SnoN functions as a tumour suppressor by inducing premature senescence (original) (raw)
Abstract
SnoN represses TGF-β signalling to promote cell proliferation and has been defined as a proto-oncogene partly due to its elevated expression in many human cancer cells. Although the anti-tumourigenic activity of SnoN has been suggested, the molecular basis for this has not been defined. We showed here that high levels of SnoN exert anti-oncogenic activity by inducing senescence. SnoN interacts with the promyelocytic leukaemia (PML) protein and is recruited to the PML nuclear bodies where it stabilizes p53, leading to premature senescence. Furthermore, overexpression of SnoN inhibits oncogenic transformation induced by Ras and Myc in vitro and significantly blocks papilloma development in vivo in a carcinogen-induced skin tumourigenesis model. The few papillomas that were developed displayed high levels of senescence and spontaneously regressed. Our study has revealed a novel Smad-independent pathway of SnoN function that mediates its anti-oncogenic activity.
Keywords: p53, PML (promyelocytic leukaemia), senescence, SnoN, tumour suppressor
Introduction
Tumour development involves inactivation of tumour suppressors and activation of proto-oncogenes. Some oncogenes have been shown to also possess anti-oncogenic activities, which makes it difficult to design targeted drugs. SnoN is a member of the Ski family of proteins identified based on sequence homology with v-Ski, the transforming protein of Sloan Kettering virus (Nomura et al, 1989; Pearson-White, 1993). It is expressed ubiquitously in most cell types with a relatively high level of expression in the embryos but at a lower level in adult cells (Pelzer et al, 1996). It was classified as an oncoprotein based on its transformation ability when overexpressed in chicken or quail embryo fibroblast (Boyer et al, 1993). The role of SnoN in mammalian tumourigenesis, however, is much more complex and even controversial. Although elevation in SnoN expression is often detected in many human carcinoma tissues and cell lines due to either gene amplification, transcriptional activation or enhanced protein stability (Imoto et al, 2001; Zhang et al, 2003; Buess et al, 2004; Edmiston et al, 2005; Zhu et al, 2006; Bravou et al, 2009; Guzman-Ayala et al, 2009), downregulation of SnoN expression has also been observed in human cancers. In tissue samples from patients with Barrett's oesophagus, a precancerous condition that associates with an increased risk for adenocarcinoma, SnoN was found at high levels in low-grade dysplasia but absent in high-grade dysplasia or adenocarcinoma (Villanacci et al, 2008). In colorectal cancer with microsatellite instability, SnoN is downregulated in 39% of samples and upregulated in 33% of samples (Chia et al, 2006). These observations suggest that high levels of SnoN, although beneficial to certain stages of tumour development, may not always be advantageous for malignant progression. More importantly, these studies enforce the hypothesis that SnoN may possess both pro-oncogenic and anti-oncogenic activities in mammalian carcinogenesis. The strongest support for a pro-oncogenic activity of SnoN comes from the observation that reducing SnoN expression in human lung and breast cancer cells inhibits cancer cell growth in vitro and in vivo (Zhu et al, 2006). However, other studies also supported an anti-oncogenic activity of SnoN. The loss of one allele of SnoN in mice results in a slightly higher incidence of tumour formation and increased susceptibility to chemical carcinogen-induced tumourigenesis (Shinagawa et al, 2000), and reducing SnoN expression in human lung and breast cancer cells enhances EMT and tumour metastasis in vivo (Zhu et al, 2006). In Drosophila, overexpressed dSnoN inhibits cell growth (Ramel et al, 2007). Finally, SnoN can cooperate with p53 in silencing of the α-fetoprotein gene, which is aberrantly overexpressed in liver cancer cells (Wilkinson et al, 2008). The mechanisms through which SnoN exerts these differential effects on malignant progression have not been defined.
SnoN is an important negative regulator of TGF-β signalling. TGF-β is a pluripotent cytokine that regulates many cellular processes, including cell proliferation, survival, cell–matrix interaction and differentiation (Schilling et al, 2008). Smad proteins are critical mediators of many TGF-β-induced signals. On the binding of TGF-β to its receptor complex, the receptor-activated Smads (R-Smad), Smad2 and Smad3, become phosphorylated by the active receptor kinase complex, form a heteromeric complex with Smad4 and translocate into the nucleus. In the nucleus, the Smad heteromeric complex binds to TGF-β responsive promoters either directly or through association with other sequence-specific transcriptional factors and regulates the expression of TGF-β responsive genes (Moustakas et al, 2001; Shi and Massague, 2003; Feng and Derynck, 2005). We and others have shown that SnoN interacts with Smad proteins and represses their transactivation activity by disrupting functional heteromeric Smad complexes, recruiting transcription co-repressor complex and blocking the binding of transcriptional co-activators to Smads, (Stroschein et al, 1999; Sun et al, 1999; Wu et al, 2002). Through these mechanisms, SnoN acts as a co-repressor of Smad proteins. Indeed, overexpression of SnoN blocks the growth inhibitory responses to TGF-β. This ability to antagonize TGF-β signalling may be responsible for the oncogenic activity of SnoN at early stages of tumourigenesis (He et al, 2003).
Although SnoN represses the activity of Smads, its own expression is also tightly regulated by R-Smads. Shortly after TGF-β stimulation, R-Smads bind to SnoN and recruit various E3 ubiquitin ligases, including the anaphase promoting complex (APC/C), Smurf2 or Arkadia to SnoN for inducing its polyubiquitinylation and degradation (Stroschein et al, 1999, 2001; Bonni et al, 2001; Wan et al, 2001; Levy et al, 2007; Nagano et al, 2007), allowing the activation of Smad-mediated transcription. Interestingly, the snoN gene itself is a transcription target of Smads and its expression is upregulated 2 h after TGF-β stimulation (Stroschein et al, 1999; Zhu et al, 2005). This later increase in SnoN expression may turn off TGF-β signalling in a negative feedback manner or regulate cell proliferation and differentiation in a TGF-β-independent manner (Zhu et al, 2005).
To elucidate the physiological function of the SnoN–Smad interaction, we generated knock-in mice substituting the endogenous snoN gene with a mutant deficient in binding to both R-Smads and Smad4. Mice expressing the mSnoN gene are resistant to chemical carcinogen-induced tumourigenesis probably due to the accumulation of senescent cells in tumours. Accordingly, mouse embryonic fibroblasts (MEFs) prepared from the knock-in mice also show premature senescence. We showed here that the ability of SnoN to promote premature senescence is dependent on p53 and PML proteins, and functions to block oncogenic transformation in vitro and tumour development in vivo. Our study, therefore, revealed a new function for SnoN in promoting senescence and provided a potential mechanism to understand the tumour suppressor functions of SnoN.
Results
Generation of knock-in mice expressing a mutant SnoN
To understand the functions of the SnoN–Smad interaction, we generated a knock-in mouse replacing the endogenous snoN gene with a snoN mutant (mSnoN) containing point mutations that alter amino-acid residues 88–92 and 267–277 to alanine through homologous recombination. These mutations disrupt the interaction of SnoN with both R-Smads and Smad4, and abolish the ability of SnoN to repress TGF-β signalling (Wu et al, 2002; He et al, 2003; Zhu et al, 2006). The mSnoN gene can be distinguished from the WT allele by the introduction of the _Swa_I and _Sph_I restriction enzyme cleavage sites with point mutations (Figure 1A). Mice carrying a targeted allele of mutant exon 1 were crossed with CMV–Cre transgenic mice to yield the knock-in mice. Expression of the knock-in allele was confirmed by isolating the exon 1 of the snoN gene from the genomic DNA by PCR followed by _Sph_I digestion (Figure 1B). Although WT snoN gene yielded a 1-kb fragment, the mutant allele produced a 0.8-kb fragment. As it is not possible to distinguish between the WT and mutant SnoN protein by western blotting, we resorted to functional assays to confirm the expression of mSnoN. MEFs from multiple E13.5 embryos were derived from both knock-in mice and WT littermates, and subjected to various assays to measure TGF-β responsiveness. As expected, the homozygous mutant (m/m) MEFs showed increased transcription responses to TGF-β in a luciferase reporter assay (Figure 1C) and were more sensitive to TGF-β-induced growth arrest (Figure 1D), consistent with the elevated Smad activity in m/m MEFs due to the lack of antagonism by SnoN.
Figure 1.
Generation of SnoN knock-in mice. (A) Targeting constructs. The _Bam_HI fragment of mouse snoN genomic DNA containing exon I and the flanking sequences were subcloned into pBluescript. The R-Smad binding site between residues 85–88 and Co-Smad binding site between residues 266–267 in exon I were mutated to alanine (shown as 85–88AAAA and 266, 267AA, respectively), and a neomycin resistance gene flanked by two LoxP sequences was inserted into intron I. These mutations also generated new _Sph_I and _Swa_I cleavage sites. (B) Genomic DNA was prepared from MEF. DNA fragment encompassing exon I was amplified by PCR and digested using _Sph_I to yield a 1-kb fragment for WT snoN and 0.8-kb fragment for mSnoN. (C) Mutant MEF showed enhanced TGF-β transcription. p3TP-Lux was transfected into the WT and m/m MEF. Luciferase activity was measured 16 h after TGF-β treatment. (D) Mutant MEFs are more sensitive to TGF-β-induced growth arrest. A total of 5 × 104 WT or m/m MEFs were cultured in the presence of TGF-β. The growth of cells was measured four days later by cell counting and compared with that of un-stimulated cells.
Knock-in mice are resistant to carcinogen-induced skin tumourigenesis
Among the pups that were born, 15.7% were homozygous for the knock-in allele, 52.4% were heterozygous and 31.9% were WT, indicating that approximately 37.2% of the homozygous embryos died before birth and 62.8% survived. The survived homozygous mice lived up to 24 months without any apparent defects. No increase in spontaneous tumour development was observed in these mice for up to 24 months (data not shown).
To determine whether the knock-in mice are more or less susceptible to chemical-induced carcinogenesis, a two-step skin tumourigenesis protocol was used (Balmain et al, 1988) (Figure 2A). WT or 8-week old m/m mice were administered with one dose of DMBA followed by twice weekly treatment of TPA for 30 weeks. Development of papilloma was monitored for 30 weeks. Under this regime, papilloma was detected first at 14–15 weeks after the initial DMBA treatment in both +/+ and m/m mice. However, although more than 80% of +/+ mice developed tumour by 30 weeks, less than 40% of m/m mice showed a tumor development (Figure 2B). In the m/m mice that developed tumours, the average number of papillomas per mouse was significantly reduced (Figure 2C). More importantly, most tumours in m/m mice ceased to grow after only a short period of time and spontaneously regressed, and few reached a size larger than 2 mm in diameter, whereas papillomas in WT mice continued to grow to be larger than 10 mm (Figure 2D). This strongly suggests that mSnoN blocked papilloma development in vivo.
Figure 2.
The snoN knock-in mice are resistant to chemical carcinogen-induced skin tumourigenesis. (A) Schematic representation of the two-step skin carcinogenesis protocol. (B) Percentage of mice that developed papilloma within a 30-week window is shown in the graph. Graph in (C) shows average number of papilloma per mouse. (D) The size distributions of papillomas from WT and m/m mice. (E) H&E staining of papilloma sections from +/+ or m/m mice (scale bar: 200 μm). (F) Lysates from papilloma samples were subjected to western blotting to measure the expression of p53, p16INK4a or phospho-ERK. Tubulin was used as a loading control. (G) Frozen papilloma sections from +/+ or m/m mice were stained using SA-β-Gal (scale bar: 100 μm). (H) Tumour sections were stained using anti-p19ARF (green; scale bar: 100 μm). The boxed areas were amplified to show the nucleolus staining of p19ARF (scale bar: 30 μm). Nuclei were stained using DAPI (blue).
Cellular senescence is a permanent non-proliferative state that can be triggered by telomere shortening or accumulation of physiological stress (Hayflick, 1965; Campisi, 2001; Collado et al, 2007). It has been shown to be an important tumour suppressive mechanism in mouse models of human cancer (Braig et al, 2005; Chen et al, 2005; Michaloglou et al, 2005; Xue et al, 2007) and can cause tumour regression. To determine whether papillomas from the m/m mice showed increased senescence, sections from papilloma sized 2 mm in diameter, collected from WT or m/m mice, were subjected to H&E analysis and staining of senescence markers. Papillomas from these mice were indistinguishable histologically from each other (Figure 2E) and showed similar levels of phospho-ERK (Figure 2F), indicating that mSnoN did not affect Ras signalling. However, although no senescence associated β-gal (SA-β-gal) staining could be detected in WT papillomas, they were readily observed in m/m papillomas (Figure 2G). In addition, the expression of another senescence maker, p19ARF, was also detected in the nucleolus in m/m tumours but not in +/+ tumours (Figure 2H). Consistent with this, p53, but not p16INK4a, expression was significantly increased in m/m tumours (Figure 2F). Thus, expression of mSnoN induces senescence in tumour tissues, and this may contribute to the resistance of the m/m mice to chemical carcinogen-induced tumourigenesis.
MEF-expressing mutant SnoN showed premature senescence
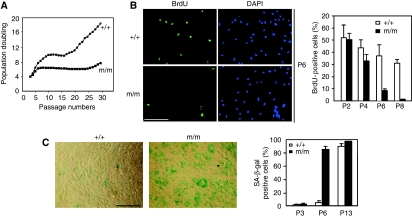
To determine the molecular mechanism underlying the senescence phenotype, we isolated primary MEFs from WT and m/m mice. During the initial passages, the m/m MEFs were indistinguishable from the WT MEFs in morphology and growth under the normal serum concentration. When cultured by a 3T9 protocol, WT MEFs gradually lost their growth capability and entered senescence at around passage 10 (P10) (Todaro and Green, 1963; Sherr and DePinho, 2000). The cells then remained in senescence for another eight passages until a small group of cells became spontaneously immortalized at around P18 (Figure 3A). The m/m MEFs proliferated and incorporated BrdU at a rate similar to WT MEFs during the first three passages (Figure 3A and B). BrdU incorporation by the m/m MEFs decreased gradually after P4, and by P6 more than 80% of the cell population was negative for BrdU (Figure 3B). At this passage, more than 80% of m/m MEFs entered senescence prematurely (Figure 3A) and were positive for SA-β-gal staining (Figure 3C). These m/m MEFs stayed in senescence for a longer duration and immortalized only after P25. This premature senescence was observed for all independently established m/m MEF lines (data not shown).
Figure 3.
MEFs from the snoN knock-in mice showed premature senescence. (A) WT or m/m MEFs were passaged according to the 3T9 protocol. Population doubling was measured by cell counting every 3 days. (B) BrdU incorporation. WT or m/m MEFs, at P6, were incubated in a medium containing BrdU followed by staining with anti-BrdU (scale bar: 40 μm). Quantification of BrdU-positive cells is shown in the graph. (C) SA-β-gal staining. WT or m/m MEFs, at P6, were subjected to staining with SA-β-gal (scale bar: 100 μm). Quantification is shown in the graph.
Premature senescence is caused by the elevated SnoN expression
The premature senescence found in m/m MEFs could be attributed to either the increased Smad activity due to the lack of repression by mSnoN or to the elevated expression of mSnoN. Previously, we have shown that interaction of SnoN with R-Smads results in polyubiquitination and degradation of SnoN (Stroschein et al, 1999, 2001; Sun et al, 1999; Bonni et al, 2001; Levy et al, 2007; Nagano et al, 2007). An mSnoN defective in Smad binding is not polyubiquitinated (Stroschein et al, 2001) and as a result, accumulated to a higher level in m/m MEFs at P6 (Figure 4A).
Figure 4.
Elevated SnoN expression is responsible for premature senescence of m/m MEFs. (A) SnoN expression is elevated in m/m MEFs. Total cell lysates from WT and m/m MEFs were subjected to western blotting with anti-SnoN. Tubulin was used as a loading control. (B) _snoN_−/− MEFs do not undergo premature senescence. MEFs prepared from _snoN_−/− or WT mice were passaged according to the 3T9 protocol. SA-β-gal staining was carried out on cells at P6 and P13 (scale bar: 100 μm). (C) A siRNA mixture targeting both Smad2 and Smad3 was transfected into m/m MEFs at P4. Levels of Smad2, Smad3 and Smad7 were assessed by western blotting 48 h after the transfection. (D) Reducing Smad signalling does not affect premature senescence of m/m MEFs. WT or m/m MEFs, expressing the siRNA mixture, were stained with SA-β-gal at P6 (scale bar: 100 μm). (E) Reducing Smad3 expression by shRNA. The expression of Smad3 and Smad7 was measured by western blotting. (F) Senescence of MEFs expressing shSmad3 was assessed by SA-β-gal staining at P6 (scale bar: 100 μm). (G) shRNA targeting SnoN or HA–SnoN cDNA in retroviral vectors was introduced into m/m MEFs. (H) SnoN expression was evaluated by western blotting, and senescence of these cells was evaluated by SA-β-gal staining at P6 (scale bar: 100 μm).
To determine whether elevated Smad activity might be responsible for the observed premature senescence, we first prepared MEFs from snoN knockout (−/−) embryos and subjected them to the 3T9 protocol. The −/− MEFs also show enhanced Smad signalling but do not express the SnoN protein. Interestingly, −/− MEFs did not show premature senescence at P6 (Figure 4B), rather, they showed a slightly reduced senescence compared with WT MEFs even at P13. The lack of premature senescence in the −/− MEFs is not due to any defect in the senescence machinery because they continue to respond to UV- and TGF-β-induced senescence (data not shown). Thus, elevated Smad signalling is not sufficient to cause premature senescence.
To directly confirm that increased Smad signalling does not induce premature senescence, a siRNA mixture, targeting both mouse Smad2 and Smad3, was introduced into WT or m/m MEFs at P4 through transfection. The transfected cells were then examined for senescence at P6. As shown in Figure 4C, the siRNA mixture effectively reduced the expression of Smad2 and partially decreased the level of Smad3. Consistent with this, the expression of Smad7, a transcription target of Smad2 and Smad3, was significantly reduced (Figure 4C). However, premature senescence in m/m MEFs continued to occur efficiently (Figure 4D), suggesting that elevated Smad activity is not responsible for premature senescence. Similarly, introduction of an shRNA specific for mouse Smad3 into m/m MEFs through retroviral infection effectively reduced the expression of Smad3 and its target Smad7 (Figure 4E), but did not affect the senescence of these cells at P6 (Figure 4F). Finally, treatment of m/m MEFs with SB431542, a pharmacological inhibitor of type I TGF-β receptor, effectively blocked Smad3 phosphorylation and activation, but did not affect premature senescence (Supplementary Figure 1). Taken together, our data suggested that elevated Smad activity in m/m MEFs is not responsible for the observed premature senescence.
To determine whether the elevated mSnoN expression is responsible for premature senescence, we introduced shRNA for murine SnoN into m/m MEFs. As shown in Figure 4G and H, when SnoN level was reduced by more than 80%, premature senescence was blocked significantly with <10% SA-β-gal-positive cells. This suggests that senescence of MEFs is sensitive to the expression level of SnoN. Consistent with this, ectopic overexpression of WT SnoN or mSnoN in WT MEFs induced premature senescence (Figures 4G–H and 6I). Thus, elevated SnoN expression in m/m MEFs, independent of its ability to antagonize Smad signalling, is responsible for the observed premature senescence.
p53 upregulation is required for SnoN-induced premature senescence
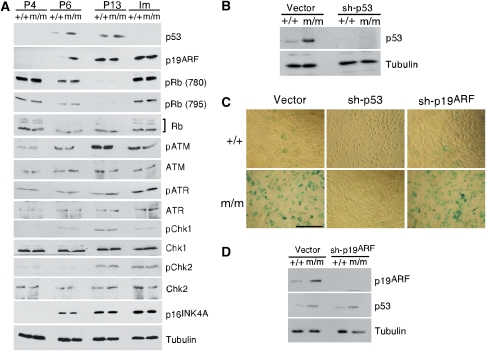
Stress-induced senescence of human fibroblasts is controlled by both p53 and Rb pathways but that of mouse fibroblasts is mostly regulated by the p53 pathway (Sharpless and DePinho, 2002; Wadhwa et al, 2004; Campisi, 2005; Rodier et al, 2007; Courtois-Cox et al, 2008). Inactivation of p53 either by homologous deletion (p53−/−) or by deleting p19ARF (p19ARF−/−) in MEFs bypasses senescence and results in spontaneous immortalization, but p16INK4A−/− MEFs undergo senescence in a manner same as WT MEFs (Sharpless et al, 2001). To determine which pathways are involved in SnoN-induced senescence, we compared the expression of p53 and p16INK4A at passages representing pre-senescence (P4), m/m senescence (P6), senescence for both (P13) and after immortalization (Im) between WT and m/m MEFs. At the pre-senescence stage, both proteins were maintained at an undetectable level. Interestingly, at P6 when most m/m MEFs had entered senescence prematurely, the level of p53 was significantly elevated in m/m MEFs but to a much lesser extent in WT MEFs, most of which had not entered senescence (Figure 5A). The elevated p53 expression was maintained through senescence and returned to background level on immortalization. Thus the profile of p53 expression correlated well with the senescence status of m/m and WT MEFs. In contrast, the level of p16INK4A did not change appreciably during this process, nor did Rb phosphorylation (at residues 780 and 795) and expression (Figure 5A). Thus, p53 seems to be the major regulator in SnoN-induced premature senescence. This is consistent with our earlier observation that p53, but not p16Ink4a, was upregulated in tumour samples derived from m/m mice (Figure 2F).
Figure 5.
p53 is required for the premature senescence of m/m MEFs. (A) p53 and p19ARF are upregulated on premature senescence of m/m MEFs. Expression levels of a variety of proteins were compared in WT and m/m MEFs at P4, P6, P13 and on immortalization (Im). This figure panel was assembled from multiple gels with identical samples. The bracket indicates total Rb. Tubulin was used as a loading control. (B) Reducing p53 expression by shRNA. p53 levels in MEFs expressing sh-p53 were measured by western blotting. (C) Senescence of m/m MEFs expressing sh-p53 or sh-p19ARF was determined by SA-β-gal staining at P6 (scale bar: 100 μm). (D) p19ARF and p53 levels in MEFs expressing sh-p19ARF were measured by western blotting.
Increase in p53 level is not due to an increase in transcription (data not shown), but most likely is due to the stabilization of p53 protein. p53 stability can be regulated by many proteins, such as p19ARF, ATM/ATR and Chk1/Chk2 kinases (Efeyan and Serrano, 2007; Rodier et al, 2007). Although activities of ATM/ATR and Chk1/Chk2, as represented by the phosphorylation of these proteins, did not change in either m/m or WT MEFs, the expression of p19ARF was elevated as soon as m/m MEFs entered senescence at P6 and remained high thereafter (Figure 5A). This increase in p19ARF expression occurred at the transcription level, as detected using RT–PCR (data not shown).
To determine whether increased p19ARF and p53 levels are necessary for SnoN-induced senescence, we introduced shRNA for p19ARF or p53 into m/m MEFs through retroviral infection at P3 and examined senescence at P6. An efficient knock down of p53 (Figure 5B) effectively blocked premature senescence of m/m MEFs at P6 (Figure 5C), confirming that p53 is indeed a critical mediator of SnoN-induced senescence. Surprisingly, reducing p19ARF expression by shRNA did not have any effect on premature senescence (Figure 5C) or p53 expression even though more than 90% of p19ARF was eliminated by the shRNA (Figure 5D). This indicates that although p19ARF is upregulated in the m/m MEFs, it is not primarily responsible for the increased p53 expression or the premature senescence of the cells. The upregulation of p19ARF expression most probably occurred as a consequence but not as a cause of premature senescence.
Taken together, our studies have established p53 as a critical mediator of SnoN-induced premature senescence.
PML mediates SnoN-induced p53 stabilization and premature senescence
To determine how SnoN induces upregulation of p53, we examined the intracellular localization of SnoN before and during senescence. In pre-senescence WT and m/m MEFs, endogenous SnoN was distributed throughout the nucleus (Figure 6A: +/+ MEFs at P3 and P6, and m/m MEFs at P3). However, upon entry into senescence, SnoN was observed to be accumulated in numerous small nuclear speckles (Figure 6A: +/+ MEFs at P13 or m/m MEFs at P6 and P13). These senescence-associated SnoN speckles are reminiscent of promyelocytic leukaemia (PML) nuclear bodies in size and morphology. Indeed, using markers of various nuclear domains, SnoN was found to co-localize only with the PML protein in PML bodies (Figure 6A), but not in heterochromatic foci or DNA damage/repair foci (data not shown). Localization of SnoN in PML bodies occurred in both WT and m/m MEFs during senescence, but not before senescence or after immortalization (Figure 6A and data not shown). Finally, in WT MEFs that undergo premature senescence due to overexpression of SnoN, SnoN also localized in PML bodies (Figure 6B). Thus, the accumulation of SnoN in PML nuclear bodies seems to correlate with the senescence status of MEFs.
Figure 6.
Interaction between SnoN and PML is required for premature senescence. (A) Endogenous SnoN (red) and PML (green) were stained with anti-SnoN or anti-PML antibodies, respectively, in WT and m/m MEFs at different passages (scale bar: 10 μm). Nuclei were stained using DAPI (Blue). (B) Flag–SnoN was introduced into WT MEFs and stained with anti-Flag. Endogenous PML was stained with anti-PML (scale bar: 10 μm). (C) Reduction of PML expression by shRNA. The levels of PML and p53 were measured by western blotting. (D) PML is required for SnoN-induced premature senescence. SA-β-gal staining was carried out at P6 in m/m MEFs expressing sh-PML (scale bar: 100 μm). (E) SnoN physically associates with PML. Interaction of His-PML with Flag–SnoN (WT or mutant) in co-transfected 293T cells was measured by immunoprecipitation with anti-Flag antibody followed by western blotting with anti-PML antibody. PML levels in the lysate were blotted using anti-PML antibody. (F) Interaction between endogenous SnoN and PML at P6 in MEFs was measured by immunoprecipitation with anti-PML antibody followed by western blotting with anti-SnoN antibody. Total PML and SnoN levels were measured by western blotting. (G) Left: schematic drawing of WT and mutant SnoN proteins. Right: interaction of Flag–SnoN with His-PML was measured as described in (E). (H) Interaction of SnoN with PML is required for SnoN-induced premature senescence. Flag-tagged WT or mutant SnoN was introduced into WT MEFs at P3 through retroviral infection. (I) At P7, expression of p53, PML and SnoN were evaluated through western blotting and senescence was measured through SA-β-gal staining (scale bar: 100 μm). (J). SnoNΔ322–366 fails to co-localize with PML. Immunofluorescence staining was performed with anti-Flag (SnoN) and anti-PML antibodies (scale bar: 10 μm).
PML nuclear bodies are discrete nuclear domains consisting of large number of proteins and have been linked to many fundamental cellular processes, including transcriptional control, anti-viral response, DNA repair, apoptosis and senescence (Pearson and Pelicci, 2001; Salomoni and Pandolfi, 2002). The PML tumour suppressor protein is essential for the formation of PML nuclear bodies and for the recruitment of diverse proteins to the nuclear domain (Goddard et al, 1995; Seeler and Dejean, 1999; Dellaire et al, 2006; Shen et al, 2006; Bernardi and Pandolfi, 2007). Of particular interest is its role in p53 stabilization in response to different stimuli. PML bodies may serve as scaffolds to increase the local concentrations of factors implicated in p53 stabilization by enhancing phosphorylation and acetylation, or regulating the ubiquitination and proteasome-dependent degradation of p53 (Pearson et al, 2000; Pearson and Pelicci, 2001; Bode and Dong, 2004; Takahashi et al, 2004). Loss of PML allows MEFs to bypass senescence due to the lack of p53 activation (Ferbeyre et al, 2000; Pearson et al, 2000; Marcotte and Wang, 2002; de Stanchina et al, 2004). Thus, it is possible that SnoN localizes to PML nuclear bodies to allow the stabilization of p53, leading to premature senescence.
To test whether PML is a critical intermediate for SnoN-induced p53 stabilization and premature senescence, we introduced shRNA for PML into m/m MEFs through retroviral infection. Interestingly, efficient reduction of PML expression (Figure 6C) completely inhibited premature senescence (Figure 6D) and prevented the stabilization of p53 in m/m MEFs (Figure 6C). Thus, PML is required for SnoN-induced p53 stabilization and premature senescence.
SnoN physically interacts with PML to promote premature senescence
As SnoN is localized in PML nuclear bodies during senescence, we hypothesized that PML may physically interact with SnoN to recruit it to the PML bodies. To test this, Flag–SnoN was co-transfected with His-PML into 293T cells and isolated by immunoprecipitation with anti-Flag antibody. The PML protein that is associated with SnoN was then detected by western blotting with anti-PML. As shown in Figure 6E, PML associated with both WT SnoN and mSnoN to the same extent, indicating that SnoN interacts with PML independently of its ability to antagonize the Smad proteins. This interaction also occurred at the endogenous level in m/m MEFs at P6 as endogenous SnoN was found to co-precipitate with PML efficiently (Figure 6F). In WT MEFs, endogenous WT SnoN also interacted with PML but to a much lesser extent, probablybecause of relatively lower level of SnoN in WT MEFs (Figure 6F).
We next mapped the domain in SnoN that is required for interaction with PML. Various SnoN deletion mutants were co-transfected into 293T cells with PML, and their ability to interact with PML was evaluated by co-immunoprecipitation assay. As shown in Figure 6G, 366 amino-terminal amino-acids of SnoN, encoded by exon 1, were sufficient for binding to PML, whereas the carboxy-terminal 367–684 fragment failed to bind. Further deletional analysis identified a short region immediately after the SAND-like domain between residues 322–366 as being critical for binding to PML. Deletion of residues 322–366 (SnoNΔ322–366) abolished the SnoN–PML interaction (Figure 6G), but did not affect the binding of SnoN to Smad4 (data not shown) nor its ability to repress TGF-β-induced transcription (data not shown). This suggests that the deletion did not disrupt the structural integrity of SnoN but specifically blocked the interaction between SnoN and PML. More importantly, binding of SnoN to PML is independent of the SnoN–Smad interaction and does not interfere with the ability of SnoN to antagonize Smad signalling.
Next we examined the ability of this deletion mutant to be recruited to PML bodies and to induce p53 stabilization and premature senescence. As shown earlier, ectopic expression of WT SnoN in WT MEFs resulted in the stabilization of p53, premature senescence and localization of SnoN in PML bodies. In contrast, ectopic expression of SnoNΔ322–266 did not lead to p53 stabilization and premature senescence (Figure 6I). Furthermore, this mutant SnoNΔ322–366 was distributed throughout the nucleocytoplasm and failed to accumulate in PML bodies (Figure 6J). These results strongly indicate that the interaction between SnoN and PML is critical for the recruitment of SnoN to PML bodies and the subsequent p53 stabilization and premature senescence.
PML expression is upregulated in m/m MEFs
During the course of our investigation, we noticed that m/m MEFs seemed to express a higher level of PML than WT MEFs. This prompted us to compare the expression of PML between WT and m/m MEFs. Using RT–PCR and western blotting, PML mRNA and protein levels were both increased in the m/m MEFs (Figure 7A and B). Interestingly, SnoN is necessary for the upregulation of PML in m/m MEFs. Knocking down SnoN by shRNA significantly decreased the level of PML (Figure 7B).
Figure 7.
SnoN upregulates PML expression to mediate p53 stabilization. PML expression is upregulated in the m/m MEFs as measured by (A) RT–PCR and (B) western blotting at P6. (B) SnoN is required for PML upregulation. The levels of PML and SnoN in control or shSnoN-expressing cells were determined by western blotting. (C) SnoN interacts with both PML and p53 in senescent cells. The levels of SnoN, PML and p53 in nuclear extracts prepared from WT or m/m MEFs at P1, P6 and P13 were measured by western blotting. Interaction of SnoN with PML and p53 was examined by immunoprecipitation with anti-SnoN antibody followed by western blotting with anti-PML or anti-p53 antibody. (D) SnoN and p53 co-localized in senescence cells. Endogenous SnoN (red) and p53 (green) were stained with anti-SnoN or anti-p53, respectively, in MEFs at P13. (E) SnoN level is not affected by PML. The levels of SnoN, PML and p53 in MEFs expressing sh-PML were determined by western blotting. (F) Model for the pro-oncogenic and anti-oncogenic functions of SnoN.
Relationship between SnoN, PML and p53 during senescence
Despite a higher level of SnoN and PML proteins in m/m MEFs, it still takes 6 passages for the cells to enter senescence. Consistent with this, the level of p53 did not peak until P6 in m/m MEFs (Figures 4A and 7C). To investigate the cause of this delay in entering senescence by m/m MEFs, we examined the expression levels of endogenous SnoN, PML and p53 as well as the interactions among these proteins in WT and m/m MEFs at P1, P6 and P13. As shown in Figures 4A and 7C, SnoN expression was observed to be at a relatively low level in m/m MEFs at early passages (P1–P3), and increased with the increase in number of passages and reached a high level at P6. In correlation with this increase in mSnoN expression, PML and p53 levels were also observed to be elevated progressively and reached maximal level at P6 in m/m MEFs. In WT MEFs, the levels of PML and p53 at P6 were still low and did not peak until P13. Interestingly, PML and p53 levels in WT MEFs at P13 were comparable with that in m/m MEFs at P6, suggesting that PML and p53 may need to accumulate to certain levels to induce cells to enter senescence. In the cells that entered senescence, SnoN and p53 were found to form a complex and co-localize in PML nuclear bodies as shown by both co-immunoprecipitation assay (Figure 7C) and by immunofluorescence staining (Figure 7D).
To determine whether the accumulation of mSnoN is required for p53 expression, we examined p53 levels in SnoN knockdown cells by western blotting (Figure 7B). When SnoN level was reduced, accumulation of PML and p53 in m/m MEFs stalled, indicating that elevation in SnoN expression precedes PML and p53 accumulation. In contrast, when PML expression was reduced by shRNA, SnoN accumulation was not affected, suggesting that SnoN acts upstream of PML (Figure 7E).
As mSnoN accumulation is a pre-requisite for the elevation of PML and p53 expression, and the peak mSnoN level is not reached until P6, senescence of m/m MEFs was not observed until P6. This gradual accumulation of mSnoN is, therefore, the rate-limiting step in inducing premature senescence and explains why it takes six passages for m/m MEFs to enter senescence. Our data so far suggest a model for how SnoN induces premature senescence (Figure 7F). In cells expressing high levels of SnoN, SnoN not only antagonizes TGF-β signalling, but also upregulates the expression of PML and more importantly, interacts directly with PML. This interaction enables SnoN to be recruited to PML nuclear bodies in which it stabilizes p53 and/or other proteins, leading to premature senescence. This ability of SnoN to induce senescence suggests an anti-oncogenic function of SnoN.
High levels of SnoN inhibit transformation of MEFs by oncogenes
As m/m MEFs show enhanced senescence, we wondered whether these cells were more resistant to transformation by oncogenes. It is known that in MEFs, although overexpression of an individual oncogene induces senescence (Serrano et al, 1997; Di Micco et al, 2007), co-expression with another oncogene results in transformation. To assess the transforming ability of m/m MEFs, we introduced a constitutively active Ras(Q61L) or c-Myc, either alone or together into WT or m/m MEFs through retroviral infection. After a brief drug selection, the infected cells were evaluated for the ability to undergo anchorage-independent growth through a soft-agar assay. When Ras(Q61L) was introduced individually into WT MEFs, it triggered senescence (Figure 8B) but not transformation (Figure 8A). However, overexpression of both Ras(Q61L) and c-Myc together resulted in oncogenic transformation (Figure 8A). In m/m MEFs that already overexpressed mSnoN, introduction of Ras(Q61L) or c-Myc alone did not result in transformation (Figure 8A and data not shown). However, Ras(Q61L) triggered a slightly more severe senescence (Figure 8B), suggesting that mSnoN did not function as an oncogene. In the presence of both Ras(Q61L) and c-Myc, m/m MEFs showed a much reduced level of transformation (Figure 8A and enlarged figures in Supplementary Figure 2). Approximately 30% of these cells still showed senescence, whereas none of the WT MEFs expressing Ras(Q61L) and c-Myc was senescent (Figure 8B). This suggests that mSnoN inhibits oncogene-induced transformation of MEFs. To confirm that this inhibition of transformation is due to the high-level expression of mSnoN but not due to the enhanced Smad activity, shRNA for SnoN or Smad3 was introduced into MEFs along with Ras(Q61L) and c-Myc. Consistent with the role of SnoN in supporting premature senescence, reducing SnoN level in m/m MEFs restored the high level of soft-agar colony formation, whereas reducing Smad3 had no effect (Figure 8A). This ability to inhibit oncogenic transformation is not unique to mSnoN but is also shown by WT SnoN. Overexpression of WT SnoN together with Ras(Q61L) and Myc in WT or m/m MEFs significantly reduced the colony formation (Figure 8A). Taken together, these results suggest that SnoN displays anti-tumourigenic activity and inhibits oncogene-induced transformation possibly through inducing senescence.
Figure 8.
Oncogenic transformation of MEFs. (A) SnoN inhibits the transformation of MEFs. WT or m/m MEFs at P3 were first infected with retroviruses expressing shRNA or HA–SnoN, and infected again with retroviruses expressing H-Ras (Q61L) and c-Myc, either individually or together, and subjected to a soft-agar assay (scale bar: 40 μm). The colonies were visualized by MTT staining and quantified (shown in the graph to the right). (B) m/m MEFs enhanced oncogene-induced senescence. WT or m/m MEFs expressing H-Ras (Q61L) either alone, or together with c-Myc or SnoNΔ322–366 were subjected to SA-β-gal staining (scale bar: 100 μm). (C) Disruption of the SnoN–PML interaction turns SnoN into a bona fide oncogene. WT MEFs at P3 were sequentially infected with retroviruses expressing SnoN (WT or SnoNΔ322–366) first and later with H-Ras (Q61L) and c-Myc either individually or together, and subjected to a soft-agar assay (scale bar: 40 μm). (D) Inactivation of p53 abolishes the tumour suppressor activity of SnoN. WT or p53−/− MEFs expressing either WT, mutant SnoN, various siRNA or shRNA were subjected to a soft-agar assay. The soft-agar colonies were visualized by MTT (left; scale bar: 40 μm) and quantified. (* enlarged figures are showed in Supplementary Figure 2).
Blocking the SnoN–PML interaction or abolishing p53 eliminates the anti-oncogenic activity of SnoN
Our results predict that the tumour suppressor activity of SnoN is dependent on the SnoN–PML interaction and on p53. Thus disruption of the SnoN–PML interaction or elimination of p53 should abolish the tumour suppressor activity of SnoN and turn it into a bona fide oncogene. To test this, we first examined the activity of SnoNΔ322–366 that is defective in interaction with PML in the soft-agar colony assay. Indeed, although WT SnoN failed to transform WT MEFs with active Ras oncogene, SnoNΔ322–366 and Ras readily induced anchorage-independent growth as effectively as Ras and Myc (Figure 8C and enlarged panels in Supplementary Figure 2). Interestingly, co-expression of SnoNΔ322–366 and Myc did not result in transformation (Figure 8C).
We also examined the ability of WT and mSnoN to induce transformation of p53−/− MEFs. In these cells, expression of the active Ras alone is sufficient for transformation (Figure 8D and enlarged panels in Supplementary Figure 2). Interestingly, expression of WT SnoN or SnoNΔ322–366 alone was sufficient for transformation of p53−/− MEFs (Figure 8D), confirming that they function as oncogenes. In contrast, mSnoN failed to induce transformation of p53−/− MEFs (Figure 8D), most probably because it cannot repress Smad signalling and therefore is defective in the pro-oncogenic branch. Consistent with this idea that repressing Smad activity is critical for the transformation of MEF by SnoN, knocking down both Smad2 and Smad3 through siRNA, or Smad3 alone by shRNA was sufficient to induce transformation of MEF when p53 is absent (Figure 8D). Although the extent of transformation induced by reducing Smads was not as strong as overexpression of WT SnoN or SnoNΔ322–366, most probably due to the incomplete knock down of Smads (Figure 4C), it nevertheless supports the idea that antagonizing Smad activity is critical for the transforming activity of SnoN. Taken together, these results indicate that SnoN possesses pro-oncogenic activity through antagonizing Smad proteins and anti-oncogenic activity through PML and p53 proteins. Disruption of the PML–p53 pathway is sufficient to inactivate the tumour suppressor activity of SnoN but preserves the oncogenic function of SnoN.
Discussion
Senescence has been recognized as an anti-tumourigenic mechanism in vitro and in vivo to prevent the accumulation of harmful oncogenic mutations. Here we have shown that elevated SnoN triggers premature senescence by binding to and co-localizing with the PML protein in PML nuclear bodies. This interaction results in the stabilization of p53 and subsequent premature senescence. The ability of SnoN to induce senescence is independent of its ability to bind to and antagonize Smad proteins but is dependent on high levels of SnoN expression. The premature senescence triggered by SnoN does not involve DNA damage responses but is reminiscent of tumour suppressor-induced senescence. Indeed, MEFs with high levels of SnoN are less susceptible to transformation by oncogenes. More importantly, knock-in mice are more resistant to carcinogen-induced skin tumourigenesis possibly because of the induction of senescence in the epidermis. Thus, we have elucidated a new tumour suppressor activity of SnoN by inducing senescence and have revealed the molecular pathway for this activity for the first time.
Although the induction of senescence by SnoN only occurs under conditions of SnoN overexpression, it is nevertheless physiologically relevant. In many human cancer cells, SnoN expression is highly elevated because of the amplification of the 3q26 amplicon, increased transcription of the snoN gene or inhibition of SnoN degradation to a level similar to that found in m/m MEFs. As SnoN has been classified as an oncogene in the past, one could argue that SnoN-induced senescence is a typical case of oncogene-induced senescence (Serrano et al, 1997; Di Micco et al, 2007). However, SnoN induced senescence is different from oncogene-induced senescence in several important aspects. First, senescence induced by oncogenes such as hyperactive Ras requires activation of the INK4A/ARF locus (Serrano et al, 1997; Sharpless, 2005; Courtois-Cox et al, 2008). However, neither p16INK4A nor p19ARF is critical for SnoN-induced senescence. Second, the DNA damage pathway is a critical mediator of oncogene-induced senescence (Bartkova et al, 2006; Di Micco et al, 2006; Mallette and Ferbeyre, 2007). Oncogenes such as activated Ras cause unbalanced DNA replication and initiate a classic p53-dependent DNA-damage response pathway. However, in SnoN-induced senescence, activation of DNA damage or check point pathways is not detected. Third, oncogenes co-operate with each other to induce transformation of MEFs. However, not only did the expression of SnoN fail to induce transformation in the presence of an active Ras or Myc, it even inhibited the transformation induced by Ras and Myc. Finally, data from mouse skin carcinogenesis model confirmed that the mSnoN functions as a tumour suppressor in vivo. Therefore, SnoN can show a tumour suppressor activity by inducing cellular senescence.
The idea that SnoN possesses both pro-oncogenic and anti-oncogenic activities is consistent with earlier observations of SnoN expression patterns in human cancer tissues and cell lines (Villanacci et al, 2008). The pro-oncogenic activity and tumour suppressor activity of SnoN are clearly mediated through two separate pathways (Figure 7F), with the pro-oncogenic activity depending on the antagonism of the TGF-β/Smad pathway (Zhu et al, 2006) and the tumour suppressor activity relying on PML and p53. Selective inactivation of the PML/p53 branch allows the oncogenic activity of SnoN to be fully manifested as in the case of SnoNΔ322–366 or the transformation of p53−/− MEFs, whereas mutation of the Smad binding activity of SnoN exposes the tumour suppressor activity of SnoN. The co-existence of both pro-oncogenic and anti-oncogenic activities in one protein is not unique to SnoN. The Ndy1 protein, a Jumonji C domain-containing histone demethylase, also harbours both pro-oncogenic activity by inhibiting senescence, and tumour suppressor activity by maintaining genomic integrity and inhibition of cell proliferation (Suzuki et al, 2006; Frescas et al, 2007; Pfau et al, 2008). This emerging group of proteins with both pro-oncogenic and anti-oncogenic activities highlights the complexity of cellular events accompanying malignant progression.
Tumour suppressors are often inactivated or deleted during malignant progression. If SnoN contains anti-tumourigenic activity, why is it upregulated in many human cancer cells? We speculate that at early stages of tumourigenesis, tumour cells may upregulate SnoN expression in an attempt to halt tumour growth by inducing senesence. Thus, SnoN upregulation may initially serve as a barrier for malignant progression. To overcome this barrier, tumour cells may specifically inactivate the SnoN senescence pathway by downregulating p53 or PML, leaving cells with high levels of SnoN, but do not undergo senescence. These high levels of SnoN may then promote tumour growth through its pro-oncogenic activity. Thus, high levels of SnoN expression may be the outcome of a complex evolving process during tumourigenesis. This model also implies that it is more advantageous for the cancer cells to maintain a high level of SnoN expression while inactivating its anti-oncogenic pathway at downstream points than to delete it. Indeed, our results that SnoN potently induces oncogenic transformation of p53−/− MEFs and that SnoNΔ322–366 functions as an oncogene support this model. Future experiments will determine whether human cancer cells with high levels of SnoN expression also harbour mutations in downstream components of the SnoN senescence pathway. This may have important implications in potential targeting of SnoN in human cancer.
Materials and methods
Mice
To generate knock-in mice, the targeting vector was introduced into the E14 mouse embryonic stem (ES) cells derived from 129P2/OlaHsd mice and injected into the blastocysts of C57BL/6 background. The resulting chimeras were crossbred with a CMV (cytomegalovirus immediate-early promoter)–cre transgenic mice to generate the knock-in mice and backcrossed to C57BL/6 for additional five generations. SnoN knockout mice in C57BL/6 background were obtained from Dr S Pearson-White. p53 knockout mice were obtained from The Jackson Laboratory. All studies conducted on the animals were approved and confirmed by the Animal Care and Use Committee at UC Berkeley.
Cells, antibodies and constructs
293T cells and Hep3B cells were maintained as described before by Zhu et al (2005). Anti-Flag antibody was purchased from Sigma; antibodies against p53 (ab26) and p19ARF (ab80) from Abcam; anti-PML antibody (Mab 3738) from Chemicon; antibodies against phosphorylated ATM, ATR, Chk1, Chk2 and Rb from Cell Signaling Technology; antibodies against p16INK4A, Smad3 (FL245), Smad7 (H79), Chk1 (FL478) and Tubulin from Santa Cruz Biotechnology.
Primary MEFs were prepared from E13.5 day embryos as described previously by Serrano et al (1997) and maintained in DMEM medium with 10% FBS. A standard 3T9 protocol was used to compare population doubling (Blasco et al, 1997) shRNAs in pSuper-Retro-puro were introduced into MEFs through retroviral infection as described previously by Zhu et al (2006). Mixture of siRNA duplex targeting mouse Smad2 and Smad3 was purchased from Dharmacon and transfected into MEFs using Nucleofector system (Amaxa). The sequences of various shRNAs are: sh-Smad3: 5′-ggccatcaccacgcagaac-3′; sh-SnoN: 5′-ctccattctgcagaggaag-3′; sh-p19ARF: 5′-caccggaatcctggaccag-3′; sh-p53: 5′-gtactctcctcccctcaat-3′; and sh-PML: 5′-cccacttagaagacaggac-3′.
RT–PCR
Total RNA extraction, cDNA preparation and PCR were carried out using the standard protocol. The primers used to amplify different genes by PCR are: p53: 5′-actgccatggaggagtcac-3′/5′-tcagtctgagtcaggcccc-3′; PML: 5′-agcaggaggcttctcagacagt-3′/5′-cttgatgatcttcctggagcaa-3′; and p19ARF: 5′-gttttcttggtgaagttcgtgc-3′/5′-tcatcacctggtccaggattc-3′.
Retroviral infection
cDNAs or shRNAs in retroviral vectors were transfected into the Phoenix-E cells to generate high titre of viral supernatants, which were used subsequently to infect the MEFs at P3. After 48 h, infected cells were selected by culturing in the presence of 2 μg/ml puromycin. Pools of puromycin-resistant cells were analysed in various experiments. For two rounds of infection, puromycin-resistant cells were infected again with retroviruses that express neomycin-resistant genes. At 48 h after infection, doubly infected cells were selected on 600 μg/ml G418 for 4 days.
Transfection, co-immunoprecipitation and luciferase assays
293T, Hep3B or MEF cells were transiently transfected using lipofectamine plus (Invitrogen). Co-immunoprecipitation assay was carried out as described previously by Zhu et al (2005). Luciferase assay was carried out as described by Zhu et al (2005).
SA-_β_-gal staining
MEFs were fixed and stained for the expression of the β-galactosidase activity at pH 6.0 (senescence-associated β-gal) according to the manufacturer's instruction (Senescence detection kit, Calbiochem). To detect senescence in mouse tissues, frozen tissue sections of 6-μm thickness were stained for SA-β-gal.
BrdU incorporation assay
Cells were incubated with medium containing 0.1 mg/ml BrdU (Roche) for 6 h. After fixation in methanol, cells were incubated with anti-BrdU working solution (Roche) for 30 min at 37°C, stained with Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) and visualized by fluorescent microscopy.
Immunofluorescence
MEFs were grown on glass cover-slips, fixed, permeabilized with 0.1% Triton X-100 for 5 min, stained with the appropriate antibodies and visualized using a Zeiss 510 confocal microscope. Cell nuclei were detected by 4,6-diamidino-2-phenylindole (DAPI) staining. Immunofluorescence for frozen tissue sections was carried out using the same procedure except that the samples were permeabilized with 0.5% Triton X-100 for 15 min.
Soft-agar assay
A total of 3 ml of growth medium containing 0.66% Bacto Agar was added to a 50-mm dish for the bottom agar layer. A total of 6 × 103 MEFs were re-suspended in 2 ml medium containing 0.4% agar and overlaid on the hardened bottom layer. A total of 2 ml of fresh medium containing 0.4% agar was added to the dish once a week for 4 weeks. The colonies were visualized by staining with 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Sigma) for 4 h at 37°C.
Skin tumourigenesis
The dorsal skins of 8-week-old mice were shaved and treated topically with 7,12-dimethylbenz(α)anthracene (DMBA; 100 μg/200 μl in acetone). One week later, 12-_O_-tetradecanoyl-phorbol-13-acetate (TPA; 20 nmol/200 μl in acetone) was applied twice weekly to the same area for 20 weeks (Balmain et al, 1988). Mice were euthanized at the end of 30 weeks. Surgically removed tumours were immediately snap-frozen in liquid nitrogen, and stored at −70°C. Alternatively, dissected skin and tumour samples were fixed in 10% neutral-buffered formalin at 4°C overnight, embedded in paraffin, cut into 6-μm thick sections, and stained using hematoxylin and eosin.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Review Process File
Acknowledgments
We thank Drs Ellen Solomon and Kun-Sang Chang for PML cDNA, Hitoshi Nishimura and Dragana Cado for generating the knock-in mice, and William Skarnes for advice. We are grateful to Dr Rosemary Akhurst and Marie Lee for assistance on carcinogen-induced skin tumourigenesis. We also thank Dr Judy Campisi for discussions. This study is supported funds from NIH RO1 CA101891, Philip Morris External Research Program grant 019016 to KL, and DOE BCRP pre-doctoral fellowship to DP.
Footnotes
The authors declare that they have no conflict of interest.
References
- Balmain A, Brown K, Akhurst RJ, Fee FM (1988) Molecular analysis of chemical carcinogenesis in the skin. Br J Cancer Suppl 9: 72–75 [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4: 793–805 [DOI] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL (2001) TGF-beta induces assembly of a Smad2–Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol 3: 587–595 [DOI] [PubMed] [Google Scholar]
- Boyer PL, Colmenares C, Stavnezer E, Hughes SH (1993) Sequence and biological activity of chicken snoN cDNA clones. Oncogene 8: 457–466 [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA (2005) Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 [DOI] [PubMed] [Google Scholar]
- Bravou V, Antonacopoulou A, Papadaki H, Floratou K, Stavropoulos M, Episkopou V, Petropoulou C, Kalofonos H (2009) TGF-beta repressors SnoN and Ski are implicated in human colorectal carcinogenesis. Cell Oncol 31: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buess M, Terracciano L, Reuter J, Ballabeni P, Boulay JL, Laffer U, Metzger U, Herrmann R, Rochlitz C (2004) Amplification of SKI is a prognostic marker in early colorectal cancer. Neoplasia 6: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J (2001) Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol 11: S27–S31 [DOI] [PubMed] [Google Scholar]
- Campisi J (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP (2005) Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia JA, Simms LA, Cozzi SJ, Young J, Jass JR, Walsh MD, Spring KJ, Leggett BA, Whitehall VL (2006) SnoN expression is differently regulated in microsatellite unstable compared with microsatellite stable colorectal cancers. BMC Cancer 6: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M (2007) Cellular senescence in cancer and aging. Cell 130: 223–233 [DOI] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cichowski K (2008) Many roads lead to oncogene-induced senescence. Oncogene 27: 2801–2809 [DOI] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW (2004) PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP (2006) Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 175: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, di Fagagna F (2007) Breaking news: high-speed race ends in arrest—how oncogenes induce senescence. Trends Cell Biol 17: 529–536 [DOI] [PubMed] [Google Scholar]
- Edmiston JS, Yeudall WA, Chung TD, Lebman DA (2005) Inability of transforming growth factor-beta to cause SnoN degradation leads to resistance to transforming growth factor-beta-induced growth arrest in esophageal cancer cells. Cancer Res 65: 4782–4788 [DOI] [PubMed] [Google Scholar]
- Efeyan A, Serrano M (2007) p53: guardian of the genome and policeman of the oncogenes. Cell Cycle 6: 1006–1010 [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R (2005) Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW (2000) PML is induced by oncogenic ras and promotes premature senescence. Genes Dev 14: 2015–2027 [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M (2007) JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450: 309–313 [DOI] [PubMed] [Google Scholar]
- Goddard AD, Yuan JQ, Fairbairn L, Dexter M, Borrow J, Kozak C, Solomon E (1995) Cloning of the murine homolog of the leukemia-associated PML gene. Mamm Genome 6: 732–737 [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M, Lee KL, Mavrakis KJ, Goggolidou P, Norris DP, Episkopou V (2009) Graded Smad2/3 activation is converted directly into levels of target gene expression in embryonic stem cells. PLoS ONE 4: e4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37: 614–636 [DOI] [PubMed] [Google Scholar]
- He J, Tegen SB, Krawitz AR, Martin GS, Luo K (2003) The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J Biol Chem 278: 30540–30547 [DOI] [PubMed] [Google Scholar]
- Imoto I, Pimkhaokham A, Fukuda Y, Yang ZQ, Shimada Y, Nomura N, Hirai H, Imamura M, Inazawa J (2001) SNO is a probable target for gene amplification at 3q26 in squamous-cell carcinomas of the esophagus. Biochem Biophys Res Commun 286: 559–565 [DOI] [PubMed] [Google Scholar]
- Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS (2007) Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol Cell Biol 27: 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Ferbeyre G (2007) The DNA damage signaling pathway connects oncogenic stress to cellular senescence. Cell Cycle 6: 1831–1836 [DOI] [PubMed] [Google Scholar]
- Marcotte R, Wang E (2002) Replicative senescence revisited. J Gerontol A Biol Sci Med Sci 57: B257–B269 [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS (2005) BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724 [DOI] [PubMed] [Google Scholar]
- Moustakas A, Souchelnytskyi S, Heldin CH (2001) Smad regulation in TGF-beta signal transduction. J Cell Sci 114: 4359–4369 [DOI] [PubMed] [Google Scholar]
- Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, Yuki K, Isogaya K, Saitoh M, Imamura T, Episkopou V, Miyazono K, Miyazawa K (2007) Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J Biol Chem 282: 20492–20501 [DOI] [PubMed] [Google Scholar]
- Nomura N, Sasamoto S, Ishii S, Date T, Matsui M, Ishizaki R (1989) Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res 17: 5489–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406: 207–210 [DOI] [PubMed] [Google Scholar]
- Pearson M, Pelicci PG (2001) PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene 20: 7250–7256 [DOI] [PubMed] [Google Scholar]
- Pearson-White S (1993) SnoI, a novel alternatively spliced isoform of the ski protooncogene homolog, sno. Nucleic Acids Res 21: 4632–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer T, Lyons GE, Kim S, Moreadith RW (1996) Cloning and characterization of the murine homolog of the sno proto-oncogene reveals a novel splice variant. Dev Dyn 205: 114–125 [DOI] [PubMed] [Google Scholar]
- Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN (2008) Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci USA 105: 1907–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel MC, Emery CS, Foulger R, Goberdhan DC, van den Heuvel M, Wilson C (2007) Drosophila SnoN modulates growth and patterning by antagonizing TGF-beta signalling. Mech Dev 124: 304–317 [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J, Bhaumik D (2007) Two faces of p53: aging and tumor suppression. Nucleic Acids Res 35: 7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP (2002) The role of PML in tumor suppression. Cell 108: 165–170 [DOI] [PubMed] [Google Scholar]
- Schilling SH, Hjelmeland AB, Rich JN, Wang X-F (2008) TGF-β: a multipotential cytokine. In The TGF-beta Family (Cold spring Harbor monograph), Derynck R, Miyazono K (eds), pp 45–78. Cold Spring Harbor, New York: Cold Spring Harbor Press [Google Scholar]
- Seeler JS, Dejean A (1999) The PML nuclear bodies: actors or extras? Curr Opin Genet Dev 9: 362–367 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Sharpless NE (2005) INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res 576: 22–38 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA (2001) Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413: 86–91 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA (2002) p53: good cop/bad cop. Cell 110: 9–12 [DOI] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP (2006) The mechanisms of PML-nuclear body formation. Mol Cell 24: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, DePinho RA (2000) Cellular senescence: mitotic clock or culture shock? Cell 102: 407–410 [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Shinagawa T, Dong H-D, Xu M, Maekawa T, Ishii S (2000) The sno gene, which encodes a component of the histone deacetylation complex, acts as a tumor suppressor in mice. EMBO J 19: 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein SL, Bonni S, Wrana JL, Luo K (2001) Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev 15: 2822–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K (1999) Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science 286: 771–774 [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA (1999) SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc Natl Acad Sci USA 96: 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG (2006) Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J 25: 3422–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H (2004) PML nuclear bodies and apoptosis. Oncogene 23: 2819–2824 [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanacci V, Bellone G, Battaglia E, Rossi E, Carbone A, Prati A, Verna C, Niola P, Morelli A, Grassini M, Bassotti G (2008) Ski/SnoN expression in the sequence metaplasia–dysplasia–adenocarcinoma of Barrett's esophagus. Hum Pathol 39: 403–409 [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Sugihara T, Taira K, Kaul SC (2004) The ARF-p53 senescence pathway in mouse and human cells. Histol Histopathol 19: 311–316 [DOI] [PubMed] [Google Scholar]
- Wan Y, Liu X, Kirschner MW (2001) The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol Cell 8: 1027–1039 [DOI] [PubMed] [Google Scholar]
- Wilkinson DS, Tsai WW, Schumacher MA, Barton MC (2008) Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming growth factor beta-mediated transcription repression. Mol Cell Biol 28: 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y (2002) Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell 111: 357–367 [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW (2007) Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lundin M, Ristimaki A, Heikkila P, Lundin J, Isola J, Joensuu H, Laiho M (2003) Ski-related novel protein N (SnoN), a negative controller of transforming growth factor-beta signaling, is a prognostic marker in estrogen receptor-positive breast carcinomas. Cancer Res 63: 5005–5010 [PubMed] [Google Scholar]
- Zhu Q, Krakowski AR, Dunham EE, Wang L, Bandyopadhyay A, Berdeaux R, Martin GS, Sun L, Luo K (2006) Dual role of SnoN in mammalian tumorigenesis. Mol Cell Biol 27: 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pearson-White S, Luo K (2005) Requirement for the SnoN oncoprotein in transforming growth factor beta-induced oncogenic transformation of fibroblast cells. Mol Cell Biol 25: 10731–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Review Process File