Cohesinopathy mutations disrupt the subnuclear organization of chromatin (original) (raw)
Nuclear morphology, chromosomal condensation, and transcriptional-mediated localization of genes to the nuclear periphery are disturbed by mutations in cohesin pathway genes.
Abstract
In Saccharomyces cerevisiae, chromatin is spatially organized within the nucleus with centromeres clustering near the spindle pole body, telomeres clustering into foci at the nuclear periphery, ribosomal DNA repeats localizing within a single nucleolus, and transfer RNA (tRNA) genes present in an adjacent cluster. Furthermore, certain genes relocalize from the nuclear interior to the periphery upon transcriptional activation. The molecular mechanisms responsible for the organization of the genome are not well understood. We find that evolutionarily conserved proteins in the cohesin network play an important role in the subnuclear organization of chromatin. Mutations that cause human cohesinopathies had little effect on chromosome cohesion, centromere clustering, or viability when expressed in yeast. However, two mutations in particular lead to defects in (a) GAL2 transcription and recruitment to the nuclear periphery, (b) condensation of mitotic chromosomes, (c) nucleolar morphology, and (d) tRNA gene–mediated silencing and clustering of tRNA genes. We propose that the cohesin network affects gene regulation by facilitating the subnuclear organization of chromatin.
Introduction
The nucleus is a spatially organized organelle with distinct domains such as the nucleolus, nuclear envelope, nuclear interior, and nuclear pores. Each of these domains associates with distinct portions of the genome. For example, transcriptionally silenced telomeres and mating type loci in both budding and fission yeast associate with the nuclear periphery. Centromeres cluster adjacent to the spindle pole body. The ribosomal DNA (rDNA) repeats are compartmentalized into the nucleolus, and tRNA genes are located adjacent to the nucleolus. The spatial organization of chromatin is also dynamic; individual genes can relocalize within the nucleus in response to environmental or developmental cues. Higher order chromatin organization has been shown to affect gene regulation as well as DNA replication, repair, and recombination (Sexton et al., 2007; for review see Ahmed and Brickner, 2007).
The evolutionarily conserved cohesin complex is responsible for chromosome cohesion during mitosis (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998). Mutations in any of the four subunits in the complex (Smc1, Smc3, Scc3, and Mcd1/Scc1) result in the precocious dissociation of sister chromatids at metaphase and missegregation of chromosomes. This function is essential for cell viability. However, several observations have suggested that the cohesin complex plays additional roles in higher order chromosome organization and transcriptional regulation. A mutation in Smc1 results in the loss of a heterochromatin boundary element at the silent mating locus in budding yeast (Donze et al., 1999). Mutations in Scc2/Nipped-B, a subunit of the cohesin loading complex, result in defects in long-range promoter–enhancer interactions in Drosophila melanogaster (Rollins et al., 1999). A mutation in the Mcd1 cohesin subunit leads to defects in cohesion, chromosome condensation, and nucleolar morphology (Guacci et al., 1997). However, it is unclear whether these phenotypes reflect a direct role for cohesin in chromatin folding and organization or result from indirect effects of the cohesion defect.
Mutations in components of the cohesin pathway cause two human diseases called Cornelia de Lange syndrome (CdLS; caused by mutations in SMC1, SMC3, and SCC2) and Roberts syndrome (caused by mutations in ESCO2; for review see Liu and Krantz, 2008). In CdLS, there is one mutant and one wild-type (WT) copy of the gene, whereas Roberts syndrome is a rare autosomal recessive developmental disorder. Mutations in Scc2 cause the most severe CdLS phenotypes (for review see Liu and Krantz, 2008). Eco1, the yeast paralogue of ESCO2, is an acetyltransferase that targets proteins in the cohesin complex and promotes cohesion (Tóth et al., 1999; Ivanov et al., 2002). The developmental phenotypes associated with such cohesinopathies suggest that they are caused by altered gene expression during early embryogenesis. Consistent with the idea that the cohesin pathway plays a role in transcriptional regulation, cohesin expression is essential for cell viability and development in postmitotic neuronal cells in flies (Pauli et al., 2008; Schuldiner et al., 2008). These results suggest that cohesin may have roles in both S phase cohesion and in regulating genome function outside of S phase.
In this study, we constructed and characterized six cohesinopathy mutations in the orthologous genes from budding yeast with the goal of uncovering the underlying molecular defects caused by these mutations. Two mutations in particular, scc2-D730V and eco1-W216G, severely disrupted the subnuclear organization of chromatin. They caused chromosomal decondensation and aberrant nucleolar morphology. Colocalization of GAL2 and tDNAs with the nucleolus was disrupted, and relocalization of GAL2 to the nuclear periphery upon activation was blocked in both of these mutants. Importantly, the cohesinopathy mutations did not significantly affect chromosome cohesion or the pattern of cohesin binding. Our results indicate that the cohesin regulators Scc2 and Eco1 significantly contribute to chromosome morphology.
Results and discussion
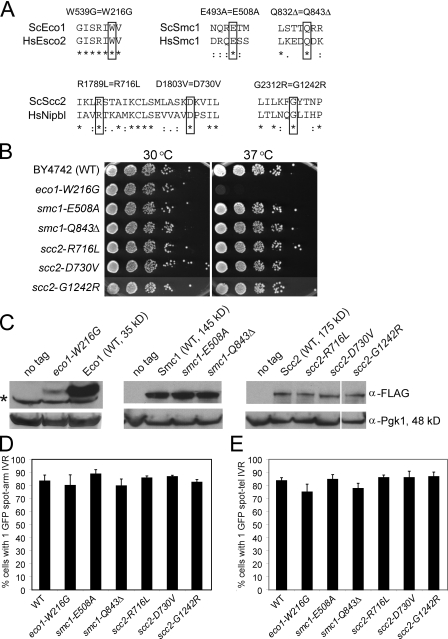
We identified amino acids in the yeast orthologues of Smc1, Scc2, and Eco1 that correspond to those mutated in the cohesinopathies (Fig. 1 A) and constructed diploid yeast strains containing the analogous mutation expressed from the native promoters at their endogenous loci. These strains were sporulated, and haploid spore clones containing the mutation were obtained and analyzed. None of the strains had detectable growth defects at 23 or 30°C, but the eco1-W216G mutant strain failed to grow at 37°C (Fig. 1 B). We made epitope-tagged WT and mutant proteins and used immunoblot analysis to confirm their expression (Fig. 1 C). The Scc2 and Smc1 mutants were expressed at levels similar to the WT protein. In contrast, the Eco1-W216G protein was expressed at a level below WT. This suggests that the levels of the mutant Eco1-W216G protein are sufficient to provide its function in cohesion. In humans, the analogous ESCO2-W539G mutant gene produces full-length protein that lacks autoacetyltransferase activity (Gordillo et al., 2008).
Figure 1.
Cohesinopathy mutations do not significantly affect chromosome cohesion. (A) The high degree of conservation between human and yeast cohesin alleles enabled the identification of several conserved residues that have previously been identified as mutated in patients afflicted with a cohesinopathy. The critical residue is indicated by a black box for each of the mutations in ECO1, SCC2, or SMC1. In the alignments, asterisks indicate identity, and colons and dots indicate similarity between amino acids. (B) Yeast strains were constructed by replacing the genomic copy of the indicated allele with a cohesinopathy allele. The indicated alleles and a WT control were serially diluted and plated onto YPD and grown at the indicated temperature. Only the Eco1 allele exhibits lethality at 37°C. (C) Strains were created in which each protein was tagged with 3× Flag at the C terminus. Equal amounts of total protein from each strain were used for Western blotting with anti-Flag antibody. The blots were reprobed with anti-Pgk1 as a loading control. The asterisk next to the Eco1 panel indicates a nonspecific background band. The Eco1-W216G-Flag protein is expressed at an ∼10-fold lower level than the WT Eco1-Flag protein. White lines indicate that intervening lanes have been spliced out. (D) Effects of cohesinopathy alleles on chromosome arm cohesion. Yeast strains were constructed containing the indicated mutation and LacO repeats on the arm of chromosome IV, and a LacI-GFP fusion was inducibly expressed in nocodazole-arrested cells at 30°C. The number of cells displaying one or two spots was counted to determine PCSC. At least three biological replicates were conducted for each strain, and at least 300 total cells were counted. None of the cohesinopathy strains demonstrate a significant defect in chromosome arm cohesion. (E) Yeast strains were constructed, grown, and analyzed similarly as in D, with the LacO repeats integrated into telomere IV-R. Error bars indicate SD.
To test whether these mutations caused defects in chromosome cohesion, an array of Lac repressor–binding sites was integrated into chromosome IV-R arm or telomere IV-R and visualized using LacI-GFP (Straight et al., 1996). Strains were arrested in metaphase with nocodazole, and the fraction of cells in the population with one or two GFP spots was counted. Two distinct spots indicate precocious sister chromatid separation (PSCS). In contrast to PSCS observed in cohesin mutants, which is typically ∼80% (Guacci et al., 1997; Michaelis et al., 1997), none of the mutant strains had significantly elevated levels of PSCS at the arm locus (Fig. 1 D), and only the eco1-W216G and smc1-Q843Δ mutants showed a slight increase (9% and 6%, respectively) at telomere IV-R at 30°C (Fig. 1 E). More significant cohesion defects were observed in some strains at 37°C (Fig. S1). We also monitored chromosome loss in several of the mutants at 30°C (Hieter et al., 1985) and did not observe significantly elevated rates (unpublished data). Because these mutants did not have strong defects in cohesion or chromosome segregation at 30°C, we hypothesized that these alleles may separate the essential function of the pathway from other cellular roles at this temperature. The remainder of experiments was performed at 30°C to minimize confounding effects caused by PSCS.
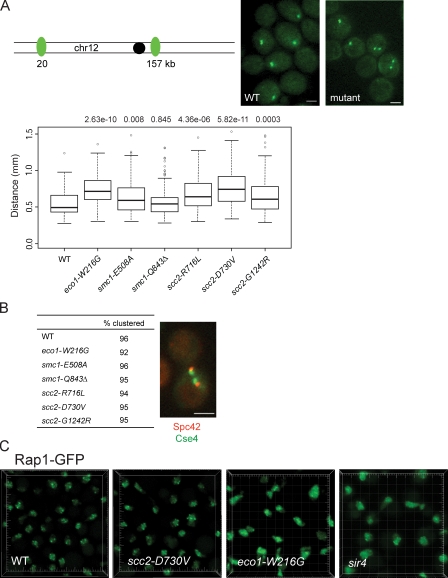
We examined the effect of the cohesinopathy mutations on chromosome condensation because a mutation in MCD1 was shown to affect condensation (Guacci et al., 1997). To determine compaction, we measured the distance between two Lac repressor arrays on chromosome XII separated by 137 kb in nocodazole-arrested cells. In WT cells, we found a mean distance of 0.55 µm. Significant defects in condensation occurred in the scc2-D730V and eco1-W216G mutants (Fig. 2 A). Distances measured were similar to published measurements for WT and condensin mutants for this interval (D’Ambrosio et al., 2008).
Figure 2.
Chromosome condensation and telomere arrangement, but not centromere clustering, is affected by the cohesinopathy mutations. (A) Strains containing 137 kb on chromosome XII (chr12) flanked by LacO sequences and the mutations indicated were imaged, and the distance between GFP spots was measured in ≥120 cells per strain. Representative images are shown. The data are shown as a box plot. Both analysis of variance and Wilcoxon tests performed to determine statistical significance had similar outcomes, and the analysis of variance results are shown. Bars (A and B), 2 µm. (B) Strains expressing Spc42-mCherry (a spindle pole body protein) and Cse4-GFP (centromeres) were imaged in live cells. A minimum of 100 cells were scored. A representative image of centromeres clustered next to the spindle pole body is shown. (C) Rap1-GFP was integrated at the TRP1 locus in a WT, eco1-W216G, scc2-D730V, and sir4Δ background. Avalanche photodiode imaging was used to visualize the telomere foci. Each large tick mark represents 2 µm.
The defects in chromosome condensation prompted us to examine other aspects of nuclear organization. Using Cse4-GFP and Spc42-mCherry to label centromeres and spindle pole bodies, respectively, we saw no difference in centromere clustering or positioning in any of the cohesinopathy mutants (Fig. 2 B). This is consistent with growth and chromosome segregation being intact (Fig. 1). In contrast, when we examined telomere clustering at the nuclear periphery using Rap1-GFP in the scc2-D730V and eco1-W216G mutants, the two mutants with the biggest condensation defects, we found that the Rap1-GFP signal was more diffuse in the eco1-W216G mutant background (Fig. 2 C). A similar diffuse signal was observed in a sir4Δ strain, which is known to have a disrupted telomere arrangement (Palladino et al., 1993). The exact nature of the telomere arrangement defect will require further characterization. Telomeres clustered into three to four foci in the scc2-D730V mutant as observed in WT (Gotta et al., 1996).
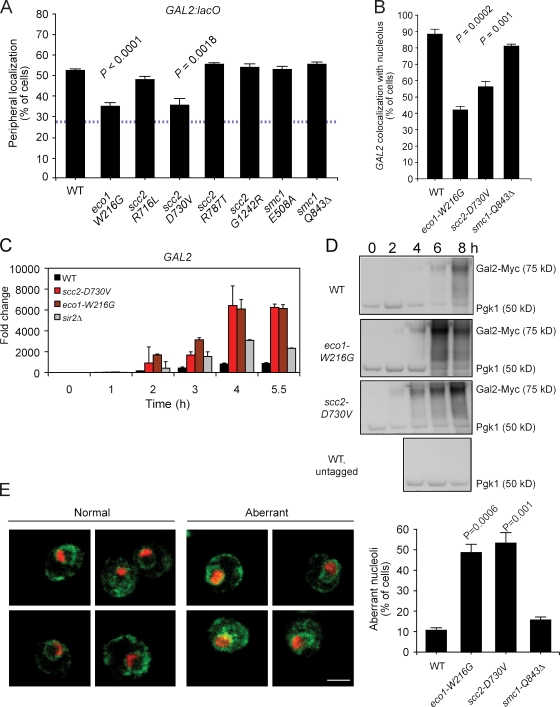
Certain highly transcribed genes in budding yeast are recruited to the nuclear periphery upon activation (for review see Ahmed and Brickner, 2007). We tested whether the subnuclear targeting of INO1, GAL1, and GAL2 was affected in our cohesinopathy mutants using LacI-GFP/Lac repressor–binding sites. The targeting of INO1 and GAL1 to the nuclear periphery upon activation was normal in all of the cohesinopathy mutants (unpublished data). However, in both the eco1-W216G and scc2-D730V mutants, the targeting of GAL2 to the nuclear periphery was impaired (Fig. 3 A). Because these particular mutants also had the strongest effect on chromosome condensation, we focused on these two strong mutants to better understand their effect on chromosome organization. For comparison, we included the smc1-Q843Δ mutant because it had no effect on condensation or GAL2 targeting to the nuclear periphery.
Figure 3.
GAL2 localization and induction and nucleolar morphology is affected by the strong cohesinopathy mutations. (A) Strains containing the indicated mutations and LacO arrays integrated near the GAL2 locus were grown overnight under inducing conditions and fixed for indirect immunofluorescence for GFP (GAL2) and the nuclear periphery (Nsp1; Brickner and Walter, 2004). 30–50 cells were scored per biological replicate; three biological replicates were performed. (B) The same strains used in A were processed for indirect immunofluoresence for GFP (GAL2) and the nucleolar marker Nop5/6. Data collection and analysis were performed as in A. (C) Cells were collected at the indicated times after transfer from YPD to YPGal. Total RNA was isolated, reverse transcribed, and the resulting cDNA was used as a template for qPCR. The value for each time point was normalized to the value for ACT1 and PGK1, genes whose transcription does not change in galactose. This time course was repeated twice with similar results. (D) Cells were collected at the indicated times after transfer from YPD to YPGal, and whole cell extract was used on a Western blot to examine levels of Gal2-13Myc. (E) Nucleolar morphology is abnormal in the strong mutants. Strains were processed for indirect immunofluoresence for the nucleolar marker Nop5/6 (red) and the nuclear periphery marker Scs2 (green). Nucleoli were scored as being either WT or aberrant. Error bars indicate SEM. The p-values are derived from an unpaired t test. Bar, 2 µm.
GAL2 is on the right arm of chromosome XII, ∼170 kb from the rDNA repeats, which constitute the bulk of the nucleolus. Recently, GAL2 was shown to localize adjacent to the nucleolus. The peripheral shift of this locus upon activation by galactose was suggested to result in part from the reduction of the nucleolar size in galactose (Berger et al., 2008). To test whether GAL2 localization to the nucleolus was affected by the cohesin pathway, we localized GAL2 with respect to the nucleolus in galactose medium. In the WT and smc1-Q843Δ mutant strains, GAL2 colocalized with the nucleolus in ∼90% of cells. However, in the eco1-W216G and scc2-D730V strains, GAL2 colocalized with the nucleolus in 40% and 55% of the cells, respectively (Fig. 3 B). These results indicate that Eco1 and Scc2 contribute to nuclear organization.
Despite the coupling of transcription to peripheral localization, loss of localization does not dramatically affect the transcriptional activity of GAL1 or GAL10 (Cabal et al., 2006; Dieppois et al., 2006; Luthra et al., 2007). In contrast, we hypothesized that the proximity of GAL2 to the nucleolus may inhibit GAL2 transcription, perhaps via Sir2, which is responsible for silencing a subset of the rDNA repeats (Bryk et al., 1997; Fritze et al., 1997; Smith and Boeke, 1997). To test this idea, we examined the transcriptional induction of GAL2 mRNA and protein by quantitative PCR (qPCR) and immunoblotting. We found that in both of the strong mutants, GAL2 mRNA and protein were induced faster and to higher levels than in a WT strain (Fig. 3, C and D). GAL2 mRNA was also more strongly induced in a sir2Δ strain, which is consistent with the idea that proximity to nucleolar Sir2 may normally curtail the activation of GAL2.
Cohesin binds to the GAL2 ORF and promoter when the gene is repressed (Glynn et al., 2004). However, upon activation, cohesin dissociates from the ORF, and a new peak of binding is detected downstream of the gene (Glynn et al., 2004; Bausch et al., 2007). We performed chromatin immunoprecipitation (ChIP) using Mcd1-13Myc and analyzed the immunoprecipitated DNA by both qPCR and whole genome microarrays to determine whether the strong mutations altered binding of cohesin at GAL2 or elsewhere in the genome. Over an entire chromosome, the binding pattern of cohesin in the strong mutants was very similar to that observed in a WT strain (Fig. S2 A, chromosome VI). However, at GAL2, we observed a reproducible decrease in binding in the eco1-W216G and scc2-D730V mutant strains by qPCR (Fig. S2 B) and microarrays (not depicted). This was especially apparent for the galactose-dependent binding downstream of GAL2, which was reduced about twofold in the mutants. Therefore, although the global pattern of cohesin binding is largely unaffected in the mutants, binding at certain loci is modestly decreased, which could account for changes in localization or expression.
Because cohesin normally binds to the nontranscribed spacer NTS2 sequence (Laloraya et al., 2000) located within the ∼150–200 copies of a 9.1-kb rDNA sequence located on chromosome XII-R, it is possible that our mutants affect nucleolar morphology. Analysis of cohesin binding to the rDNA using ChIP/qPCR showed no change in the scc2-D730V mutant but reduced levels in the eco1-W216G mutant (Fig. S2 C). Using indirect immunofluoresence microscopy, we examined the morphology of the nucleolus. Nucleoli were scored as being either WT (e.g., small sphere or cup shaped with a diameter less than one third of the nuclear volume) or aberrant (e.g., enlarged cup diameter expanded to whole nuclear volume or aberrant shape based on nuclear volume; Stone et al., 2000). Both eco1-W216G and scc2-D730V mutant strains showed a fivefold increase in abnormal nucleolar shape (Fig. 3 E). These data argue that Scc2 and Eco1 have roles in the maintenance of higher order chromatin structures, such as the nucleolus, that are separable from their essential roles in cohesion for the purposes of chromosome segregation.
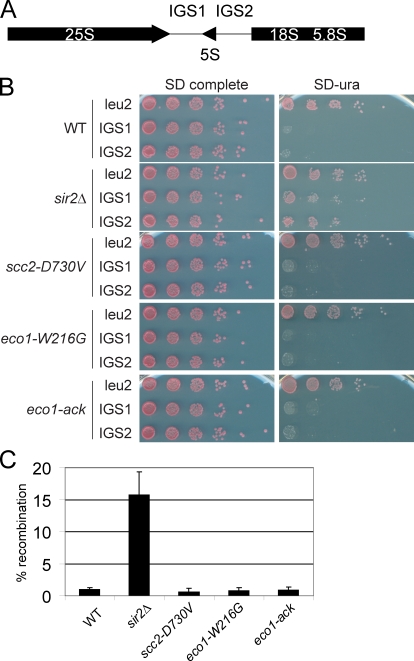
Given the strong disruption of nucleolar morphology, we checked silencing and stability of the rDNA repeats in the strong mutants. Silencing was monitored using strains in which URA3 has been inserted into IGS1, IGS2, or the non-rDNA locus LEU2 (Fig. 4 A). Growth on synthetic defined ura medium indicates that the URA3 gene is expressed. As previously shown, deletion of SIR2 relieves silencing of URA3 within IGS1 or IGS2, allowing growth (Gottlieb and Esposito, 1989; Mekhail et al., 2008). Surprisingly, the scc2-D730V and eco1-W216G mutations did not affect silencing of IGS1 or IGS2 (Fig. 4 B) or transcription of the 25S and 18S rRNA genes (not depicted).
Figure 4.
Transcription and recombination of RDN1 is unaffected in the strong mutants. (A) Schematic of the RDN1 locus. (B) Silencing of the URA3 gene inserted at IGS1, IGS2, or LEU2. (C) Recombination was monitored by loss of the ADE2 gene inserted in the RDN1 locus. Error bars indicate SD.
We also measured recombination using strains in which the ADE2 gene has been inserted into the rDNA. We found that the scc2-D730V and eco1-W216G mutations did not increase instability of the rDNA, whereas deletion of SIR2 significantly elevated instability (Fig. 4 C; Gottlieb and Esposito, 1989; Mekhail et al., 2008). The eco1-ack allele, another mutation that disrupts acetyltransferase activity (Brands and Skibbens, 2005), also had no effect on silencing or recombination at the rDNA. Thus, although the two strong mutations disrupt nucleolar morphology, they did not affect transcription or stability of the rDNA.
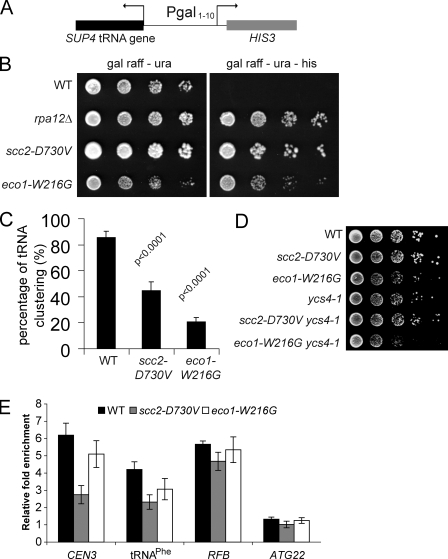
Although transcription of RDN1 by RNA polymerase I was unaffected in the strong mutants (Fig. 4), GAL2, an RNA polymerase II–transcribed gene that is nucleolar associated, shows a large change in its induction profile (Fig. 3). tRNA genes, which are transcribed by RNA polymerase III, also cluster near the nucleolus (Thompson et al., 2003) and can silence a neighboring RNA polymerase II–transcribed gene, which is a phenomenon known as tRNA gene–mediated (tgm) silencing (Hull et al., 1994). Mutations in condensin, which may be loaded by Scc2 (D’Ambrosio et al., 2008), disrupt tRNA gene clustering and silencing (Haeusler et al., 2008). We investigated whether tgm silencing of HIS3 was disrupted by the strong mutations (Fig. 5 A). Deletion of RPA12 served as a control (Wang et al., 2005). Both strong mutations relieve tgm silencing (Fig. 5 B). We further examined tDNA clustering by FISH using a tRNALeu(CCA) probe, which should detect seven tRNA genes. In the WT strain, these tDNAs clustered near the nucleolus in 86% of the cells. In contrast, tDNA clustering is reduced to 45% in the scc2-D730V cells and 21% in the eco1-W216G cells (Fig. 5 C). Thus, the repression of HIS3 and GAL2 appears to correlate with proximity to the nucleolus. The colocalization of tDNAs and GAL2 with the nucleolus is disrupted by mutations in Scc2 and Eco1.
Figure 5.
Strong cohesinopathy mutants relieve tgm silencing. (A) A schematic representation showing a tgm-silencing test construct in which the SUP4 tRNA gene silences the transcription of HIS3. (B) Strong cohesinopathy mutants grow on medium lacking histidine (Gal raffinose-ura-His). (C) tRNA gene clustering was detected by FISH. In most WT cells, the tRNALeu(CCA) genes cluster near or within the nucleolus. However, in the strong cohesinopathy mutants, the signal of tRNALeu(CCA) genes was dispersed and could not be detected above the background. Three independent experiments were performed for each strain, and at least 150 total cells were scored. The p-values from Fisher’s exact test are indicated. (D) The eco1-W216G mutation in combination with the ycs4-1 mutation causes synthetic sickness. (E) ChIP/qPCR for Smc4-Pk9 at three condensin-binding sites, CEN3, replication fork blocking (RFB), and tRNAPhe, and at a site where condensin does not bind, ATG22. Error bars indicate SD.
Because the clustering of tRNA genes is dependent on condensin and Scc2 promotes the loading of both cohesin and condensin, we asked whether the strong mutants genetically interacted with the condensin mutant ycs4-1 (Bhalla et al., 2002). Although eco1-W216G had a growth defect in combination with ycs4-1, scc2-D730V did not (Fig. 5 D). We further examined whether condensin binding was affected at several different loci in the strong mutants using ChIP/qPCR. At the replication fork–blocking site in rDNA (Johzuka and Horiuchi, 2009), we found no difference in binding. However, at CEN3 and tRNAPhe, we found that condensin binding was decreased in the scc2-D730V mutant and, to a lesser extent, in the eco1-W216G mutant (Fig. 5 E). Therefore, loading of condensin onto some but not all sites is affected by these two mutations.
In summary, we find that two mutations associated with human cohesinopathies, scc2-D730V and eco1-W216G, exhibit several phenotypes in haploid yeast that suggest roles in chromosome organization. Both mutants displayed enhanced GAL2 induction, disrupted tDNA clustering, and aberrant nucleolar morphology, which may all derive from the defects in chromatin condensation. However, these mutants also displayed distinctive phenotypes. The eco1-W216G mutation disrupted telomere organization, whereas the scc2-D730V mutant was more deficient in condensin binding. We propose that Eco1 and Scc2 have related but distinct molecular functions in chromosome organization that are separable from their essential role in chromosome cohesion/segregation. In the case of Scc2, mutations may affect the loading or dynamics of cohesin, condensin, or both. Eco1 regulates proteins in the cohesin network through acetylation, including Smc3 and Mcd1, but it is likely that additional targets have yet to be discovered. Future studies directed at how the cohesin network contributes to the organization of chromatin will add to our understanding of higher order chromosome structure and its effect on gene regulation and human disease.
Materials and methods
Yeast strains
To construct the cohesinopathy allele strains, the SMC1, SCC2, and ECO1 genes, including promoter regions, were amplified from yeast genomic DNA and cloned into pRS316. The genes were sequence verified and subjected to site-directed mutagenesis. All point mutations were verified by sequence analysis. To introduce the cohesinopathy alleles, fusion PCR was performed to link a hygromycin B resistance cassette 100-bp 3′ to the gene and the Phusion polymerase. Strains BY4743 and W303a were transformed with the resulting PCR product using standard transformation protocol. Positive transformants were identified by PCR, and individual mutations were verified by sequencing. Table S1 lists all strains used in this study.
Cohesion assays
Strains for the one-spot/two-spot assay were constructed by mating strains containing cohesinopathy alleles to SLJ1988 and SLJ1989 (provided by S. Jaspersen, Stowers Institute for Medical Research, Kansas City, MO) and dissecting haploids. Logarithmically growing cells were arrested with 13.2 µg/ml nocodazole, fixed with ethanol, and stained with DAPI before cell counting. Aliquots of cells were removed for cytometric analysis to verify mitotic arrest.
Microscopy
Methods for indirect immunofluorescence have been previously described (Brickner and Walter, 2004). Live confocal microscopy for Rap1-GFP was performed on an inverted microscope (LSM 510 Axiovert; Carl Zeiss, Inc.) with a 100× Plan Apochromat 1.46 NA oil objective with avalanche photodiodes using AIM software (Carl Zeiss, Inc.). To detect GFP, cells were suspended in PBS at room temperature, and a power-attenuated 30-mW argon gas 488-nm laser was used. Standard conditions for pinhole size, brightness, and contrast were used for capturing the images. The background value of the signal level outside the cell (∼15% maximum) was subtracted using Imaris. Image capture and background subtraction have been uniformly performed on all images for comparison purposes.
Methods for FISH have been previously described (Haeusler et al., 2008). FISH probes are as follows: the sequence of the U14 snoRNA probe was 5′-T*ATCCAAGGAAGGT*AGTTGCCAACAT*AAGACTTTCTGGT*GGAAACTACGAAT*T-3′, with Oregon green 488 conjugated at the T sites followed by asterisks. The sequence of the tRNALeu(CCA) gene probe was 5′-GGT*TGTTTGGCCGAGCGGT*CTAAGGCGCCT*GATTCAAGAAATAT*C-3′, with CY3 conjugated at the T sites followed by asterisks.
Online supplemental material
Fig. S1 shows cohesion at 37°C at arm and telomere locations for all six cohesinopathy mutants, and Fig. S2 shows ChIP data for the strong mutants. Table S1 lists all of the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906075/DC1.
Acknowledgments
We thank the Stowers Molecular Biology and Microscopy groups for excellent technical assistance and the Biological Imaging Facility at Northwestern University for access to confocal microscopes. We also thank F. Uhlmann, D. Engelke, F. Stutz, and R. Li for strains and reagents.
We thank the Hudson Foundation and the Stowers Institute for Medical Research for financial support of this project. J.H. Brickner is supported by the National Institutes of Health (grant GM080484) and the Baldwin Fund for Biomedical Research.
Footnotes
Abbreviations used in this paper:
CdLS
Cornelia de Lange syndrome
ChIP
chromatin immunoprecipitation
PSCS
precocious sister chromatid separation
qPCR
quantitative PCR
rDNA
ribosomal DNA
tgm
tRNA gene mediated
WT
wild type
References
- Ahmed S., Brickner J.H. 2007. Regulation and epigenetic control of transcription at the nuclear periphery. Trends Genet. 23:396–402 10.1016/j.tig.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Bausch C., Noone S., Henry J.M., Gaudenz K., Sanderson B., Seidel C., Gerton J.L. 2007. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:8522–8532 10.1128/MCB.01007-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A.B., Cabal G.G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J.C., Gadal O., Zimmer C. 2008. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 5:1031–1037 10.1038/nmeth.1266 [DOI] [PubMed] [Google Scholar]
- Bhalla N., Biggins S., Murray A.W. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 13:632–645 10.1091/mbc.01-05-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands A., Skibbens R.V. 2005. Ctf7p/Eco1p exhibits acetyltransferase activity—but does it matter? Curr. Biol. 15:R50–R51 10.1016/j.cub.2004.12.052 [DOI] [PubMed] [Google Scholar]
- Brickner J.H., Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255–269 10.1101/gad.11.2.255 [DOI] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., Nehrbass U. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C., Schmidt C.K., Katou Y., Kelly G., Itoh T., Shirahige K., Uhlmann F. 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22:2215–2227 10.1101/gad.1675708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26:7858–7870 10.1128/MCB.00870-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams C.R., Rine J., Kamakaka R.T. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698–708 10.1101/gad.13.6.698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze C.E., Verschueren K., Strich R., Easton Esposito R. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495–6509 10.1093/emboj/16.21.6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:E259 10.1371/journal.pbio.0020259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M., Vega H., Trainer A.H., Hou F., Sakai N., Luque R., Kayserili H., Basaran S., Skovby F., Hennekam R.C., et al. 2008. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum. Mol. Genet. 17:2172–2180 10.1093/hmg/ddn116 [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363 10.1083/jcb.134.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R.E. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 56:771–776 10.1016/0092-8674(89)90681-8 [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 91:47–57 10.1016/S0092-8674(01)80008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler R.A., Pratt-Hyatt M., Good P.D., Gipson T.A., Engelke D.R. 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22:2204–2214 10.1101/gad.1675908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R.W. 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 40:381–392 10.1016/0092-8674(85)90152-7 [DOI] [PubMed] [Google Scholar]
- Hull M.W., Erickson J., Johnston M., Engelke D.R. 1994. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 14:1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Schleiffer A., Eisenhaber F., Mechtler K., Haering C.H., Nasmyth K. 2002. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12:323–328 10.1016/S0960-9822(02)00681-4 [DOI] [PubMed] [Google Scholar]
- Johzuka K., Horiuchi T. 2009. The cis element and factors required for condensin recruitment to chromosomes. Mol. Cell. 34:26–35 10.1016/j.molcel.2009.02.021 [DOI] [PubMed] [Google Scholar]
- Laloraya S., Guacci V., Koshland D. 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151:1047–1056 10.1083/jcb.151.5.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Krantz I.D. 2008. Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 9:303–320 10.1146/annurev.genom.9.081307.164211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986–1997 10.1101/gad.12.13.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., Corbett A.H. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 282:3042–3049 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- Mekhail K., Seebacher J., Gygi S.P., Moazed D. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 456:667–670 10.1038/nature07460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45 10.1016/S0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 75:543–555 10.1016/0092-8674(93)90388-7 [DOI] [PubMed] [Google Scholar]
- Pauli A., Althoff F., Oliveira R.A., Heidmann S., Schuldiner O., Lehner C.F., Dickson B.J., Nasmyth K. 2008. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell. 14:239–251 10.1016/j.devcel.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins R.A., Morcillo P., Dorsett D. 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 152:577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O., Berdnik D., Levy J.M., Wu J.S., Luginbuhl D., Gontang A.C., Luo L. 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell. 14:227–238 10.1016/j.devcel.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Schober H., Fraser P., Gasser S.M. 2007. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 14:1049–1055 10.1038/nsmb1324 [DOI] [PubMed] [Google Scholar]
- Smith J.S., Boeke J.D. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241–254 10.1101/gad.11.2.241 [DOI] [PubMed] [Google Scholar]
- Stone E.M., Heun P., Laroche T., Pillus L., Gasser S.M. 2000. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr. Biol. 10:373–382 10.1016/S0960-9822(00)00413-9 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608 10.1016/S0960-9822(02)70783-5 [DOI] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R. 2003. Nucleolar clustering of dispersed tRNA genes. Science. 302:1399–1401 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320–333 10.1101/gad.13.3.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Haeusler R.A., Good P.D., Thompson M., Nagar S., Engelke D.R. 2005. Silencing near tRNA genes requires nucleolar localization. J. Biol. Chem. 280:8637–8639 10.1074/jbc.C500017200 [DOI] [PMC free article] [PubMed] [Google Scholar]