Interferons Direct an Effective Innate Response to Legionella pneumophila Infection (original) (raw)
Abstract
Legionella pneumophila remains an important opportunistic pathogen of human macrophages. Its more limited ability to replicate in murine macrophages has been attributed to redundant innate sensor systems that detect and effectively respond to this infection. The current studies evaluate the role of one of these innate response systems, the type I interferon (IFN-I) autocrine loop. The ability of L. pneumophila to induce IFN-I expression was found to be dependent on IRF-3, but not NF-κB. Secreted IFN-Is then in turn suppress the intracellular replication of L. pneumophila. Surprisingly, this suppression is mediated by a pathway that is independent of Stat1, Stat2, Stat3, but correlates with the polarization of macrophages toward the M1 or classically activated phenotype.
Legionella pneumophila, the causative agent of Legionnaires' Disease, remains an important cause of human morbidity (1). Like other pulmonary pathogens, L. pneumophila subverts host immunity through its capacity to replicate within macrophages. Genetic studies have mapped L. pneumophila virulence and immune evasion to the Dot-Icm type IV secretion system (reviewed in Refs. 2–4). Subsequent studies identified “effector” proteins, which the Dot-Icm system injects into host cells, enabling the bacteria to avoid lysosomal targeting and establish a unique replicative compartment (5).
The ability of murine macrophages to resist L. pneumophila infection has been attributed to several redundant innate sensor and response systems. For example, the unique susceptibility of macrophages from A/J mice to L. pneumophila infection has been mapped to Naip5/Birc1e (2, 6). Subsequent studies determined that the Naip5 sensor functioned upstream of Ipaf in its capacity to recognize L. pneumophila flagellin, culminating in inflammasome activation (7–10). The inflammasome directs the caspase 1-dependent secretion of interleukin (IL)2-1β and IL-18, as well as promoting pyroptosis (7, 11, 12). Additional studies have implicated TLR2 and MyD88, as well as a subsequent secretion of TNF, in the innate response to L. pneumophila (9, 13–17).
Both type I (e.g. IFN-α/β) and type II (IFN-γ) IFNs play an important role in the innate response to intracellular microbes (18). Although IFN-γ's ability to classically activate macrophages toward an M1 phenotype has been intimately associated with antibacterial activity, more recent studies have also ascribed antibacterial activity to IFN-I (19–22).
Characterization of IFN's antiviral activity led to the identification of the IFN-α receptor (IFNAR), the IFN-γ receptor (IFNGR), and the JAK-STAT signaling paradigm, where STATs (Signal Transducers and Activators of Transcription) are transcription factors and JAKs (Janus Kinases) the receptor-associated kinases that activate them (23). Specifically, IFN-γ directs the formation of a single transcription factor, the Stat1 homodimer, whereas IFN-I directs the activation of both Stat1 homodimers and ISGF-3 (IFN-stimulated gene factor 3; Stat1 + Stat2 + IRF9). Recent studies have underscored an important role for IRF-3 and IRF-7 in promoting activation of an IFN-I autocrine loop. This entails a sequential expression of IFN-β and IFN-α, and is dependent on IFNAR. This autocrine loop plays an important role in innate immunity (23–25).
Both IFN-γ and IFN-Is suppress L. pneumophila growth in murine macrophages (13, 26, 27). Our studies highlight a role for the IFN-I autocrine loop in the innate response to this bacterium. The ability of L. pneumophila to induce IFN-I expression was found to depend on IRF-3, yet be independent of the flagellin-Naip5 axis, as well as p50/cRel. Additionally, in contrast to the critical role Stat1 plays in the antibacterial response of IFN-γ, the ability of IFN-α to suppress L. pneumophila growth was found to be independent of both Stat1 and Stat2. Finally, the Stat1-independent protection afforded by IFN-α correlated with the induction of classically activated M1 macrophage markers.
EXPERIMENTAL PROCEDURES
Mice
129, C57Bl/6J and A/J strains of mice were purchased from Jackson Laboratories and bred in a specific pathogen free facility. Homozygous Stat1(−/−), Stat2(−/−), IFNAR1(−/−), IFNGR1(−/−), and Stat3-fl/fl × Rosa26-ER(T2)-Cre (Stat3-cko) mice were either in a 129 or C57Bl/6J background, as noted (19, 28–32). p50/cRel double knock-out (dko), IRF-3(−/−), and TBK-1/TNFR1 dko mice have been previously described (33–36). Columbia University review committees approved all relevant studies.
Cell Culture
Primary murine macrophages were prepared by culturing bone marrow (BM) cells in RPMI 1640 (Invitrogen), 10% fetal calf serum (Hyclone, Logan, UT), P/S (GIBCO), and 20% L929-conditioned medium for 5–7 days, as previously reported (32, 37). L-cell-conditioned medium was collected from L929 cells (ATCC, Manassas, VA) cultured in Dulbecco's modified Eagle's medium (GIBCO) plus 10% fetal calf serum for 7 days. Cells were stimulated with IFN-αA/D (1000 units/ml; PBL, Piscataway, NJ), active on both human and murine cells, or murine IFN-γ (50 units/ml; PBL). Some cells were pretreated with LY294002 (50 μm; 30 min; Calbiochem, La Jolla, CA) or SB203580 (50 μm; 30 min; Calbiochem). Stat3 conditional knock-out (cko) BM cells were treated with 250 nm 4-hydroxytamoxifen (Sigma-Aldrich) on culture days 1 and 5 to delete Stat3.
L. pneumophila
The L. pneumophila strain JR32 (restriction-defective Philadelphia-1, streptomycin-resistant) and dotA strains in the JR32 background were grown in AYE broth or on CYE plates, previously described (38, 39). GFP-L. pneumophila was created through the introduction of pXDC-31, and Fla− (Flagellin-deficient) strain through standard techniques (38–40).
Growth Curves
Ex vivo bacterial growth was evaluated by infecting (MOI = 0.25) day 6 bone marrow-derived murine macrophages (BMMs) with post-exponential phase L. pneumophila in 24-well plates (2.5 × 105 BMMs per well; (41)). Specifically, BMMs were infected with bacteria resuspended in RPMI. The zero hour, collected after 2 h of absorption, as well as all other time points were collected after washing infected BMMs with sterile water (10 s) and then lysing them in 1 ml of sterile water (5 min). Recovered bacteria were serially diluted in water and plated on CYE agar. Three days later, colony counts were enumerated. All infections were carried out in triplicate, and verified in at least three independent studies.
RT-PCR
Total RNA was prepared from day 6 BMMs before or after L. pneumophila infection (MOI = 10) through lysis in Trizol (Invitrogen, Carlsbad, CA). 3–5 μg of total RNA was reverse-transcribed (M-MLV; Invitrogen), as previously reported (32, 37). For semi-quantitative analysis, cDNA was amplified for 28 cycles, fractionated on a 0.8% agarose gel and stained with ethidium bromide. For quantitative (Q) analysis, cDNA was amplified in an ABI Prism 7500 with SYBR green master mix (Applied Biosystems, Foster City, CA) and specific primers (see supplemental Table S1). Gene expression was normalized to a β-actin control. For each primer set, Ct values and standard curves were generated by plotting log DNA concentration versus Ct values from 1:5 serial dilutions with SDS1.9.1 software (Applied Biosystems).
FACS Analysis
Cells were stained with different combinations of fluorochrome-coupled antibodies (32). Anti-CD86 (GL1-PE), anti-I-Ab (AF6–120), and anti-Strepavidin-APC were from BD Pharmingen (San Diego, CA). BM macrophage cultures were infected with fluorescent bacteria. After 1–2 days of growth, cells were stained, and data collected on an LSRII (Beckton Dickinson, Franklin Lakes, NJ). Flow data were analyzed with FlowJo software (TreeStar, Ashland, OR), as previously reported (32).
Biochemical Studies
Whole cell extracts were prepared from IFN-αA/D (1000 u/ml; PBL)-treated cells and evaluated by immunoblotting, as reported (20). Membranes were probed with antibodies specific for Phospho-Stat3 (Cell Signaling Tech., Beverly, MA), Stat3 (Cell Signaling Tech), and Stat1 (20).
NO Assay
48 h supernatants were collected from BMMs and evaluated for NO production by a modified Griess Assay (37). Macrophages were pretreated with IFN-αA/D (1000 units/ml, 2 h; PBL), murine IFN-γ (50 units/ml, 2h; PBL) and/or infected with L. pneumophila (MOI = 1). In some cases, cells were treated with L-NIL (100 μm, Sigma) or L-NMMA (1 mm, Sigma) at 30 min after infection.
RESULTS
IFN-α and IFN-γ Protect Macrophages from Infection with L. pneumophila
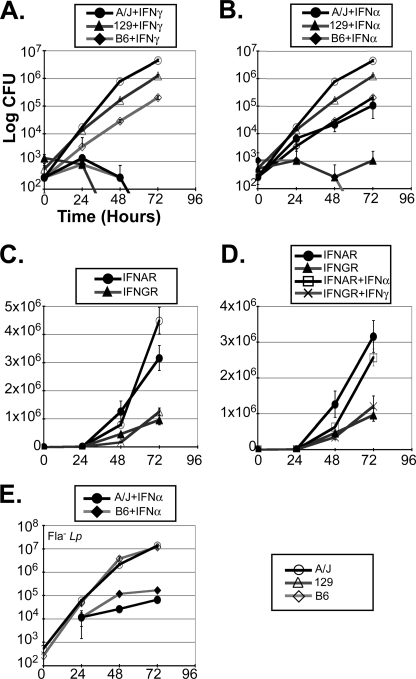
To more carefully explore the ability of IFNs to suppress L. pneumophila replication in mice, BMMs were prepared from three distinct murine backgrounds, A/J, 129 and C57Bl/6J, as well as IFNGR and IFNAR knock-out mice (28). As anticipated, L. pneumophila replicated more effectively in A/J than C57Bl/6J BMMs. 129 macrophages exhibited an intermediate phenotype (see Fig. 1, A and B and Refs. 13, 26, 27). Consistent with previous reports, IFN-γ pretreatment rendered all three genotypes, but not IFNGR(−/−) BMMs, resistant to infection (Fig. 1, A and D and Refs. 26, 42, 43). IFN-α also significantly suppressed L. pneumophila growth in all three genotypes, but not in IFNAR(−/−) macrophages (Fig. 1, B and D). Although IFN-α tended to be a bit less potent, (Fig. 1, A versus B, see also Figs. 4 and 7), it effectively suppressed the growth of both wild type and flagellin null (Fla−, panel E) L. pneumophila, in all genotypes (panels B and D; data not shown). This provided evidence that the response to IFN-Is is independent of Naip5 and the inflammasome (13). Intriguingly, L. pneumophila replicated significantly more efficiently in IFNAR(−/−) than the parental 129 macrophages (Fig. 1C), suggesting that L. pneumophila may stimulate IFN-I autocrine activity (see below and Refs. 13, 27). The lack of an analogous response in IFNGR(−/−) BMMs, likely reflects the inability of macrophages to secrete autocrine IFN-γ (data not shown).
FIGURE 1.
L. pneumophila infection is sensitive to IFNs. L. pneumophila (JR-32; WT) growth was evaluated over 3 days (t = 0, 24, 48, and 72 h) by colony forming unit (CFU) assay of infected (MOI = 0.25) day 6 BMMs (2.5 × 105/well of 24-well plate) from A/J, 129-SvEv (129) and C57Bl/6J (B6) mice (A–E). In some studies A/J, 129 and C57Bl/6J BMMs were pretreated with a single dose of IFN-γ (A, 50 units/ml) or IFN-αA/D (B, 1000 units/ml) 2 h prior to L. pneumophila infection. C and D, control studies revealed significantly enhanced L. pneumophila replication in IFN-α receptor (IFNAR) knock-out BMMs (p < 0.0097 _versus_ 129 BMMs), but not in IFN-γ receptor (IFNGR) knock-out BMMs (_p_ > 0.1 versus 129 BMMs). E, Fla− L. pneumophila growth was evaluated in A/J and C57Bl/6J BMMs with or without IFN-αA/D (1000 units/ml) pretreatment. Differences in L. pneumophila growth were statistically significant in A/J, C57Bl/6J, and 129 cells (p < 0.005), when compared with IFN-α-treated samples. Each infection was carried out in triplicate. Data are representative of more than three independent experiments.
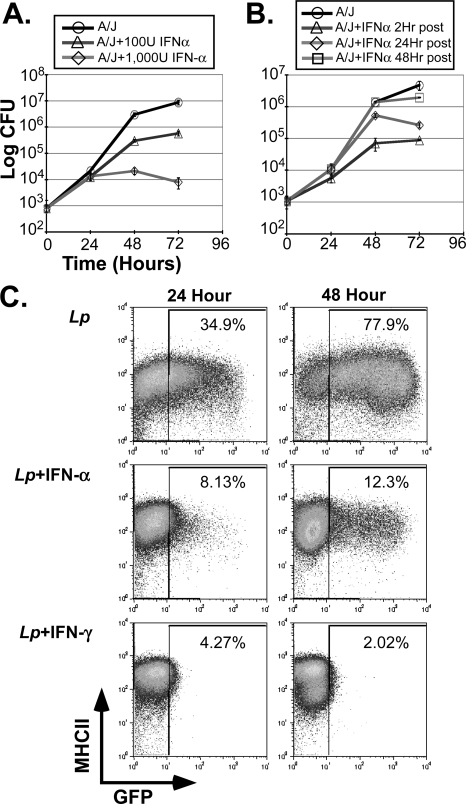
FIGURE 4.
Characterization of IFN-α-dependent response to L. pneumophila infection. A, L. pneumophila (JR-32) growth was evaluated in d6 A/J BMMs, after 2 h of pretreatment with 100 units/ml or 1000 units/ml of IFN-αA/D, as in Fig. 1. B, L. pneumophila growth was evaluated in A/J BMMs treated with IFN-αA/D (1000 units/ml) either at 2, 24, or 48 h after infection, as indicated. C, GFP-_Fla_− L. pneumophila growth in d6 C57Bl/6J BMMs was evaluated by FACS, 24 and 48 h after infection (MOI = 1). Macrophages were pretreated (2 h before infection) with one dose of IFN-αA/D (500 units/ml), or IFN-γ (50 units/ml). Cells were harvested, stained for MHC-II (I-Ab + anti-Strepavidin-APC; BD Pharmingen) and analyzed with LSRII and FlowJo software. Each study is representative of more than three independent experiments.
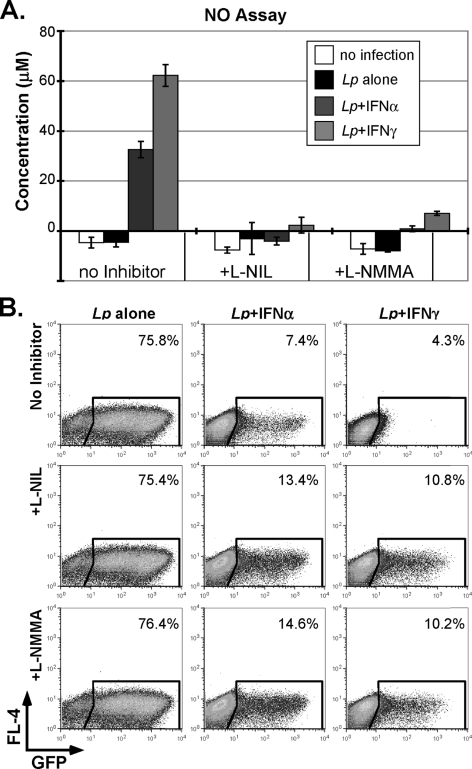
FIGURE 7.
NO production in IFN-stimulated and _L. pneumophila_-infected macrophages. A, culture supernatants were evaluated for NO production 48 h after infection of day 6 C57Bl/6J BMMs with L. pneumophila (GFP Fla−, MOI = 1). In some cases, native, IFN-αA/D- or IFN-γ-pretreated BMMs (as in Fig. 4C) were also treated with l-NIL (100 μm) or l-NMMA (1 mm) at 30 min after infection. B, C57Bl/6J BMMs, treated as in panel A, were evaluated 48 h after infection by FACS, as detailed in Fig. 4. The percentage of GFP-positive cells is indicated. These studies are representative of three independent experiments.
L. pneumophila Infection Stimulates the Secretion of IFN-I
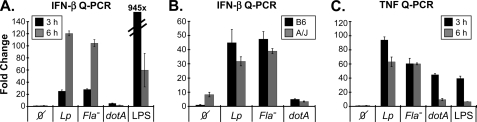
To determine whether L. pneumophila stimulated autocrine IFN-I production, IFN-β expression was evaluated by a sensitive Q-PCR assay (22, 44). Both WT (denoted Lp) and Fla− L. pneumophila stimulated a robust 50–100-fold induction of IFN-β in C57Bl/6J macrophages (Fig. 2A). This response was maximal at 6 h after infection. However, dotA mutants, defective in type IV secretion and intracellular replication, did not stimulate IFN-β expression (Fig. 2A). Similar results were obtained in A/J macrophages, indicating that the flagellin-Naip5 axis does not contribute to this response (Fig. 2B and Ref. 13). Control studies revealed that WT (Lp) and Fla− L. pneumophila stimulated strong TNF induction at both 3 and 6 h, whereas the response to dotA was considerably more transient (Fig. 2C). Moreover, this was associated with considerably lower quantities of secreted TNF (supplemental Fig. S1_A_ and Ref. 13, 45)). Likewise, only WT L. pneumophila stimulated robust IL-1β secretion (supplemental Fig. S1_B_). These results underscore the need for sustained intracellular growth to stimulate a strong inflammatory response (Fig. 2 and supplemental Fig. S1). Moreover, differences in the pattern and kinetics of cytokine expression suggested that IFN-β and TNF expression are directed by distinct L. pneumophila sensor systems, which are also independent of the Naip5-Ipaf axis.
FIGURE 2.
Cytokine expression during L. pneumophila infection. A, fold induction of IFN-β expression was evaluated by Q-PCR (t = 3 & 6 h; see supplemental Table S1 for primers) on RNA recovered from C57Bl/6J BMMs infected (MOI = 10) with wild type (JR-32; Lp), Fla−, or dotA L. pneumophila. Expression was normalized to β-actin. Stimulation with LPS (1 μg/ml) served as a positive control. B, fold induction IFN-β expression 6-h after L. pneumophila (JR-32 = Lp; Fla−; or dotA) infection of C57Bl/6J and A/J BMMs, as in panel A. C, fold induction TNF expression (t = 3 and 6 h; see supplemental Table S1 for primers) evaluated by Q-PCR, as in panel A. Results are each representative of three independent experiments.
L. pneumophila IFN-β Expression Is IRF-3-dependent
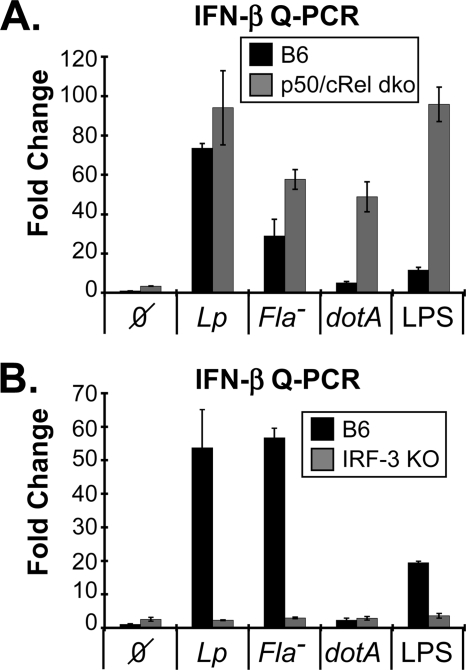
Two transcription factors, NF-κB and IRF-3, play an acute role in stimulating the expression of IFN-β (35, 46–49). Electrophoretic mobility shift assays (EMSAs) revealed that p50 and cRel are the major NF-κB transcription factors activated by LPS and L. pneumophila in murine macrophages (supplemental Fig. S2, A–C). Moreover, dotA-dependent NF-κB activation was considerably more transient than with either WT or Fla− L. pneumophila (supplemental Fig. S2_C_). To explore the potential role NF-κB subunits may play in IFN-I expression, p50 and cRel double knock-out BMMs were exploited (35). Upon infection with L. pneumophila, p50/cRel double knock-out macrophages exhibited a pattern of IFN-β expression that was analogous to control C57Bl/6J cells (Fig. 3A). Curiously, the p50/cRel double knock-out macrophages displayed increased IFN-β induction when they were infected with dotA or stimulated with LPS, underscoring a potentially complex counter regulatory role recently highlighted for NF-κB (13, 45, 50, 51). Moreover, L. pneumophila replicated normally in p50/cRel double knock-out BMMs (supplemental Fig. S2_D_). These observations indicate that NF-κB does not significantly contribute to _L. pneumophila_-dependent IFN-I response.
FIGURE 3.
L. pneumophila induced IFN-I expression in p50/cRel and IRF-3 knock-out BMMs. A, fold induction IFN-β expression in L. pneumophila infected C57Bl/6J and p50/cRel dko BMMs 6-h after infection, as in Fig. 2. B, fold induction IFN-β expression in _L. pneumophila_-infected C57Bl/6J and IRF3(−/−) BMMs 6-h after infection, as above. Both sets of data are representative of three independent studies.
Next, the potential role IRF-3 plays in directing the L. pneumophila dependent expression of IFN-β was evaluated. This analysis exploited IRF3(−/−) BMMs, thereby avoiding the more limited sensitivity of biochemical assays for IRF-3 activity (34). As above, WT and Fla− L. pneumophila both stimulated a robust induction of IFN-β expression in control C57Bl/6J BMMs (Fig. 3B). However, IFN-β expression was absent in IRF3(−/−) BMMs, underscoring the important role this transcription factor plays in regulating the IFN-I autocrine loop (14, 22, 34, 49). Notably, _L. pneumophila_-dependent TNF expression was not affected by the loss of IRF-3 (supplemental Fig. S2_E_).
IFN-α Stimulates a Unique Innate Immune Response
Next, we turned our attention to understanding how IFN-Is suppress L. pneumophila growth. Pretreatment with either 100 or 1000 units/ml of IFN-α rendered A/J BMMs resistant to L. pneumophila growth, in a dose-dependent manner (Fig. 4A). Similar results were obtained in 129 or C57Bl/6J macrophages (not shown). Moreover, IFN-α treatment impeded L. pneumophila replication, even when applied 24-h after infection, suggesting that intracellular bacteria represent the critical target of IFN-I activity (Fig. 4B). Consistent with this, IFN-α pretreatment did not affect the uptake of GFP-labeled L. pneumophila (not shown). Likewise, when GFP-L. pneumophila were exploited in a FACS-based assay, their replication was sensitive to IFN treatment (Fig. 4C). Consistent with growth curve studies (see Fig. 1), a modest level of GFP-L. pneumophila replication was observed in IFN-α-treated BMMs, whereas IFN-γ completely suppressed GFP-L. pneumophila growth at 24 and 48 h (Fig. 4C).
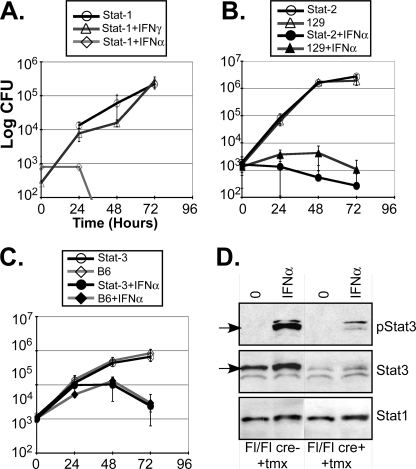
Stat1 has been ascribed a critical role in directing the antimicrobial activity associated with IFN-γ, or “classically activated” (i.e. M1) macrophages (19–21, 52, 53). To determine whether Stat1 played a similarly important role in the ability of IFN-α to suppress L. pneumophila replication, BMMs from Stat1(−/−) mice in the 129 background were evaluated. As anticipated, L. pneumophila growth was resistant to IFN-γ treatment in Stat1(−/−) BMMs (Fig. 5A). In striking contrast, L. pneumophila remained sensitive to IFN-α treatment in Stat1(−/−) BMMs. Likewise, IFN-α was able to effectively suppress bacterial growth in Stat2(−/−) BMMs (Fig. 5B), excluding an important role for either Stat1 homodimers or ISGF-3 (Stat1 + Stat2 + IRF-9, Ref. 23) in this response. Next, a potential role for Stat3, which is also activated by IFN-Is (54), was evaluated. IFN-α was still able to suppress L. pneumophila growth in Stat3-cko BMMs, albeit with the slower kinetics associated with the parental C57Bl/6J background (Fig. 5C). These results reveal that IFN-Is do not direct their protective response toward L. pneumophila through canonical Stat1-, Stat2-, or Stat3-dependent pathways, but rather through a distinct pathway.
FIGURE 5.
The role of Stat1, Stat2, and Stat3 on IFN suppression of L. pneumophila growth. A, L. pneumophila (JR-32; Lp) growth was evaluated in d6 Stat1(−/−) BMMs, as in Fig. 1, either with or without IFN-αA/D (1000 units/ml) or IFN-γ (50 units/ml) pretreatment (2 h before infection). B, L. pneumophila (JR-32; Lp) growth was evaluated in d6 Stat2(−/−) and 129 BMMs after IFN-αA/D pretreatment, as in panel A. C, L. pneumophila (JR-32; Lp) growth was evaluated in d6 Stat3-cko and C57Bl6/J BMMs (see below), after IFN-αA/D pretreatment, as in panel A. D, Stat3-cko macrophages were generated through the addition of 250 nm tamoxifen to day 1 and day 5 BM cultures from Stat3-fl/fl × Rosa26-ER(T2)-Cre (Stat3-cko) mice. Control BMMs were generated from Stat3-fl/fl mouse. Whole cell extracts were prepared after IFN-αA/D (1000 units/ml; 30 min) treatment and evaluated by immunoblotting with antibodies for phospho-Stat3, Stat3, and Stat1. These results are each representative of more than three independent experiments.
IFNs Activate M1 Macrophages
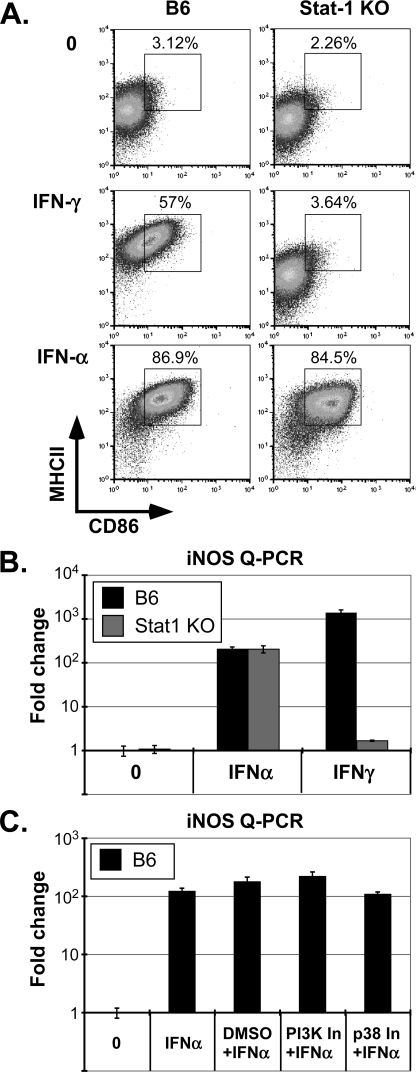
The antibacterial response to IFN-γ correlates directly with the development of M1 or “classically activated” macrophages ((52); see Fig. 6), whereas polarization toward alternatively activated M2 macrophages had no effect on L. pneumophila growth (data not shown). As previously reported, this response was associated with an up-regulation of MHC-II and CD86 surface expression, as well as induction of iNOS (53, 55). Likewise, the ability of IFN-γ to direct MHC-II, CD86, and iNOS expression was fully dependent on Stat1 (see Fig. 6, A and B). In contrast, the equally effective ability of IFN-α to induce these M1 markers was fully independent of Stat1 (Fig. 6B). Moreover, IFN-α directed M1 polarization was also independent of Stat2 (not shown). The close correlation between IFN-I stimulated, STAT-independent suppression of L. pneumophila growth and M1 polarization, suggests that the classical activation of macrophages is important for this innate response. These observations also highlight an unexpected functional dichotomy in the ability of type I and II IFNs to polarize macrophages toward the M1 phenotype and suppress L. pneumophila growth.
FIGURE 6.
IFN-α directs STAT independent M1 polarization of macrophages. A, IFN-αA/D (500 units/ml; 48 h)- or IFN-γ (50 units/ml; 48 h)-treated C57Bl/6J and Stat1(−/−) d6 BMMs were stained for MHC-II (I-Ab + anti-Strepavidin-APC) and anti-CD86-PE and evaluated by FACS, as in Fig. 4. Percentages of double positive cells, delineated by box are indicated. B, fold induction iNOS expression was evaluated by Q-PCR (see supplemental Table S1 for primers) from IFN-αA/D (1000 units/ml; 24 h) or IFN-γ (50 units/ml; 24 h)-treated d6 C57Bl/6J and Stat1(−/−) BMMs, as in Fig. 2. C, fold induction iNOS expression was evaluated in IFN-αA/D (1000 units/ml; 10 h) stimulated C57Bl/6J BMMs, as in panel A. Macrophages were either pretreated with DMSO (solvent control), a PI3K inhibitor (LY294002, 50 μm; 30 min), or a p38 inhibitor (SB203580, 50 μm; 30 min). Each study is representative of at least three independent experiments.
The relatively rapid, IFN-α-stimulated and Stat1-independent induction of iNOS, a characteristic M1 marker, provided an opportunity to explore the potential role noncanonical signaling pathways may play in IFN-I response. Based on previous evidence of a role for p38 and PI-3 kinases in STAT-independent IFN-I response (56, 57), the ability of IFN-α to induce iNOS expression was evaluated in the presence of two specific inhibitors, SB203580 and LY94002, respectively (Fig. 6C and Ref. 56). Neither inhibitor blocked IFN-α stimulated expression of iNOS, raising the possibility that these two kinases also do not contribute to the IFN-I-dependent M1 polarization associated with L. pneumophila suppression. Consistent with this possibility, biochemical studies suggest that IFN-I does not effectively activate these kinases in primary murine macrophages (supplemental Fig. S4).
Nitric Oxide Accounts for Phenotypic Differences between Type I and II IFNs
NO production has not been ascribed a significant role in the innate response to L. pneumophila infection (26, 58). To determine whether NO might contribute to the noncanonical IFN-I response associated with L. pneumophila suppression, additional studies were carried out. As previously reported, L. pneumophila failed to effectively stimulate NO production (26, 58). However, pretreatment of cells with IFN-γ and to a lesser extent with IFN-α led to a robust induction of NO (Fig. 7A). Moreover, this was blocked through the addition of either the iNOS-specific inhibitor, L-NIL, or a more general NOS inhibitor, l-NMMA (Fig. 7A). These observations raised the possibility that the more robust NO response stimulated by IFN-γ might account for the modestly enhanced ability of IFN-γ (versus IFN-α) to suppress L. pneumophila growth (e.g. Figs. 1, 4C, and 7B). Consistent with this, the level of intracellular L. pneumophila replication was modestly increased when NO production was blocked in IFN-pretreated macrophages (Fig. 7B). Moreover, the change in L. pneumophila growth was considerably more striking in IFN-γ-pretreated macrophages, suggesting that NO production is likely to account for the more effective ability of IFN-γ (versus IFN-α) to suppress L. pneumophila replication. These studies also suggest that M1-associated responses that are independent of iNOS play a far more important role in the ability of IFNs to impede L. pneumophila growth.
DISCUSSION
Pattern recognition sensors systems evolved to detect the presence of pathogen associated molecular patterns (PAMPs) and initiate effective innate responses (24, 59, 60). Exploiting the murine system, investigators have identified two cytosolic sensors, Naip5/Birc1e and Ipaf, which specifically initiate an innate response to L. pneumophila flagellin (2, 7–9, 11). This entails activation of the inflammasome, leading to caspase1 dependent secretion of IL-1β and IL-18, as well as the induction of pyroptosis (7, 11, 12). Curiously however, IL-1β and IL-18 do not appear to directly regulate L. pneumophila growth in macrophages (3, 13). Moreover, the Naip5-Ipaf sensor system does not seem to participate in the secretion of other cytokines that have been associated with L. pneumophila infection (13, 27).
Evidence that L. pneumophila replicated more robustly in IFNAR1(−/−) BMMs raised the possibility that the IFN-I autocrine loop may contribute to the innate response in WT macrophages, as recently suggested (13, 27). Consistent with this, both WT and Fla− L. pneumophila potently stimulated IFN-β expression, analogous to what has been reported for L. monocytogenes, F. tularensis, as well as other microbial pathogens (34, 44, 61, 62). Moreover, this response is largely independent of TLRs (13, 14), IPS-1/MAVs (63), the Naip5-Ipaf-flagellin axis (Fig. 2 and Ref. 13), IFNAR1, and Stat1 (supplemental Fig. S3_A_). Mechanistic studies determined that L. pneumophila stimulated IFN-β expression was dependent on IRF-3, but not p50/c-Rel (Fig. 3). Consistent with this, IFN-β expression also appeared to be dependent on TBK-1, the kinase that activates IRF-3 (see supplemental Fig. S3_B_ and Ref. 44). Previous RNAi-based studies in human epithelial cells, which do not support robust L. pneumophila growth, have also implicated IRF-3, as well as IPS-1 in _L. pneumophila_-stimulated IFN-I expression (64).
Genetic studies excluding a role for IPS-1 in _L. pneumophila_-dependent IFN-β expression raised the possibility that novel host receptors may sense the cytoplasmic accumulation of bacterial nucleic acids (14, 63, 65–69). However, the μg quantities of bacterial nucleic acids required to stimulate IFN-β expression and our inability to detect even traces of L. pneumophila nucleic acids in the cytoplasm of infected macrophages (data not shown) do not support this model. Thus, the identification of the innate sensor that promotes activation of the IFN-I autocrine loop during a L. pneumophila infection remains an important goal for future studies.
Studies exploring how IFNs antagonize L. pneumophila growth revealed a correlation with polarization of macrophages to the inflammatory M1, but not M2 phenotype (Figs. 4 and 6; not shown, Ref. 52). Curiously, even though Stat1 was shown to play a critical role in both IFN-γ-dependent M1 polarization and L. pneumophila suppression, this was not the case for IFN-α. Rather, the ability of IFN-I to impede L. pneumophila growth was found to be independent of Stat1, Stat2, and Stat3. Intriguingly, IFN-I-dependent M1 polarization was also found to be independent of these canonical STAT signaling pathways (Fig. 5), raising the possibility that these two responses are functionally related. Additional studies exploiting the more rapid kinetics of IFN-α-stimulated iNOS expression, an archetypal M1 response, revealed a similar noncanonical IFN-I dependent signaling pathway. Moreover, the relatively rapid kinetics of IFN-I stimulated iNOS expression provided an opportunity to largely exclude an important role two well characterized STAT independent signals, p38 and PI-3 kinases, in this noncanonical IFN-I response. Consistent with the possibility that these 2 kinases may not contribute to M1 polarization or L. pneumophila suppression, biochemical studies failed to reveal robust activation of p38 and PI-3 kinases in IFN-α treated macrophages. Future studies will employ additional knock-out models to more rigorously explore the nature of IFN-I stimulated signals in M1 polarization and the suppression of L. pneumophila replication.
In summary, the innate response to L. pneumophila infection entails the activation of multiple pattern recognition systems that direct the secretion of several inflammatory cytokines, including IL-1β, IL-18, TNF, and IFN-β (7, 11, 13). Although it seems likely that these pathways synergize to mount an effective innate response to L. pneumophila, IFN-I remains the single most effective cytokine in suppressing L. pneumophila growth (12, 13).
Supplementary Material
Supplemental Data
Acknowledgments
We thank Dr. K. de Felipe for generating the Fla− mutant, and Dr. X. Charpentier for providing GFP-L. pneumophila, and Dr. Genhong Cheng for providing TBK-1 knockout cells.
Note Added in Proof
Two recently published studies suggest that RNA polymerase III directs a RIG-I–IPS-1-dependent response to microbial DNA, including that of L. pneumophila, that culminates in IFN-β expression (70, 71).
*
This work was supported, in whole or in part, by National Institutes of Health Grants AI 058211 (to C. S.), HL 55413 (to C. S.), 5T32 AI07525 (to C. R. P.), 5T32 DK007328 (to C. R. P.), AI 059715 (to A. A. B.), and AI23549 (to H. A. S.). This research was also supported by the Austrian Science Foundation (FWF) Grant P20522-B05 (to T. D.).
2
The abbreviations used are:
IL
interleukin
IFN
interferon
JAK
Janus kinases
STAT
Signal Transducers and Activators of Transcription
WT
wild type
MOI
multiplicity of infection
GFP
green fluorescent protein
IFNAR
IFN-α receptor
IFNGR
IFN-γ receptor
BMM
bone marrow-derived murine macrophages.
REFERENCES
- 1.Sabrià M., Campins M. (2003) Am. J. Respir. Med. 2, 235–243 [DOI] [PubMed] [Google Scholar]
- 2.Fortier A., Diez E., Gros P. (2005) Trends Microbiol. 13, 328–335 [DOI] [PubMed] [Google Scholar]
- 3.Neild A. L., Roy C. R. (2004) Cell Microbiol. 6, 1011–1018 [DOI] [PubMed] [Google Scholar]
- 4.Segal G., Feldman M., Zusman T. (2005) FEMS Microbiol. Rev. 29, 65–81 [DOI] [PubMed] [Google Scholar]
- 5.Ninio S., Roy C. R. (2007) Trends Microbiol. 15, 372–380 [DOI] [PubMed] [Google Scholar]
- 6.Losick V. P., Stephan K., Smirnova II, Isberg R. R., Poltorak A. (2009) Infect. Immun. 77, 196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lightfield K. L., Persson J., Brubaker S. W., Witte C. E., von Moltke J., Dunipace E. A., Henry T., Sun Y. H., Cado D., Dietrich W. F., Monack D. M., Tsolis R. M., Vance R. E. (2008) Nat. Immunol. 9, 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molofsky A. B., Byrne B. G., Whitfield N. N., Madigan C. A., Fuse E. T., Tateda K., Swanson M. S. (2006) J. Exp. Med. 203, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren T., Zamboni D. S., Roy C. R., Dietrich W. F., Vance R. E. (2006) PLoS Pathog. 2, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutterwala F. S., Ogura Y., Zamboni D. S., Roy C. R., Flavell R. A. (2006) J. Endotoxin. Res. 12, 251–256 [DOI] [PubMed] [Google Scholar]
- 11.Zamboni D. S., Kobayashi K. S., Kohlsdorf T., Ogura Y., Long E. M., Vance R. E., Kuida K., Mariathasan S., Dixit V. M., Flavell R. A., Dietrich W. F., Roy C. R. (2006) Nat. Immunol. 7, 318–325 [DOI] [PubMed] [Google Scholar]
- 12.Amer A., Franchi L., Kanneganti T. D., Body-Malapel M., Ozören N., Brady G., Meshinchi S., Jagirdar R., Gewirtz A., Akira S., Núñez G. (2006) J. Biol. Chem. 281, 35217–35223 [DOI] [PubMed] [Google Scholar]
- 13.Coers J., Vance R. E., Fontana M. F., Dietrich W. F. (2007) Cell Microbiol. 9, 2344–2357 [DOI] [PubMed] [Google Scholar]
- 14.Stetson D. B., Medzhitov R. (2006) Immunity 24, 93–103 [DOI] [PubMed] [Google Scholar]
- 15.Archer K. A., Roy C. R. (2006) Infect. Immun. 74, 3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawn T. R., Smith K. D., Aderem A., Skerrett S. J. (2006) J. Infect. Dis. 193, 1693–1702 [DOI] [PubMed] [Google Scholar]
- 17.Hawn T. R., Berrington W. R., Smith I. A., Uematsu S., Akira S., Aderem A., Smith K. D., Skerrett S. J. (2007) J. Immunol. 179, 6981–6987 [DOI] [PubMed] [Google Scholar]
- 18.Schindler C., Plumlee C. (2008) Semin Cell Dev. Biol. 19, 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meraz M. A., White J. M., Sheehan K. C., Bach E. A., Rodig S. J., Dighe A. S., Kaplan D. H., Riley J. K., Greenlund A. C., Campbell D., Carver-Moore K., DuBois R. N., Clark R., Aguet M., Schreiber R. D. (1996) Cell 84, 431–442 [DOI] [PubMed] [Google Scholar]
- 20.Park C., Li S., Cha E., Schindler C. (2000) Immunity 13, 795–804 [DOI] [PubMed] [Google Scholar]
- 21.Filipe-Santos O., Bustamante J., Chapgier A., Vogt G., de Beaucoudrey L., Feinberg J., Jouanguy E., Boisson-Dupuis S., Fieschi C., Picard C., Casanova J. L. (2006) Semin. Immunol. 18, 347–361 [DOI] [PubMed] [Google Scholar]
- 22.Decker T., Müller M., Stockinger S. (2005) Nat. Rev. Immunol. 5, 675–687 [DOI] [PubMed] [Google Scholar]
- 23.Schindler C., Levy D. E., Decker T. (2007) J. Biol. Chem. 282, 20059–20063 [DOI] [PubMed] [Google Scholar]
- 24.Pichlmair A., Reis e Sousa C. (2007) Immunity 27, 370–383 [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001) Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 26.Akamine M., Higa F., Haranaga S., Tateyama M., Mori N., Heuner K., Fujita J. (2007) Microbiol. Immunol. 51, 279–287 [DOI] [PubMed] [Google Scholar]
- 27.Schiavoni G., Mauri C., Carlei D., Belardelli F., Pastoris M. C., Proietti E. (2004) J. Immunol. 173, 1266–1275 [DOI] [PubMed] [Google Scholar]
- 28.Müller U., Steinhoff U., Reis L. F., Hemmi S., Pavlovic J., Zinkernagel R. M., Aguet M. (1994) Science 264, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 29.Park C., Lecomte M. J., Schindler C. (1999) Nucleic Acids Res. 27, 4191–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3801–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vooijs M., Jonkers J., Berns A. (2001) EMBO Rep. 2, 292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao W., Cha E. N., Lee C., Park C. Y., Schindler C. (2007) J. Immunol. 179, 463–471 [DOI] [PubMed] [Google Scholar]
- 33.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 34.Stockinger S., Reutterer B., Schaljo B., Schellack C., Brunner S., Materna T., Yamamoto M., Akira S., Taniguchi T., Murray P. J., Müller M., Decker T. (2004) J. Immunol. 173, 7416–7425 [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Hussain S., Wang E. J., Wang X., Li M. O., García-Sastre A., Beg A. A. (2007) J. Immunol. 178, 6770–6776 [DOI] [PubMed] [Google Scholar]
- 36.Perry A. K., Chow E. K., Goodnough J. B., Yeh W. C., Cheng G. (2004) J. Exp. Med. 199, 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song L., Bhattacharya S., Yunus A. A., Lima C. D., Schindler C. (2006) Blood 108, 3237–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J., de Felipe K. S., Clarke M., Lu H., Anderson O. R., Segal G., Shuman H. A. (2004) Science 303, 1358–1361 [DOI] [PubMed] [Google Scholar]
- 39.Chien M., Morozova I., Shi S., Sheng H., Chen J., Gomez S. M., Asamani G., Hill K., Nuara J., Feder M., Rineer J., Greenberg J. J., Steshenko V., Park S. H., Zhao B., Teplitskaya E., Edwards J. R., Pampou S., Georghiou A., Chou I. C., Iannuccilli W., Ulz M. E., Kim D. H., Geringer-Sameth A., Goldsberry C., Morozov P., Fischer S. G., Segal G., Qu X., Rzhetsky A., Zhang P., Cayanis E., De Jong P. J., Ju J., Kalachikov S., Shuman H. A., Russo J. J. (2004) Science 305, 1966–1968 [DOI] [PubMed] [Google Scholar]
- 40.Hovel-Miner G., Pampou S., Faucher S. P., Clarke M., Morozova I., Morozov P., Russo J. J., Shuman H. A., Kalachikov S. (2009) J. Bacteriol. 191, 2461–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hilbi H., Segal G., Shuman H. A. (2001) Mol. Microbiol. 42, 603–617 [DOI] [PubMed] [Google Scholar]
- 42.Deng J. C., Tateda K., Zeng X., Standiford T. J. (2001) Infect. Immun. 69, 6382–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nash T. W., Libby D. M., Horwitz M. A. (1988) J. Immunol. 140, 3978–3981 [PubMed] [Google Scholar]
- 44.Hiscott J., Lin R., Nakhaei P., Paz S. (2006) Trends Mol. Med. 12, 53–56 [DOI] [PubMed] [Google Scholar]
- 45.Abu-Zant A., Jones S., Asare R., Suttles J., Price C., Graham J., Kwaik Y. A. (2007) Cell Microbiol. 9, 246–264 [DOI] [PubMed] [Google Scholar]
- 46.Escalante C. R., Shen L., Thanos D., Aggarwal A. K. (2002) Structure 10, 383–391 [DOI] [PubMed] [Google Scholar]
- 47.Schneider K., Loewendorf A., De Trez C., Fulton J., Rhode A., Shumway H., Ha S., Patterson G., Pfeffer K., Nedospasov S. A., Ware C. F., Benedict C. A. (2008) Cell Host Microbe 3, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turer E. E., Tavares R. M., Mortier E., Hitotsumatsu O., Advincula R., Lee B., Shifrin N., Malynn B. A., Ma A. (2008) J. Exp. Med. 205, 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiscott J. (2007) J. Biol. Chem. 282, 15325–15329 [DOI] [PubMed] [Google Scholar]
- 50.Losick V. P., Isberg R. R. (2006) J. Exp. Med. 203, 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Wang X., Hussain S., Zheng Y., Sanjabi S., Ouaaz F., Beg A. A. (2007) J. Immunol. 178, 6777–6788 [DOI] [PubMed] [Google Scholar]
- 52.Timmer A. M., Nizet V. (2008) J. Exp. Med. 205, 1255–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fong C. H., Bebien M., Didierlaurent A., Nebauer R., Hussell T., Broide D., Karin M., Lawrence T. (2008) J. Exp. Med. 205, 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao W., Lee C., Piganis R., Plumlee C., de Weerd N., Hertzog P. J., Schindler C. (2008) J. Immunol. 180, 5483–5489 [DOI] [PubMed] [Google Scholar]
- 55.Hagemann T., Lawrence T., McNeish I., Charles K. A., Kulbe H., Thompson R. G., Robinson S. C., Balkwill F. R. (2008) J. Exp. Med. 205, 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gimeno R., Lee C. K., Schindler C., Levy D. E. (2005) Mol. Cell Biol. 25, 5456–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uddin S., Majchrzak B., Woodson J., Arunkumar P., Alsayed Y., Pine R., Young P. R., Fish E. N., Platanias L. C. (1999) J. Biol. Chem. 274, 30127–30131 [DOI] [PubMed] [Google Scholar]
- 58.Gebran S. J., Yamamoto Y., Newton C., Klein T. W., Friedman H. (1994) Infect. Immun. 62, 3197–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trinchieri G., Sher A. (2007) Nat. Rev. Immunol. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 60.Kawai T., Akira S. (2006) Nat. Immunol. 7, 131–137 [DOI] [PubMed] [Google Scholar]
- 61.Henry T., Brotcke A., Weiss D. S., Thompson L. J., Monack D. M. (2007) J. Exp. Med. 204, 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tam M. A., Sundquist M., Wick M. J. (2008) Cell Microbiol. 10, 1517–1529 [DOI] [PubMed] [Google Scholar]
- 63.Sun Q., Sun L., Liu H. H., Chen X., Seth R. B., Forman J., Chen Z. J. (2006) Immunity 24, 633–642 [DOI] [PubMed] [Google Scholar]
- 64.Opitz B., Vinzing M., van Laak V., Schmeck B., Heine G., Günther S., Preissner R., Slevogt H., N′Guessan P. D., Eitel J., Goldmann T., Flieger A., Suttorp N., Hippenstiel S. (2006) J. Biol. Chem. 281, 36173–36179 [DOI] [PubMed] [Google Scholar]
- 65.Kanneganti T. D., Ozören N., Body-Malapel M., Amer A., Park J. H., Franchi L., Whitfield J., Barchet W., Colonna M., Vandenabeele P., Bertin J., Coyle A., Grant E. P., Akira S., Núñez G. (2006) Nature 440, 233–236 [DOI] [PubMed] [Google Scholar]
- 66.Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K. L., Superti-Furga G. (2009) Nat. Immunol. 10, 266–272 [DOI] [PubMed] [Google Scholar]
- 67.Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009) Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. (2009) Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts T. L., Idris A., Dunn J. A., Kelly G. M., Burnton C. M., Hodgson S., Hardy L. L., Garceau V., Sweet M. J., Ross I. L., Hume D. A., Stacey K. J. (2009) Science 323, 1057–1060 [DOI] [PubMed] [Google Scholar]
- 70.Chiu Y. H., Macmillan J. B., Chen Z. J. (2009) Cell 138, 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K. A., Hornung V. (2009) Nat. Immunol. 10, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data