_-regulatory effects on gene expression in bone cells using the average of two biological replicates as the trait value for the 17,080 genes tested. Of those, we detected 994 _cis_-eQTLs (see Supplemental Table 2 for the complete list) at P < 3.5 × 10−8 corresponding to 235 genes that were associated with at least one SNP. We note that the use of both biological replicates increased the number of hits at this significance level by ∼60% as compared to the use of either replicate alone. Using the less conservative false discovery rate (FDR) approach (Benjamini and Hochberg 1995) for multiple testing of expression traits we detected 4937 _cis_-eQTLs at a 0.05 FDR level and 2931 at a 0.01 FDR level, respectively.
In a comparison study of publicly available data from cortical brain tissue and PBMCs with a similar experimental design and statistical power (Heinzen et al. 2008), we examined the total number of high-confidence, independent _cis_-eQTLs (defined as probes mapped to RefSeq transcripts and associated with at least one SNP). In total, five _cis_-eQTLs using cortical brain tissues and 18 using PBMCs were reported at P < 5 × 10−9. At the same _P_-value cutoff, we found 112 and 139 _cis_-eQTLs in the first and second biological replicate, respectively, indicating that the numbers of detected _cis_-eQTLs in HObs were more than an order of magnitude higher (>10-fold) than in the complex tissues. The higher number of _cis_-eQTLs identified in HObs could, in principle, be driven by a higher false discovery rate in our study. To exclude this possibility we did comparative analyses of _P_-values corresponding to 0.01 FDR in the three data sets (including all transcript level association tests by us and Heinzen et al. [2008]) and found that the corresponding _P_-values were 1.4 × 10−5 for HObs, 9.5 × 10−6 for PBMCs, and 2.0 × 10−6 for brain samples. The higher _P_-value corresponding to the same FDR cutoff in primary cells versus complex tissues suggests that the above estimate for a higher discovery rate of _cis_-eQTLs in HOb data may even be conservative.
We have made the osteoblast _cis_-eQTL results available through an interactive web-based browser (http://www.regulatorygenomics.org).
Integration of _cis_-eQTLs with disease associations
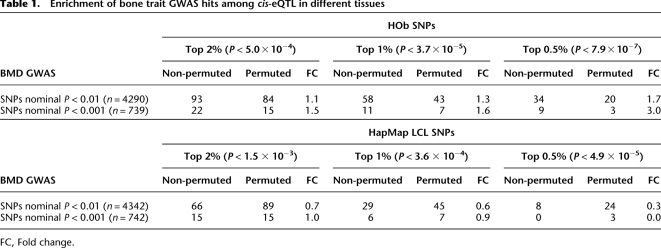
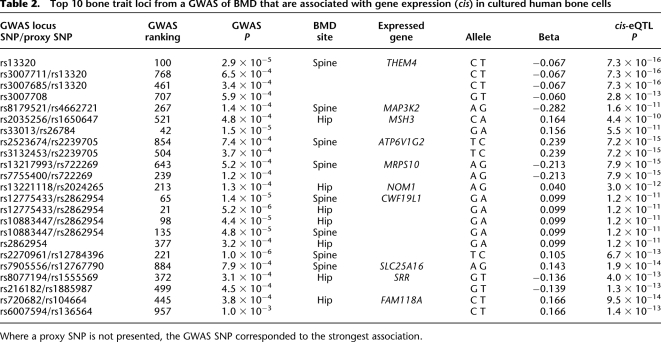
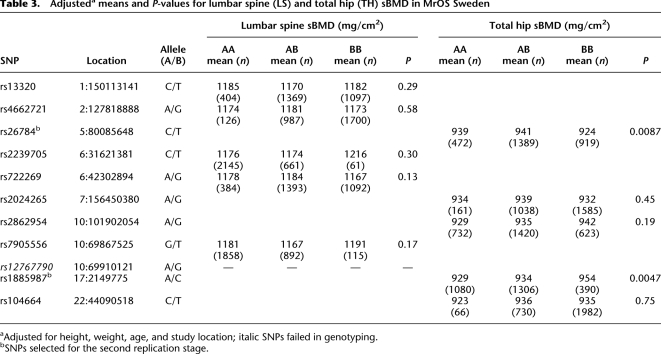
Recently a GWAS (Illumina Hap300K) of BMD (Styrkarsdottir et al. 2008) published all SNPs associated either with hip or spine BMD. We examined the relative levels of replication of significant SNPs (P < 0.01) from the full set of ∼300K SNPs with gene expression using (1) our data set of 94 HObs and (2) publicly available data using 60 HapMap LCL data (Stranger et al. 2007). We used a frequency-binning algorithm (see Methods) where SNPs were grouped into classes depending on MAF (10%–50%) to account for the higher likelihood of detecting positive associations in all data sets for more common alleles. Converging SNPs (i.e., significant in both studies) at various _P_-value thresholds were counted and compared with the same list that was permuted 10 times (Table 1), which represents the overlap expected by chance in MAF-adjusted data. Using the top significant HOb _cis_-eQTLs, we observed an enrichment ratio of 3 for converging _cis_-eQTLs in HObs for BMD GWAS hits at _P_ < 0.001. For BMD-associated SNPs no enrichment of _cis_-eQTLs in LCLs was observed. Furthermore, in a pairwise comparison using identically sized LCL and HOb sample sets measured by the same microarray technology we observed a significant enrichment of SNPs overlapping BMD hits in HObs (permutation significance <0.05) but no enrichment in LCLs (Supplemental Table 3). We then examined the top 1000 GWAS hip and spine BMD hits, respectively, using the Illumina 300K chips (Styrkarsdottir et al. 2008) as well as their best proxy SNP (defined as D′ > 0.8, MAF > 0.1, and mapping ±50 kb from the GWAS hit) included on the Illumina 550K chip for their association with gene expression (Supplemental Table 4). The top 10 GWAS loci with SNPs showing strong _cis_-effects on gene expression in HObs are presented in Table 2. Notably, the putative functional SNPs were ranked 21st–957th (median 377th) in the original GWAS list. We selected the top SNP from each locus for further validation studies using a two-step replication strategy using Caucasian male subjects of European origin including (1) the MrOS Cohort, Sweden (n = 3014), and (2) the Rotterdam Study, Netherlands (n = 2090). Cohort characteristics are presented in Supplemental Table 5. We tested all 10 SNPs for association with hip or spine BMD as indicated by the original data from the GWAS in the MrOS cohort (Table 3). Using a Bonferroni cutoff of P = 5.0 × 10−3 (0.05/10) we identified one significant SNP (rs1885987, P = 4.7 × 10−3) and one suggestive SNP (rs26784, P = 8.7 × 10−3), respectively. We pursued replication of the rs1885987 (SRR) and rs26784 (MSH3) associations with hip BMD in 2090 elderly men included in the Rotterdam Study. With genotype frequencies of CC = 0.12, CA = 0.47, and AA = 0.41 (MAF, C = 0.35, H-W, P = 0.20), we found a significant association of the rs1885987 SNP with hip BMD (P = 9.3 × 10−4, beta = 0.11). However, we were unable to replicate the previous findings of the rs26784 SNP with hip BMD (P = 0.44). The association of rs1885987 with BMD in males yielded a combined _P_-value of 5.6 × 10−5 (χ2 = 24.7, Fisher's combined probability test). We note that no interaction between rs1885987 genotypes and sex was observed in eQTL tests, indicating sex as an independent effect of the SRR locus on gene expression. We thus pursued replication of the rs1885987 SNP with femoral neck BMD in 2731 female subjects from the Rotterdam Study and found a similar trend for association to BMD although not significant (P = 0.14, beta = 0.04). However, the combined analysis of male and female subjects from the Rotterdam Study (n = 4821) showed a significant association of rs1885987 genotypes with femoral neck BMD (P = 0.001, beta = 0.07).
Table 1.
Enrichment of bone trait GWAS hits among _cis_-eQTL in different tissues
Table 2.
Top 10 bone trait loci from a GWAS of BMD that are associated with gene expression (cis) in cultured human bone cells
Table 3.
Adjusteda means and _P_-values for lumbar spine (LS) and total hip (TH) sBMD in MrOS Sweden
Fine-mapping and validation of functional SNPs in a novel BMD locus: Serine racemase (17p13.3)
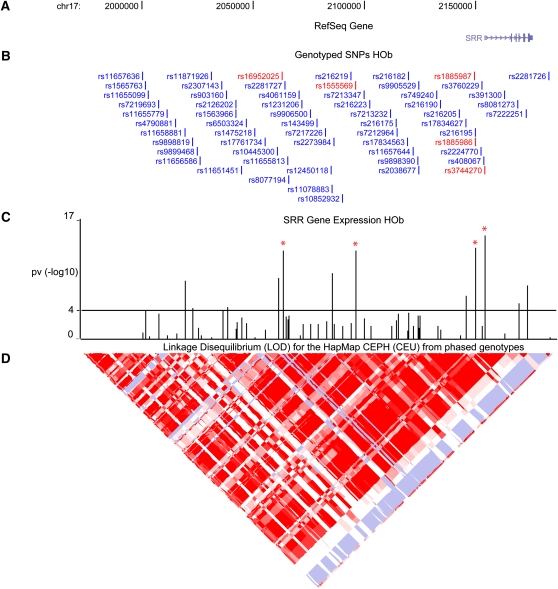
We followed-up the novel disease-associated locus, the serine racemase (SRR) gene located on chromosome 17p13.3, in greater detail. We fine-mapped (chr17:2,000,670–2,183,155) and re-sequenced (chr17:2,148,922–2,155,389) the candidate region in the HOb samples followed by further genotyping to allow delineation of the causal variant(s) (see Supplemental Table 6 for complete list of genotyped SNPs). We performed linear regression analysis and found that SNPs in close linkage disequilibrium (LD) with rs1885987 were strongly associated with SRR expression, with the rs3744270 SNP in the promoter region explaining most of the variance (_r_2 = 0.5; Fig. 2). We confirmed the significance and direction of the association analysis of the SRR SNPs by quantitative RT-PCR (Supplemental Fig. 3).
Figure 2.
Fine-mapping and validation of the serine racemase (17p13.3) locus. (A) The serine racemase (SRR) gene located on chromosome 17 was fine-mapped and re-sequenced; (B) the SNPs were associated with expression levels of SRR in 94 cultured human osteoblasts. (C) The _P_-values from linear regression analysis represented as –log10(_P_-value) are shown as vertical bars with a horizontal line indicating a cutoff of P = 10−4. SNPs in close LD with rs1885987 showing the most significant association with SRR expression are marked in red. (D) The HapMap PhaseII CEU LD blocks are shown as a diamond-shaped plot using log odds (LOD) measures.
We note that the regulatory potential of the rs3744270 region in vitro was suggested in earlier studies (Morita et al. 2007). We tested three haplotypes containing the rs3744270 SNP in a pGL3 basic vector (promoter activity) by transient transfection assays in five cell types but the results were inconclusive: We could not replicate the previously published transfection results in HeLa cells, and in the other cell types studied patterns of promoter haplotype on reporter gene expression varied between cell types (Supplemental Fig. 4).
Functional validation of new sequence variants associated with BMD
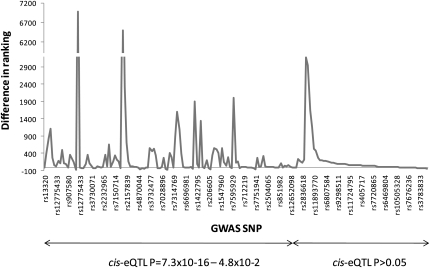
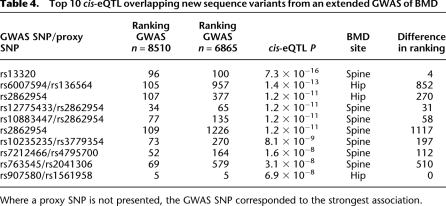
Following our initial selection of BMD-_cis_-eQTL overlaps, an extension of the original GWAS (Styrkarsdottir et al. 2008) for BMD in Icelandic population was published, increasing the study size from 6865 to 8510 subjects (Styrkarsdottir et al. 2009). The larger study changed the rank order of the BMD associations significantly. Therefore, we examined the new top ∼200 ranked SNPs for spine and hip BMD as well as their best proxy SNP (defined as D′ > 0.8, MAF > 0.10, and mapping ±50 kb from the GWAS hit) included on the Illumina 550K chip for their association with gene expression (Supplemental Table 7), and 12% of those were associated with cis gene expression at a nominal _P_-value of <10−4 (range _P_ = 7.3 × 10−16 − 7.1 × 10−5). We looked at the difference in ranking of the hip and spine SNPs before and after the GWAS was extended and found that SNPs associated with gene expression at _P_ < 10−4 corresponded to a median improvement in ranking of 112; while SNPs associated with gene expression at _P_ > 10−4 had a median improvement of only 34. The ranking differences of all SNPs sorted by the _P_-value from gene expression associations are graphically presented in Figure 3. If restricted to SNPs whose ranking improved by more than 100 upon sample size increase, the SNPs with overlapping _cis_-eQTLs (P < 10−4) were observed in 65% (15/23) of cases whereas SNPs _cis_-eQTLs with _P_ > 10−4 were significantly (χ2 = 7.8, P = 0.005) less likely to show this level of improvement in GWA rank as it was observed in only 35% (61/174) of cases. We then looked at the top 10 GWAS loci from the list of new sequence variants with the strongest association to gene expression in greater detail and found three examples among the top 10 where ranking improved by more than 500 upon sample size increase (Table 4). Taken together, this suggests that functional SNPs have higher a priori likelihood of associating with disease. We note, however, that the SRR locus was not among the top 10 listed in Table 4 but the SRR marker (rs216182) included in the GWAS was only weakly associated with gene expression (P = 1.6 × 10−4; Supplemental Table 4), as compared to SNPs close by and in moderate LD (rs1885987, P = 1.3 × 10−13; _r_2 = 0.2, D′ = 0.8), suggesting that the bone trait GWAS may have lacked adequate power to reveal highly significant association between hip BMD and rs216182.
Figure 3.
Difference in ranking of BMD GWAS SNP following sample size increase. The top ∼200 ranked SNPs that were identified in an extended GWAS (from 6865 to 8510 Icelandic subjects) of BMD were functionally validated using HOb _cis_-eQTLs. The rank order differences, i.e., ranking in original GWAS-ranking in extended GWAS, (_y_-axis) of each SNP (_x_-axis) are connected with a line. SNPs are sorted based on _cis_-eQTL _P_-value where the SNPs with the smallest values are presented to the left and vice versa. The specific rs-number of every 7th SNP is presented on the _x_-axis but the line chart includes data points from all 200 SNPs.
Table 4.
Top 10 _cis_-eQTL overlapping new sequence variants from an extended GWAS of BMD
Discussion
Previous studies on the genetic effect of gene expression using complex tissues or immortalized cell lines have either used single replicates or technical replicates separating the samples at the RNA level for the detection of _cis_- and _trans_-regulatory variants (Stranger et al. 2007; Schadt et al. 2008). To our knowledge, this is the first study evaluating population genomics in independently derived primary cell lines using biological replicates. Several lines of evidence suggest that our desire to biologically control eQTL analysis was successful. The _cis_-eQTLs were highly reproducible in the biological replicates based on simple power calculations accounting for effect size detected in one replicate, suggesting that, even if cell culture-induced confounders play a role in gene expression of individual lines (Choy et al. 2008), the detected _cis_-eQTLs are true down to P < 10−5 level. Consequently, significant improvement in detection of _cis_-eQTLs was achieved by combining the data from independent replicates. In contrast, t_rans_-eQTLs were detected at much lower rates than expected based on identical power calculations and, even for _trans_-eQTLs with large effect sizes, <40% could be replicated in the corresponding biological replicate. This indicates that most of _trans_-eQTLs detected herein are false-positives and larger sample sizes are needed in order to find SNPs with true _trans_-regulatory effects on gene expression. We compared the data with publicly available _cis_-eQTLs from primary tissues and cells using similar experimental designs that were recently published (Heinzen et al. 2008) and found that the numbers of detected _cis_-eQTLs were almost an order of magnitude higher in our cultured osteoblasts than in the complex tissues. While differences in FDR could be excluded as the source of this observation, the differences in genetic architecture of eQTLs between cell types could partly be a possible explanation to the observed difference in numbers of detected _cis_-eQTLs. Nevertheless, our observations raise the possibility that population genomic analyses targeted to assign function to genetic variants detected in association studies of clinical traits can benefit from cell versus complex tissue-based disease models. If the primary goal is to develop biomarkers and generate gene networks across cell types, then tissue-based population genomics is most likely more appropriate (Emilsson et al. 2008; Schadt et al. 2008).
In our previous studies of cultured osteoblasts from individual donors (Grundberg et al. 2008) or from panels of donors (Kwan et al. 2009) we showed that the osteoblast transcriptome expresses many key genes and pathways involved in bone remodeling, e.g., genes in the insulin-like growth factor (IGF), Wnt/beta-catenin and bone morphogenetic protein (BMP) signaling pathways, respectively. On the other hand, the LCL transcriptome showed expression pattern of genes included in B-cell receptor signaling, NFKB signaling, and natural killer cell signaling, which are all involved in the immune response system and are typically functions associated with lymphocytic cell types. Taken together, this suggests that the cell model should target the particular disease or phenotype of interest as the population panels of osteoblasts do for the analysis of bone diseases.
In this study, we provide multiple lines of evidence that _cis_-eQTLs detected in appropriate cell models can be used to prioritize GWAS hits. First, we show statistically significant enrichment of BMD-associated variants from GWAS at P < 0.001 (i.e., top 1000 SNPs from GWAS list) among _cis_-eQTLs detected in bone cells compared to LCLs. We also show that, by increasing the sample size of GWAS (Styrkarsdottir et al. 2008, 2009), thus improving the statistical power, SNPs with a _cis_-regulatory effect, as shown in our panel of osteoblasts, will considerably increase its ranking, suggesting that the intersection of GWAS and _cis_-eQTL data may allow more efficient selection of SNPs for follow-up. It is notable that, in cases where there was preferential improved ranking of functional SNPs in GWAS hits of a clinical trait, the data sets originated from the same population alone. More commonly, meta-analyses increasing the sample size for GWAS are achieved through combination of data from different populations, which can only highlight population-independent associations. Therefore, functional prioritization for one primary GWAS may identify different hits than large meta-analyses including the same SNP.
Another advantage of the prioritization strategy is the possibility to assure the identification of factors influencing a specific pathway. Genuine genetically determined differences in BMD across individuals may be the results of factors acting through both bone-specific and bone-independent pathways. Factors underlying the associations identified through this strategy are already proven to exert their influence on bone biology (osteoblasts) instead of other extra-skeletal influences (e.g., co-morbidity, weight change, muscle load, etc.).
Regulatory variants controlling expression of a novel candidate gene, the serine racemase (SRR) gene on chromosome 17p13.3, emerged through our prioritization analysis yielding replication of the BMD association in two moderately large population-based cohorts. The SRR protein was purified a decade ago (Wolosker et al. 1999) and shown to convert L-serine to D-serine, the endogenous ligand of the glycine site of NMDA receptors in the brain, and since then the focus has been on its association to neurological disorders (Yamada et al. 2005; Morita et al. 2007). However, certain subunits of NMDA receptors are known to be expressed in osteoblasts and chondrocytes (Chenu et al. 1998; Laketic-Ljubojevic et al. 1999; Hinoi et al. 2003), and recently it was shown that SRR negatively regulates chondrocyte differentiation through the inhibition of the SOX9 transcriptional activity (Takarada et al. 2008). SOX9 regulates COL2A1 expression and other genes involved in endochondral bone formation, and thus is considered as a key player in chondrogenesis. Here, we show an association of the SRR rs1885987 G allele with low expression of the gene itself while the same allele was associated with high BMD in both our studies supporting a negative effect of SRR in bone cells as shown by Takarada et al. (2008).
The large size-effect of the _cis_-eQTL detected for SRR (_r_2 = 0.5) allowed efficient fine-mapping to the proximal promoter along with validation by independent expression methods. The fact that in vitro transient transfection assays failed to recapitulate validated expression differences in the reporter gene system could either reflect that the true functional SNP was not included in our genotyped sites but is in strong LD with them or that the requirement of additional regulatory elements (i.e., proper sequence and chromatin context) or transcription factors were not present in the reporter gene system used. The latter possibility is highlighted by recent studies observing disconnection between in vitro transfection assay results and in vivo determined genetically controlled expression (Cirulli and Goldstein 2007; Wang et al. 2008).
In conclusion, we show that the full power of genetical genomics in understanding human disease can only be realized with cell models tailored for the trait in question as demonstrated here by enrichment of converging _cis_-eQTLs for BMD-associated SNPs in osteoblasts compared to LCLs. We identified a potential candidate gene, SRR, for a metabolic bone disorder comprising a strong _cis_-eQTL that was ranked too low in the original GWAS and thus did not meet the criteria for follow-up studies, indicating that in parallel with meta-analyses approaches for GWAS follow-up functional prioritization may increase the yield of true disease associations.
Methods
Cell culture
Human trabecular bone from the proximal femoral shaft was collected from 95 donors undergoing total hip replacement at the Uppsala University Hospital, Uppsala, Sweden. The bone samples from each donor were thoroughly minced and cultured in three biological replicates. The cells were grown in medium containing α-MEM (SigmaAldrich) supplemented with 2 mmol/l L-Glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin (National Veterinary Institute of Sweden), and 10% fetal bovine serum (SigmaAldrich) at 37°C with 5% CO2. At 70%–80% confluence, the cells were passaged and subcultured in six-well plates (100,000 cells/well) for 12 d after which the cells were harvested and stored at −70°C until RNA and DNA extraction. Oral informed consent was obtained for all subjects and the study was approved by the local ethics committee (Dnr Ups 03-561).
RNA extraction
RNA was extracted from cell lysates resuspended in 600 μL RLT lysis buffer (Qiagen) using the RNeasy Mini Kit (Qiagen). High RNA quality was confirmed for all samples using the Agilent 2100 BioAnalyzer (Agilent Technologies) and the concentrations were determined using Nanodrop ND-1000 (NanoDrop Technologies).
DNA extraction
DNA was extracted from cell lysates resuspended in 200 μL PBS using the GenElute DNA Miniprep Kit (SigmaAldrich). Concentrations were determined using the Quant-iT PicoGreen kit (Molecular Probes).
Expression profiling
Expression profiling was performed using the Illumina HumRef-8v2 BeadChips (Illumina Inc.) where 200 ng of total RNA was processed according to the protocol supplied by Illumina. The biological replicates were always separated and hybridized on different BeadChips in order to avoid that results are confounded by technical effects. Any sample or array that showed an abnormal distribution of signal intensities as visualized in box plots was removed or rehybridized. The raw data were imported to Bioconductor (Gentleman et al. 2004) using the R 2.5.0 package for variance-stabilizing transformation and robust spline normalization to obtain normalized mean expression values. The microarray data have been deposited in the Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo, accession no. GSE15678).
Whole-genome genotyping
Genotyping was performed using the Illumina HapMap 550k Duo chip (Illumina Inc.) according to the protocol provided by the manufacturer. The total number of SNPs included on the chip was 561,303. Individuals with low genotyping rate (<70%) and SNPs showing significant deviation from Hardy-Weinberg equilibrium (P < 0.05) were excluded. Similarly, low frequency SNPs (MAF < 0.10) and SNPs with high rates of missing data (<95%) were excluded.
Genotyping of additional HapMap SNPs
Selected HapMap SNPs in the SRR (Supplemental Table 6) gene were genotyped using the Sequenom MassARRAY iPLEX Gold technology (Sequenom Inc.). Briefly, ∼40 SNPs were multiplexed per individual reaction and a single base primer extension step was performed. The primer extension products were then analyzed using MALDI-TOF mass spectrometry.
Re-sequencing and genotyping of non-HapMap SNPs
To identify all genetic polymorphisms potentially responsible for the differences in gene expression, 6.2 kb of the SRR candidate region was re-sequenced using standard Sanger sequencing with ABI Big Dye chemistry and capillary electrophoresis on an ABI 3730 sequencer (Applied Biosystems). A total of eight individuals were selected based on the candidate SNPs (four samples from each homozygous genotype group of the top SNPs), and overlapped regions were amplified and sequenced using primers listed in Supplemental Table 7. Non-HapMap SNPs and/or novel SNPs that were homozygous in all eight samples were genotyped in the complete panel of 94 HObs using Sanger sequencing.
Gene expression validation by RT-PCR
The effect of SRR genetic variation on gene expression in HObs was verified by quantitative RT-PCR. The same sources of total RNA used in the microarray experiments were used for the data validation by RT-PCR. Aliquots of the different RNA from the biological replicates were each annealed to 250 ng of random hexamers (Invitrogen Corp.) at 70°C in 10 min. First-strand cDNA synthesis was performed using SuperScript II reverse transcriptase (Invitrogen Corp.) according to manufacturer's instructions.
For each biological replicate, the SRR gene as well as the GAPDH housekeeping gene were analyzed in duplicates as well as a calibration curve from a twofold dilution series of nonstimulated trabecular bone cells cDNA and nontemplate control (NTC) samples . The RT-PCR assays were performed on the Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Sciences) using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen Corp.) according to manufacturer's recommendation. The cycling conditions on the Rotor-Gene 6000 real-time rotary analyzer were: 4 min at 95°C, 40 cycles × (20 sec at 95°C, 30 sec at 55°C, and 30 sec at 72°C) followed by the dissociation protocol at 72°C.
Results of the experimental samples were analyzed using the comparative CT method. The CT mean and standard deviation value of each technical replicate sample were calculated and the mean CT value was then normalized to the GAPDH mean CT value. The delta CT was then averaged from the biological replicate samples.
Primer design
Primers were designed using the Primer3 v. 0.4.0 software (http://frodo.wi.mit.edu/) and all primer sequences can be found in Supplemental Table 8.
In vitro transient transfection assays
To test for allelic activity, haplotype-specific constructs for rs3744270 (chr17:2,153,816–2,154,275) were subcloned into a pGL3 vector containing a firefly luciferase reporter gene according to a previously published method (Belanger et al. 2005). All constructs were tested into five different human immortalized cell lines. The cervical cancer (HeLa), choriocarcinoma (Jeg3), hepatocellular liver carcinoma (HepG2), osteosarcoma (MG63), and T-cell lymphoblast-like (Jurkat) cell lines were transfected using lipofectamine 2000 (Invitrogen Corp.) according to the manufacturer's instructions. To control for transfection efficiency, the measurement of the firefly luciferase was normalized to the measurement of the Renilla luciferase. Experiments were performed in quadruplicate, the activities of the two luciferases were measured 24 h after transfection, and allelic haplotypes for each SNP were compared. Statistical significance (_P_-value) was determined using an unpaired Student's _t_-test.
Population-based cohorts and BMD measurements
The effect of the identified SNPs on clinical bone phenotypes was tested in two population-based studies involving Caucasian male subjects of European origin: the MrOS Cohort, Sweden (MrOS), and the Rotterdam Study, Netherlands (RS).
The MrOS study is a multicenter prospective fracture epidemiology investigation involving elderly men from different sites around the world including the US, Hong Kong, and Sweden. The Swedish part consists of 3014 men aged 69–81 yr that were randomly selected from the population registry (Mellstrom et al. 2006). BMD of the lumbar spine (L1–L4) and total hip was measured using dual energy X-ray absorptiometry (DXA) using either a Hologic QDR 4500/A-Delphi (QDR 4500 W, Hologic Inc.) or a Lunar Prodigy DXA (GE Lunar Corp.). To allow pooling of DXA measurements performed with different equipment, standardized BMD (sBMD) was calculated using previously reported algorithms (Genant et al. 1994; Hui et al. 1997). DNA samples from 2880 individuals were included in the study and all SNPs were genotyped using the Sequenom MassARRAY iPLEX Gold technology (Sequenom Inc.) including single-base primer extension and MALDI-TOF mass spectrometry.
The RS is a prospective population-based cohort study of chronic disabling conditions in Dutch elderly individuals aged 55 yr and older (Hofman et al. 1991, 2007). BMD measurements at the lumbar spine (L1–L4) and the femoral neck were performed by DXA following standard manufacturer protocols (GE-Lunar Corp. or Hologic Inc.). The rs1885987 and rs26784 SNP information (Little et al. 2009) was obtained from microarray genotyping performed in the complete RS using the Illumina Infinium II HumanHap550K Genotyping BeadChip version 3 following manufacturer's protocol (Richards et al. 2008).
Statistical analysis
Of the 22,184 Illumina probes (corresponding to 18,144 genes), those with SNPs (dbSNP build 126) within them were excluded. We tested for association of the expression levels to SNPs using a linear regression model implemented in the PLINK software package (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al. 2007) with year of birth and sex as covariates. _Cis_-regulatory effects were tested by including SNPs mapping ±250 kb flanking the gene, and _trans_-regulatory effects were defined as SNPs being located on a different chromosome than the gene whose expression level the SNP was associated with. We also performed surrogate variable analysis (SVA) in order to identify unmodeled sources of heterogeneity in our data as described previously (Leek and Storey 2007) using the R package (sva) v 2.6 (http://www.R-project.org) and with sex as our primary variable. We reanalyzed the linear regression models with sex, year of birth, and the surrogate variable identified in the SVA as covariates.
The _cis_-eQTL results are available through an interactive web-based browser (http://www.regulatorygenomics.org). Under the detailed view in the genome browser, tracks showing _cis_-eQTL data are selected as follows: GRID→Allelic expr.→Genotype-Expression assoc.
In order to study if there was an enrichment of functional SNPs identified in HOb or LCL eQTL studies (www.sanger.ac.uk/humgen/genevar/) (Stranger et al. 2007), among the GWAS hits, a frequency binning algorithm was used grouping SNPs into classes depending on MAF (15, 20, 25,…, 50%). Converging SNPs (i.e., significant in both studies' MAF groups) at various _P_-value thresholds were counted and compared with the same list that was permuted 10 times, which represented the MAF-adjusted expected overlaps by chance. Power calculations were performed using the R package (pwr.f2) v 2.6 (http://www.R-project.org), and combined _P_-values were obtained using the Fisher's combined probability test (−2Sum[ln(pi)]) taking into consideration effect direction.
All statistical analyses for the clinical associations in the MrOS Study were performed using SAS 9.1 software (SAS institute Inc.). A general linear model was used to study the effect of genotype on phenotypes (lumbar spine and hip BMD) adjusted for age, weight, height, and study center. The Rotterdam Study used age- and weight-adjusted standardized residuals of both lumbar spine and femoral neck BMD analyzed under an additive (per allele) genetic model using PLINK version 1.05 (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al. 2007).
Acknowledgments
We thank research nurses and assistants, especially Jessica Pettersson and Anna-Lena Johansson, at the Departments of Surgical and Medical Sciences, Uppsala University, Sweden for large-scale collection and culture of bone samples. The authors are grateful to the study participants, the staff from the MrOS Sweden Study and the Rotterdam Study and the participating general practitioners and pharmacists. We also thank Pascal Arp, Mila Jhamai, Dr. Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database for the Rotterdam Study. Dr. David B. Goldstein is thanked for providing access to full _P_-value distributions from the Heinzen et al. (2008) study. This work was supported by Genome Quebec, Genome Canada, and the Canadian Institutes of Health Research (CIHR). T.P. is a recipient of a Canada Research Chair. E.G. is supported by the Swedish Research Council. The generation and management of GWAS genotype data for the Rotterdam Study are supported by the Netherlands Organization of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2) and the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project no. 050-060-810. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, and the Ministry for Health, Welfare.
Footnotes
References
- Belanger H, Beaulieu P, Moreau C, Labuda D, Hudson TJ, Sinnett D. Functional promoter SNPs in cell cycle checkpoint genes. Hum Mol Genet. 2005;14:2641–2648. doi: 10.1093/hmg/ddi298. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat Soc. 1995;57:289–300. [Google Scholar]
- Chenu C, Serre CM, Raynal C, Burt-Pichat B, Delmas PD. Glutamate receptors are expressed by bone cells and are involved in bone resorption. Bone. 1998;22:295–299. doi: 10.1016/s8756-3282(97)00295-0. [DOI] [PubMed] [Google Scholar]
- Chio A, Schymick JC, Restagno G, Scholz SW, Lombardo F, Lai SL, Mora G, Fung HC, Britton A, Arepalli S, et al. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:1524–1532. doi: 10.1093/hmg/ddp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, et al. Genetic analysis of human traits in vitro: Drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 2007;16:1931–1939. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Ducy P, Schinke T, Karsenty G. The osteoblast: A sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Xie X. Common polymorphic transcript variation in human disease. Genome Res. 2009;19:567–575. doi: 10.1101/gr.083477.108. [DOI] [PubMed] [Google Scholar]
- Genant HK, Grampp S, Gluer CC, Faulkner KG, Jergas M, Engelke K, Hagiwara S, Van Kuijk C. Universal standardization for dual X-ray absorptiometry: Patient and phantom cross-calibration results. J Bone Miner Res. 1994;9:1503–1514. doi: 10.1002/jbmr.5650091002. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Brandstrom H, Lam KC, Gurd S, Ge B, Harmsen E, Kindmark A, Ljunggren O, Mallmin H, Nilsson O, et al. Systematic assessment of the human osteoblast transcriptome in resting and induced primary cells. Physiol Genomics. 2008;33:301–311. doi: 10.1152/physiolgenomics.00028.2008. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: Implications for the study of complex traits. PLoS Biol. 2008;6:e1000001. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E, Fujimori S, Yoneda Y. Modulation of cellular differentiation by N-methyl-d-aspartate receptors in osteoblasts. FASEB J. 2003;17:1532–1534. doi: 10.1096/fj.02-0820fje. [DOI] [PubMed] [Google Scholar]
- Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: The Rotterdam Elderly Study. Eur J Epidemiol. 1991;7:403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- Hofman A, Breteler MM, van Duijn CM, Krestin GP, Pols HA, Stricker BH, Tiemeier H, Uitterlinden AG, Vingerling JR, Witteman JC. The Rotterdam Study: Objectives and design update. Eur J Epidemiol. 2007;22:819–829. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui SL, Gao S, Zhou XH, Johnston CC, Jr, Lu Y, Gluer CC, Grampp S, Genant H. Universal standardization of bone density measurements: A method with optimal properties for calibration among several instruments. J Bone Miner Res. 1997;12:1463–1470. doi: 10.1359/jbmr.1997.12.9.1463. [DOI] [PubMed] [Google Scholar]
- Jonsson KB, Frost A, Nilsson O, Ljunghall S, Ljunggren O. Three isolation techniques for primary culture of human osteoblast-like cells: A comparison. Acta Orthop Scand. 1999;70:365–373. doi: 10.3109/17453679908997826. [DOI] [PubMed] [Google Scholar]
- Kwan T, Grundberg E, Koka V, Ge B, Lam KC, Dias C, Kindmark A, Mallmin H, Ljunggren O, Rivadeneira F, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genetics. 2009;8:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laketic-Ljubojevic I, Suva LJ, Maathuis FJ, Sanders D, Skerry TM. Functional characterization of N-methyl-d-aspartic acid-gated channels in bone cells. Bone. 1999;25:631–637. doi: 10.1016/s8756-3282(99)00224-0. [DOI] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE statement. Eur J Epidemiol. 2009;24:37–55. doi: 10.1007/s10654-008-9302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellstrom D, Johnell O, Ljunggren O, Eriksson AL, Lorentzon M, Mallmin H, Holmberg A, Redlund-Johnell I, Orwoll E, Ohlsson C. Free testosterone is an independent predictor of BMD and prevalent fractures in elderly men: MrOS Sweden. J Bone Miner Res. 2006;21:529–535. doi: 10.1359/jbmr.060110. [DOI] [PubMed] [Google Scholar]
- Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP. Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci. 2008;105:617–622. doi: 10.1073/pnas.0705554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Ujike H, Tanaka Y, Otani K, Kishimoto M, Morio A, Kotaka T, Okahisa Y, Matsushita M, Morikawa A, et al. A genetic variant of the serine racemase gene is associated with schizophrenia. Biol Psychiatry. 2007;61:1200–1203. doi: 10.1016/j.biopsych.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Pocock NA, Eisman JA, Hopper JL, Yeates MG, Sambrook PN, Eberl S. Genetic determinants of bone mass in adults. A twin study. J Clin Invest. 1987;80:706–710. doi: 10.1172/JCI113125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: A genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson G, Halldorsson BV, Styrkarsdottir U, Kristjansson K, Stefansson K. Impact of genetics on low bone mass in adults. J Bone Miner Res. 2008;23:1584–1590. doi: 10.1359/jbmr.080507. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Snorradottir S, Center JR, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- Takarada T, Hinoi E, Takahata Y, Yoneda Y. Serine racemase suppresses chondrogenic differentiation in cartilage in a Sox9-dependent manner. J Cell Physiol. 2008;215:320–328. doi: 10.1002/jcp.21310. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadee W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112:1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosker H, Sheth KN, Takahashi M, Mothet JP, Brady RO, Jr, Ferris CD, Snyder SH. Purification of serine racemase: Biosynthesis of the neuromodulator D-serine. Proc Natl Acad Sci. 1999;96:721–725. doi: 10.1073/pnas.96.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Ohnishi T, Hashimoto K, Ohba H, Iwayama-Shigeno Y, Toyoshima M, Okuno A, Takao H, Toyota T, Minabe Y, et al. Identification of multiple serine racemase (SRR) mRNA isoforms and genetic analyses of SRR and DAO in schizophrenia and D-serine levels. Biol Psychiatry. 2005;57:1493–1503. doi: 10.1016/j.biopsych.2005.03.018. [DOI] [PubMed] [Google Scholar]