Hypoxia Up-regulates CD36 Expression and Function via Hypoxia-inducible Factor-1- and Phosphatidylinositol 3-Kinase-dependent Mechanisms (original) (raw)
Abstract
Neovascular and degenerative diseases of the eye are leading causes of impaired vision and blindness in the world. Hypoxia or reduced oxygen tension is considered central to the pathogenesis of these disorders. Although the CD36 scavenger receptor features prominently in ocular homeostasis and pathology, little is known regarding its modulation by hypoxia. Herein we investigated the role and regulation of CD36 by hypoxia and by the major hypoxia effector, hypoxia-inducible factor (HIF)-1. In vivo, hypoxia markedly induced CD36 mRNA in corneal and retinal tissue. Subsequent experiments on human retinal pigment epithelial cells revealed that hypoxia time-dependently increased CD36 mRNA, protein, and surface expression; these responses were reliant upon reactive oxygen species production. As an important novel finding, we demonstrate that hypoxic stimulation of CD36 is mediated by HIF-1; HIF-1α down-regulation abolished CD36 induction by both hypoxia and cobalt chloride. Sequence analysis of the human CD36 promoter region revealed a functional HIF-1 binding site. A luciferase reporter construct containing this promoter fragment was activated by hypoxia, whereas mutation at the HIF-1 consensus site decreased promoter activation. Specific binding of HIF-1 to this putative site in hypoxic cells was detected by a chromatin immunoprecipitation assay. Interestingly, inhibition of the phosphatidylinositol 3-kinase pathway blocked the hypoxia-dependent induction of CD36 expression and promoter activity. Functional ramifications of CD36 hypoxic accumulation were evinced by CD36-dependent increases in scavenging and anti-angiogenic activities. Together, our findings indicate a novel mechanism by which hypoxia induces CD36 expression via activation of HIF-1 and the phosphatidylinositol 3-kinase pathway.
Hypoxia, a reduction in cellular oxygen tension, is a key determinant of tissue pathology and survival during tumor development and ischemic diseases including retinopathies, myocardial infarction, and atherosclerosis. In response to hypoxia, mammalian cells express a variety of gene products important for erythropoiesis, angiogenesis, and glycolysis, thereby improving tissue oxygenation and facilitating metabolic demands (1, 2). These adaptive responses require the concerted activation of various transcription factors including hypoxia-inducible factor-1 (HIF-1),4 which is generally considered the master regulator of oxygen homeostasis (2–5). HIF-1 is a heterodimer comprised of an oxygen-regulated α subunit (HIF-1α) and the ubiquitous aryl hydrocarbon receptor nuclear translocator (ARNT or HIF-1β) (4, 6, 7). HIF-1α protein turnover in normoxia is very rapid due to the action of prolyl hydroxylases. These oxygen-dependent enzymes hydroxylate two conserved proline residues of HIF-1α, promoting binding of the Von Hippel-Lindau protein, ubiquitination, and subsequent proteosomal degradation. Under hypoxic conditions the prolyl hydroxylases are inhibited, thereby allowing stabilization and accumulation of the HIF-1α protein (4–6, 8, 9).
It has become increasingly evident that reactive oxygen species (ROS) and the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathway play a crucial role in regulating HIF-1 activity (10–15). Because hypoxia, ROS production, and angiogenesis typically occur concurrently, it is conceivable that scavenger receptors, specifically CD36, may contribute toward hypoxia-induced adaptive responses. As such, CD36 is an important participant of oxidative, angiogenic, and inflammatory processes (16, 17), particularly those pertaining to ocular neovascular disease (18–22).
CD36 is a heavily glycosylated transmembrane protein that belongs to an evolutionarily conserved family of scavenger receptors (16). This multifunctional receptor is expressed on the surface of microvascular endothelial cells, macrophages, dendritic cells, platelets, smooth muscle cells (16), and specialized epithelial cells of the cornea (18) and retina (23). CD36 functions in scavenger recognition of oxidized low density lipoproteins (oxLDL), apoptotic cells, and photoreceptor outer segments (POS) (16, 23, 24), in addition to having established roles as a fatty acid transporter (25, 26) and inhibitor of angiogenesis (16, 18, 27). Despite the multiple roles ascribed to CD36, limited data exists regarding its regulation by hypoxia, albeit several lines of evidence would support this inference.
It was recently described that CD36 expression was up-regulated during chronic lung hypoxia, with the identification of a putative hypoxia response element (HRE) in the Cd36 murine gene (28). Consistent with this, we have reported that inflammatory corneal neovascularization induces elevated levels of CD36 (18), whereas others have documented an up-regulation of CD36 during cerebral ischemia (29) and an attenuation of its expression and activity following antioxidant treatment (30, 31). Collectively, these studies led us to hypothesize that hypoxia modulates CD36 expression and function, with the objective of characterizing the mechanisms behind its regulation. Indeed our findings describe a novel mechanism in which hypoxia induces CD36 expression and promoter activity by stimulating ROS production in a HIF-1- and PI3K-dependent manner.
EXPERIMENTAL PROCEDURES
Compounds and Antibodies
Actinomycin D (ActD), allopurinol, angiotensin II, cobalt chloride (CoCl2), fluorescein isothiocyanate (FITC), myxothiazol, rotenone, isotype control antibodies anti-human IgA, anti-mouse IgG1, and anti-mouse IgM (Sigma); cycloheximide (CHX) and rapamycin (Calbiochem); monoclonal β-actin, CD36 monoclonal antibody (mAb) clone SMO, and anti-HIF-1α antibody (Abcam); CD36 rabbit polyclonal antibody, PI 3-kinase p85α mouse monoclonal antibody, and horseradish peroxidase-linked IgG (Santa Cruz Biotechnology); monoclonal CD36-FITC (Serotec, Oxford, UK); CD36 mAb clone JC63.1 (aCD36 (oxLDL)) and YC-1 (Cayman Chemical, Ann Arbor, MI); U74389G (Biomol); Tempol (Fluka Biochemica); 1, 1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI)-oxidized LDL (DiI-oxLDL; Intracel, Frederick, MD); wortmannin and LY294002 (Alomone Labs Ltd.); CD36 mAb clone FA6-152 (aCD36 (TSP-1) (Beckman Coulter, Fullerton, CA)); 4′,6-diamidino-2-phenylindole, dichlorofluorescein diacetate, annexin V-FITC/PI staining (Molecular Probes, Eugene, OR); CellTracker Green 5-fluoromethylfluorescein diacetate (Cambrex Corp.); and Lipofectamine 2000 (Invitrogen Corp.) were used.
Animals and in Vivo Hypoxia Model
Six-week-old male C57BL/6 mice purchased from Charles River (St. Constant, Quebec, Canada) were used according to a protocol approved by the Research Center of CHU Sainte-Justine Animal Care Committee. Mice were exposed to ambient room air or placed in an OxyCycler (BioSpherix, Ltd.) and exposed to hypoxia (8% O2) for 6 h. At the end of the experiments, animals were immediately sacrificed and tissues were processed for quantitative real time-PCR (qRT-PCR).
Cell Culture and Hypoxia
Human dermal microvascular endothelial cells, human pulmonary artery smooth muscle cells (Cambrex, Walkersville, MD), human retinal pigment epithelial (ARPE-19), and human promyelocytic leukemia (HL60) cells (ATCC, Manassas, VA) were maintained according to standard procedures. For the majority of experiments, ARPEs were used because of their relevance to our ocular studies. For hypoxia exposure, culture dishes were placed in a hypoxia chamber (Billups Rothenburg Inc.) allowing the establishment of a hypoxic environment of 2% O2, 5% CO2, and 93% N2; unless otherwise stated, this hypoxic level (24 h) was used in all experiments. The airtight incubator was kept at 37 °C for preset time periods, whereas normoxic cells were placed at 37 °C in a 21% O2, 5% CO2, and 74% N2 humidified incubator.
RNA Isolation and qRT-PCR
Total RNA was extracted using the standard TRIzol RNA isolation protocol (Invitrogen) and treated with DNase I (Qiagen, Hilden, Germany). cDNA was synthesized from 1 μg of RNA with Moloney murine leukemia virus reverse transcriptase (Promega Corp.) according to the manufacturer's instructions and amplified using SYBR Green I (Stratagene) in a sequence detection system (MxPro 3000 QPCR systems, Stratagene). Primers for CD36, HIF-1α, and β-Actin were synthesized by Invitrogen as follows: human CD36, forward 5′-TCTTTCCTGCAGCCCAATG-3′, reverse 5′-AGCCTCTGTTCCAACTGATAGTGA-3′; human HIF-1α, forward 5′-TGCTTGGTGCTGATTTGTGA-3′, reverse 5′-GGTCAGATGATCAGAGTCCA-3′; human β-actin, forward 5′-GGGRCAGAAGGATTCCTATG-3′, reverse 5′-GGTCTCAAACATGATCTGGG-3′; mouse CD36, forward 5′-TCCTCTGACATTTGCAGGTCTATC-3′, reverse 5′-AAAGGCATTGGCTGGAAGAA-3′; and mouse β-actin, forward 5′-ACTATTGGCAACGACCGGTTT-3′, reverse, 5′-AAGGAAGGCTGGAAAAGAGGG-3′. Primers for PI3K p85α were supplied by Santa Cruz Biotechnology. PCR amplification protocol involved 40 cycles of denaturation at 95 °C for 30 s, primer annealing at 55 °C, and primer extension at 72 °C for 60 s. Each sample was analyzed in triplicate along with RT and no template controls.
Down-regulation of HIF-1α and PI3K p85α with Silencing RNA
ARPEs were grown to 40% confluence and transfected using Lipofectamine 2000 with scrambled Silencer Negative Control 1 silencing RNA (siRNA; Ambion) or sequence-specific siRNA targeting _PI3K p85_α (Santa Cruz Biotechnology) or _HIF-1_α (Ambion; pre-designed siRNA ID 106498 and 106500 both showed similar knock-down effectiveness and the former was used in all experiments). Cells were incubated with _HIF-1_α or _PI3K p85_α siRNA for 24 h after which they were subjected to hypoxia followed by analysis by qRT-PCR, Western blot, or luciferase assay.
Determination of Total Protein Expression by Western Blot Analysis
Protein extraction from cells and Western blots were performed as previously described (19) using 40 μg of total protein and anti-CD36 (1:400), anti-p85α (1:300), anti-HIF-1α (1:250), or anti-β-Actin (1:10000) antibodies. Protein expression was quantified via densitometry (Image-Pro Plus software, version 4.1; Media Cybernetics, Silver Spring, MD).
Determination of CD36 Surface Expression
ARPEs at 70% confluence were exposed to normoxia or hypoxia followed by analysis of CD36 expression via fluorescence-activated cell sorting (FACS) (FACScan; BD Biosciences) using an anti-CD36-FITC antibody. A minimum of 10,000 cells/sample were assessed. Data were acquired and analyzed using CellQuest software.
Measurement of mRNA Stability, mRNA Translation, and Protein Stability
ARPEs were pre-exposed to 21 or 2% O2 followed by addition of ActD (4.5 μg/ml) to block transcription and subsequent return to normoxia or hypoxia. Cells were harvested at various time points after ActD treatment and processed for qRT-PCR. CD36 mRNA stability was determined as the percentage of initial mRNA remaining after ActD exposure. To elucidate the contribution of ongoing protein synthesis and protein stability, CHX was added to block translation before (25 μm) or after (100 μm) hypoxia. CD36 protein stability was assessed as the proportion of the initial protein remaining after CHX exposure.
ROS and 8-Isoprostane Measurements
Intracellular ROS generation was measured using the fluorescent probe dichlorofluorescein diacetate. ARPEs were cultured in 24-well plates and exposed to hypoxia at the indicated time points or angiotensin II (100 nm) for 45 min. Cells were subsequently incubated with dichlorofluorescein diacetate (10 μm) for 30 min at 37 °C followed by measurements using a multiwell fluorescent plate reader (Wallac 1420 VICTOR Multilabel Counter) set at 485 nm excitation and 535 nm emission wavelengths. 8-Isoprostanes (8-Iso-prostaglandin F2α) were measured in normoxia and hypoxia were exposed ARPEs by enzyme immunoassay (Cayman Chemical, Ann Arbor, MI) as previously described (18).
Generation of Promoter Constructs, Transient Transfection, and Reporter Gene Assays
A human bacterial artificial chromosome (clone RP11-542H16 from chromosome 7) carrying the human gene for the CD36 promoter (NCBI accession number DD115563) was obtained from the BACPAC Resources Center (Children's Hospital Oakland Research Institute, Oakland, CA). The identity of the DNA clone was verified by restriction digest. A fragment containing the 5′-untranslated region (∼3.5 kbp) of the putative CD36 promoter was generated from the bacterial artificial chromosome by PCR using the following primers: forward, 5′-GCGAGCTCCTGAGGAATACAATTGGGGATTGAG-3′ and reverse, 5′-GCGCTCGAGGCAGTCCTCAGTACATAAATGCGT-3′. This product was cloned into the SacI and XhoI sites of the pGL3-basic and pGL3-promoter vectors (Promega Corp.), and the generated plasmids were designated pCD36-luc1 and pCD36- luc2, respectively. In the pCD36-mutHRE construct, the putative HRE of pCD36-luc1 was replaced from 5′-CGTG-3′ to 5′-AAAG-3′ using the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing. ARPEs at about 70% confluence in 12-well plates were transiently transfected with reporter plasmid (1 μg) using Lipofectamine 2000 according to the manufacturer's directions. To correct for variable transfection efficiency, cells were co-transfected with pcDNA3.1 LacZ (0.5 μg), which encodes for the β-galactosidase gene. Transfected cells were allowed to recover for 24 h in fresh medium, and then treated with CoCl2 (100 μm) or subjected to normoxia or hypoxia. Cells were lysed and luciferase activity determined using the Luciferase Reporter Assay system (Promega) via a multiwell luminescence reader (Wallac 1420 VICTOR Multilabel Counter).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation was performed according to the chromatin immunoprecipitation assay kit protocol (Upstate Biotechnology, Lake Placid, NY). Briefly confluent cells were grown on 15-cm dishes and exposed to CoCl2, normoxia, or hypoxia (4% O2, 1 h). Cells were immediately fixed with 1% formaldehyde/phosphate-buffered saline, lysed, and sonicated to obtain 500 to 1000-base pair DNA fragments. Chromatin was immunoprecipitated using 5 μg of anti-HIF-1α (overnight, 4 °C) or rabbit IgG as a negative control. Immunoprecipitated DNA complex was amplified by PCR with primers for the CD36 promoter (forward, 5-TGAGTGTGTGAGAATTAAGGTTGA-3; and reverse, 5-GAAATGACCATGTGCAATCTCT-3) flanking the HIF-1 binding site. PCR products (∼239 bp) were separated by electrophoresis through 1.8% agarose gels and visualized by ethidium bromide staining.
Uptake of DiI-labeled oxLDL
Analysis of DiI-oxLDL uptake was performed according to the manufacturer's protocol with minor modifications. Briefly, cells were pre-treated for 2 h with IgA or aCD36 (oxLDL) (clone JC63.1, 10 μg/ml) prior to normoxia or hypoxia exposure. Next, cells were incubated for 6 h with DiI-oxLDL (20 μg/ml) in serum-free medium at 37 °C, washed extensively with phosphate-buffered saline, 0.1% bovine serum albumin, pelleted, and resuspended in 400 μl of FACS flow for immediate FACS analysis.
Preparation of Apoptotic HL60 Cells
HL60 cells were pre-labeled with 5 μm CellTracker Green for 45 min at 37 °C according to the manufacturer's instructions. Cells were subsequently rendered apoptotic after treatment with 5 μm camptothecin (4 h), washed with media, and cultured for an additional 6 h before use. Apoptosis was confirmed by DNA laddering and annexin V-FITC/PI staining (>50% annexin V positive).
Preparation of Photoreceptor Outer Segments
POS were isolated on 25–60% sucrose gradients from porcine eyes obtained fresh from the slaughterhouse according to established protocols (24, 32–34). Before use, POS were labeled with 10 μg/ml FITC for 1 h at room temperature in the dark, washed, and resuspended in Dulbecco's modified Eagle's medium as previously described (24, 34).
Phagocytosis Assays and Immunofluorescence
We used modified versions of established phagocytosis assay protocols (24, 32, 35). ARPEs were seeded onto 15-mm glass coverslips and pre-treated for 2 h with IgA or aCD36 (oxLDL) (10 μg/ml) followed by hypoxic exposure. 5 × 105 apoptotic cells or 106 POS were allowed to incubate for 3 h on ARPE monolayers in serum-free medium. After incubation, unbound cells were removed by extensive washing with medium. Cells were fixed in ice-cold methanol (15 min at room temperature), washed with phosphate-buffered saline, and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (1:3000, 5 min). Bound or ingested cells were detected by their green fluorescence and observed using a Nikon eclipse E800 epifluorescent microscope with a Nikon DXM 1200 digital camera.
Aortic Ring Angiogenesis Assay
This assay was performed as described previously (18). The aortic ring culture media was changed on day 4 with addition of the following test compounds: aCD36 (TSP-1) (clone FA6-152, 10 μg/ml), a CD36 agonist (clone SMO, 5 μg/ml), and their respective isotype controls IgG1 and IgM. Photographs were taken before (day 4) and after treatment (day 5) using an inverted microscope (Eclipse TE300; Nikon). The area of neovessel formation was determined using Image Pro Plus software.
Statistical Analysis
All experiments were repeated at least three times and values are presented as mean ± S.E. Data were analyzed by Student's t test, one-way or two-way analysis of variance followed by post-hoc Bonferroni tests for comparison among means. Statistical significance was set at p < 0.05.
RESULTS
Systemic Hypoxia Increases CD36 Expression in Vivo
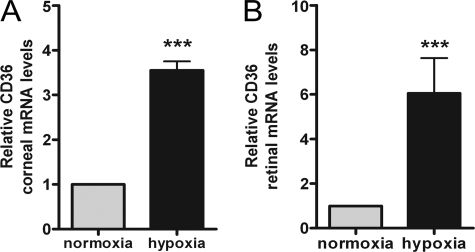
Based on previous reports eluding to the hypoxic regulation of CD36 (28, 36), we first investigated the effects of hypoxia on CD36 expression in vivo. CD36 mRNA levels were determined in corneal and retinal tissue following mouse subjection to ambient room air or whole body hypoxia (8% O2, 92% N2 for 6 h). We focused our attention on the cornea and retina due to their abundant CD36 expression (18, 19, 23) and relevance to neovascular eye diseases. As shown in Fig. 1, CD36 expression was markedly increased by 3.6- and 6.0-fold in corneal and retinal tissue derived from hypoxic mice.
FIGURE 1.
Effect of hypoxia on CD36 mRNA levels in vivo. C57BL/6 mice were subjected to ambient air or hypoxia (constant flow of 8% O2, 92% CO2 in an oxycycler) for 6 h (n = 6 animals per group). At the end of the experiment, animals were sacrificed, corneal (A) and retinal (B) tissue was harvested and processed for quantitative real-time PCR to determine relative CD36 mRNA levels. Values depicted as mean ± S.E. (three independent experiments) were normalized to β-actin and expressed relative to normoxia (set to 1). ***, p < 0.001 versus normoxia by Student's t test.
Hypoxia Up-regulates CD36 mRNA, Protein, and Surface Expression in Vitro
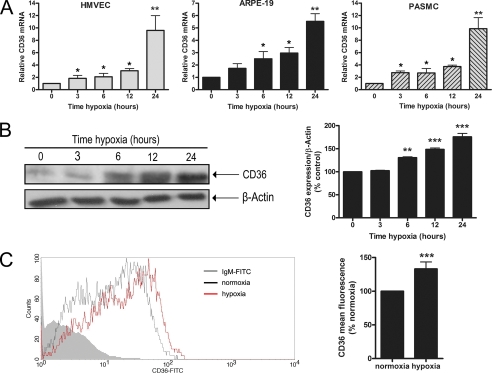
Given the ability of hypoxia to potently induce CD36 mRNA levels in vivo, we explored the kinetics of the hypoxic regulation of CD36 in various cell types that are known to highly express CD36. Human dermal microvascular endothelial cells, ARPEs, and pulmonary artery smooth muscle cells were exposed to hypoxia (2% O2) from 0 to 24 h and CD36 expression was assessed by qRT-PCR. Exposure to hypoxia resulted in a time-dependent increase in CD36 mRNA in all cell types that was evident at 3 h and sustained up to 24 h (Fig. 2A). Based on these findings, and the demonstrated induction of CD36 in hypoxic retinal tissue, subsequent experiments were conducted using ARPEs. Western blot analysis on ARPEs exposed to 2% O2 from 0 to 24 h yielded parallel increases in CD36 protein expression (Fig. 2B). The immunoblot data were confirmed by flow cytometry, which revealed a significant increase in CD36 surface expression under hypoxia (Fig. 2C, p < 0.001).
FIGURE 2.
Effect of hypoxia on CD36 expression in vitro. A, human dermal microvascular endothelial cell (HMVEC), ARPE, and pulmonary artery smooth muscle (PASMC) cells were grown under normoxia (21% O2) or hypoxia (2% O2) for the indicated times up to 24 h and CD36 mRNA was determined by qRT-PCR expressed relative to control (set to 1). Representative images are shown of ARPEs exposed to 21% O2 or 2% O2 for up to 24 h and analyzed by Western blot (B) or flow cytometry (C), with values in histograms depicted as a percentage of the appropriate control (set to 100%). For each experiment, results shown represent the mean ± S.E. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus 0 h, normoxia, or control by Student's t test or one-way analysis of variance.
Implication of Translational Activity in the Hypoxic Regulation of CD36
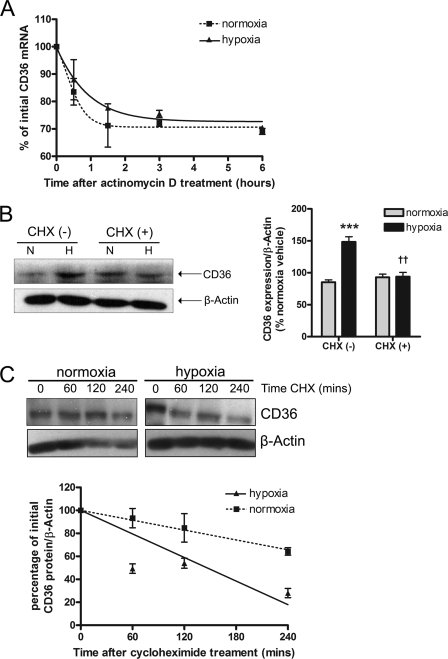
Accumulation of CD36 during hypoxia could result from increases in mRNA stability, mRNA translation, or protein stability. To test whether CD36 mRNA was post-transcriptionally stabilized after hypoxia, ARPEs were maintained in normoxia or hypoxia for 24 h, transcription was blocked with ActD (4.5 μg/ml), and the cells were harvested at different time points for qRT-PCR analysis. CD36 mRNA stability was unchanged by hypoxia; the percentage of initial CD36 mRNA was 68.8 ± 2.6 (21% O2) and 70.5 ± 0.9% (2% O2) following 6 h of ActD treatment (Fig. 3A, p > 0.05). To elucidate the importance of ongoing protein synthesis, ARPEs were pretreated with the translation inhibitor CHX (25 μm) before hypoxic exposure. CHX potently abrogated the hypoxia-induced up-regulation of CD36 protein (2% O2, 148 ± 8 versus 2% O2 + CHX, 94 ± 6%; Fig. 3B, p < 0.01). The effect of hypoxia on CD36 protein stability was determined by investigating the time course of CD36 protein decay in normoxia or hypoxia pre-exposed cells treated with CHX (100 μm). Hypoxia dramatically reduced CD36 protein levels such that after 240 min, 28 ± 4% of the initial protein remained compared with 64 ± 3.2% under normoxia (Fig. 3C).
FIGURE 3.
Effect of hypoxia on CD36 mRNA stability, protein synthesis, and protein stability. A, ARPEs were subjected to normoxia (N, 21% O2) or hypoxia (H, 2% O2) for 24 h followed by treatment with actinomycin D (4.5 μg/ml) and harvesting of cells at the indicated times. CD36 mRNA was analyzed by qRT-PCR. The value obtained at time 0 h was set at 100% and used to determine the percentage of CD36 mRNA remaining after ActD addition. ARPEs were preincubated in: B, the absence (−) or presence (+) of cycloheximide (CHX, 25 μm, 1 h) and exposed to 21% O2 or 2% O2 for 16 h, or C, pre-exposed to 21% O2 or 2% O2 for 24 h, followed by incubation with CHX (100 μm) for the indicated times. In B and C total protein lysates were analyzed by Western blot for CD36 expression with β-actin serving as loading control. Representative blots and histograms demonstrate CD36 protein levels expressed as percentage of normoxia CHX (−) in B or as percentage of the point before CHX addition in C. In each case, experiments were repeated at least 3 times and values are expressed as mean ± S.E. ***, p < 0.001 versus normoxia CHX (−); ††, p < 0.01 versus hypoxia CHX (−).
ROS Are Involved in the Hypoxic Induction of CD36 Transcripts
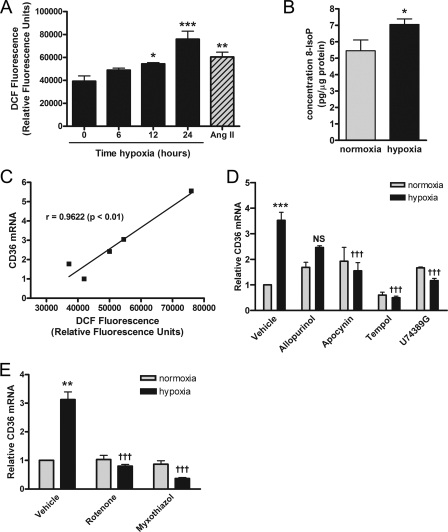
Studies have demonstrated that ROS generated during cerebral ischemia are markedly attenuated in Cd36 null mice and that treatment with antioxidant peptides down-regulates ischemia-induced CD36 expression (29, 31). Consequently, we sought to determine whether ROS activity was implicated in the hypoxic regulation of CD36. Given the controversial relationship between ROS production and hypoxia (1, 37, 38), we first measured levels of ROS and 8-isoprostanes, well established markers of lipid peroxidation (39–41), in normoxia and hypoxia pre-exposed cells. As shown in Fig. 4, hypoxia significantly increased the levels of intracellular ROS (Fig. 4A, p < 0.001) and 8-isoprostanes (Fig. 4B, p < 0.05). Moreover, correlation analysis revealed that CD36 mRNA levels were significantly correlated with ROS levels during hypoxia (r = 0.9622; Fig. 4C, p < 0.01). Consistent with this, the superoxide dismutase mimetic tempol (1 mm), and inhibitors of lipid peroxidation (U74389G, 50 μm), and the superoxide generating enzyme NADPH oxidase (apocynin, 50 μm), abrogated the hypoxia-induced increase in CD36 mRNA (Fig. 4D). Inhibition of xanthine oxidase (allopurinol, 50 μm), another major ROS generating enzyme, had no significant effect (Fig. 4D).
FIGURE 4.
Role of ROS in hypoxic up-regulation of CD36. Levels of ROS (A) (positive control angiotensin II, 100 nm) and 8-isoprostanes (B) were evaluated using the peroxide-sensitive fluorescent probe 2′,7′-dichlorofluorescein (DCF) diacetate or enzyme immunoassay, respectively, in ARPEs cultured under normoxic (21% O2) or hypoxic (2% O2) conditions. C, representative plot of a correlation analysis (Pearson, r) between CD36 mRNA and ROS levels as indicated by DCF fluorescence generated during hypoxia. Prior to the 24-h hypoxic challenge and quantitative real time PCR analysis, ARPEs were incubated for 1 h with: D, cellular (allopurinol, 50 μm; apocynin, 50 μm; U74389G, 50 μm; tempol, 1 mm); or E, mitochondrial electron transport chain complex I (rotenone, 100 nm) and III (myxothiazol, 200 nm) inhibitors. For each experiment, results are expressed as mean ± S.E. of three to six independent experiments performed in quadruplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus 0 h or normoxia; †††, p < 0.001 versus hypoxia vehicle; NS, nonsignificant.
Recent evidence has implicated mitochondria in the O2 sensing underlying functional responses to hypoxia (1) and has shown that the rate of mitochondrial ROS production is elevated during hypoxia (42). Accordingly, inhibition of mitochondrial electron transport complexes I and III with rotenone (100 nm) and myxothiazol (200 nm), respectively, caused marked suppression of CD36 mRNA levels during hypoxia (Fig. 4E).
HIF-1-dependent Hypoxic Regulation of CD36 Expression
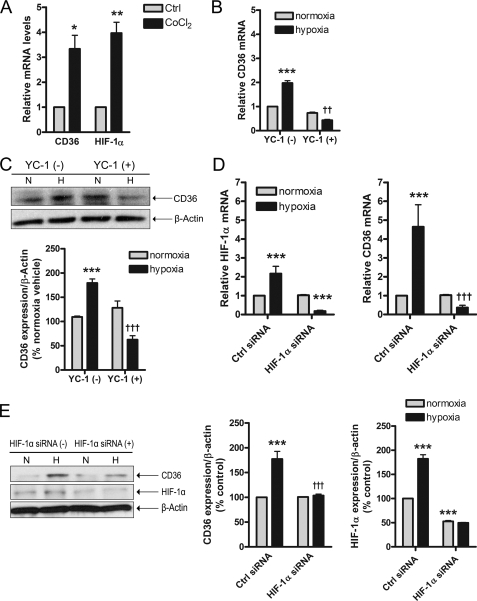
Important insights into the mechanism of oxygen sensing have been garnered using pharmacological hypoxia mimetics such as CoCl2 (43), which is documented to result in up-regulation of HIF-1, a ubiquitous transcription factor responsible for the hypoxic activation of multiple genes (3, 44). Our experiments focused on HIF-1α, a key component of HIF-1 whose expression determines the cellular HIF-1 activity. When ARPEs were treated with CoCl2 (200 μm, 6 h), a 3.3-fold rise in CD36 mRNA was observed, which was comparable with HIF-1α whose expression was induced by 4-fold (Fig. 5A). To further pursue the involvement of HIF-1, ARPEs were exposed to 2% O2 in the presence or absence of the HIF-1 inhibitor YC-1 (5 μm) (45–47), or _HIF-1_α siRNA (50 nm). YC-1 potently suppressed the hypoxic induction of CD36 expression at both the mRNA (Fig. 5B, p < 0.01) and protein (Fig. 5C, p < 0.001) levels. Importantly, these observations were corroborated when knock-down of _HIF-1_α by siRNA abolished the induction of CD36 mRNA (Fig. 5D, p < 0.001) and protein (Fig. 5E, p < 0.001).
FIGURE 5.
Involvement of HIF-1α in CD36 hypoxic induction. ARPEs were incubated with cobalt chloride (A) (200 μm, 3 h) subsequent to analysis of CD36 and HIF-1α mRNA expression by qRT-PCR, or exposed to normoxia (N, 21% O2) or hypoxia (H, 2% O2, 24 h) in the absence (−) or presence (+) of YC-1 (B and C) (5 μm), or _HIF-1_α siRNA (D and E) (50 nm). Representative blots and histograms depicting CD36 and/or HIF-1α protein levels expressed as percentage of control (Ctrl) are shown. In each scenario, results represent the mean ± S.E. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus Ctrl or normoxia; ††, p < 0.01; †††, p < 0.001 versus hypoxia Ctrl.
Hypoxia Activates CD36 Promoter Activity
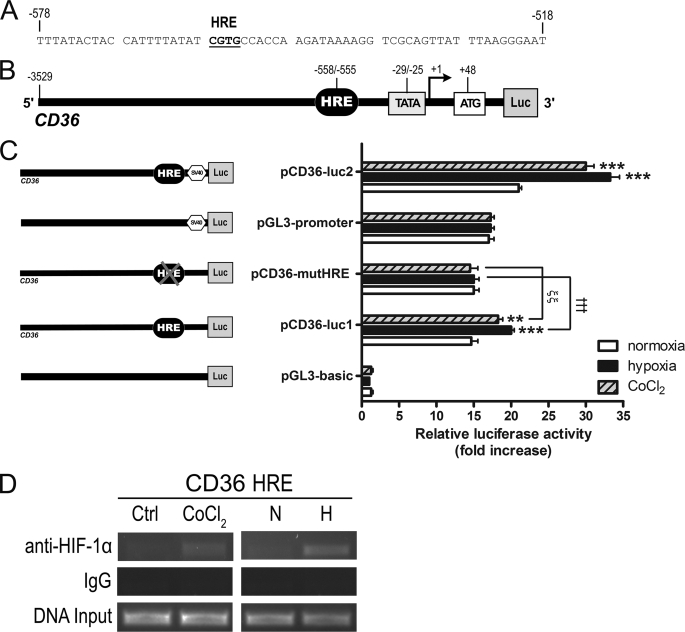
To further investigate the transcriptional modulation of CD36, we cloned a putative CD36 promoter region (∼3.5 kb) with the goal of determining whether it possessed promoter activity, and if so whether the region mediated CD36 activation in response to hypoxia (Fig. 6A). We constructed reporter plasmids in which the CD36 5′-flanking sequence was fused to the firefly luciferase basic (pCD36-luc1) or promoter (pCD36-luc2) vectors (Fig. 6, B and C). These constructs were transiently transfected into ARPEs and their luciferase activities under normoxia assessed. Data reveal that the luciferase activities of pCD36-luc1 and pCD36-luc2 were enhanced 13.4- and 4.0-fold, respectively, compared with the pGL3-basic or pGL3-promoter vectors, which served as their respective negative controls (Fig. 6C). This suggested that the cloned region possessed promoter activity and regulatory elements that were potentially modulated by hypoxia. Based on this precedence, we subjected the constructs to either 21% O2, 2% O2 (1 h), or CoCl2 (100 μm, 3 h) prior to luciferase measurements. Interestingly, hypoxia and CoCl2 stimulated the pCD36-luc1 reporter construct by 5.3- and 3.6-fold, respectively (Fig. 6C). Likewise, transfection with pCD36-luc2 caused 12.3- and 9.0-fold increases in luciferase activity following hypoxia and CoCl2, respectively (Fig. 6C).
FIGURE 6.
CD36 is a target gene of HIF-1. Schematic representation of the 5′ CD36 promoter region illustrating (A) the sequence of the putative HRE as indicated in bold and underlined text, and (B) the potential regulatory elements. Numbering refers to the position relative to the predicted transcription start site. ARPEs were co-transfected with the indicated reporter constructs containing the wild-type CD36 promoter fused to (C) the pGL3-basic (pCD36-luc1) or pGL3-promoter (pCD36-luc2) vectors together with pcDNA3.1 LacZ as a transfection control (Ctrl). pCD36-mutHRE is the mutant form of pCD36-luc1 with point mutations at the putative core HRE. Transfected cells were exposed to normoxia (N, 21% O2), hypoxia (H, 2% O2), or cobalt chloride (CoCl2, 200 μm) for 1 h before measurement of luciferase activity. Promoter activity was expressed as the fold-increase in relative luciferase activity (mean ± S.E. of three independent experiments performed in quadruplicate, **, p < 0.01; ***, p < 0.001 versus normoxia; †††, p < 0.001 versus pCD36-luc1 hypoxia; ξξ, p < 0.01 versus pCD36-luc1 CoCl2). D, chromatin immunoprecipitation was carried out with ARPEs exposed to either 21% O2, 2% O2, or CoCl2 (200 μm) for 1 h. Immunoprecipitation was performed with an anti-HIF-1α antibody. Co-precipitated genomic DNA fragments were evaluated by PCR using primers flanking the HRE-containing region of the CD36 promoter. The gel is representative of three independent experiments.
HIF-1 binds to a conserved HRE located in the regulatory sequences of target genes (3, 6, 7, 48). Sequence analysis of the 5′-flanking region of the human CD36 promoter revealed the presence of a putative HRE with the characteristic motif 5′-RCGTG-3′. This element was located between −558 and −555 bp upstream of a predicted (Berkeley Drosophila Genome Project software) transcription start site (Fig. 6, A and B). A mutant plasmid was generated from pCD36-luc1 by introducing a 4-bp substitution through site-directed mutagenesis at the putative HRE (pCD36-mutHRE). Following transient transfection into ARPEs and exposure to either 21% O2, 2% O2 (1 h), or CoCl2 (100 μm, 3 h), pCD36-mutHRE remained insensitive to hypoxia (Fig. 6C, p > 0.05).
HIF-1 Binding to the Putative HRE in the CD36 Promoter
Chromatin immunoprecipitation analysis was next performed to explore whether HIF-1 interacts with the HRE consensus sequence identified in the CD36 promoter. In vivo binding of HIF-1α to the CD36 promoter was detected in ARPEs following incubation in either 2% O2 (1 h) or CoCl2 (100 μm, 3 h) (Fig. 6D). As expected, HIF-1α was not significantly associated with the CD36 promoter in cells maintained in 21% O2.
Hypoxic Activation of CD36 and HIF-1α Is Linked to the PI3K/mTOR Pathway
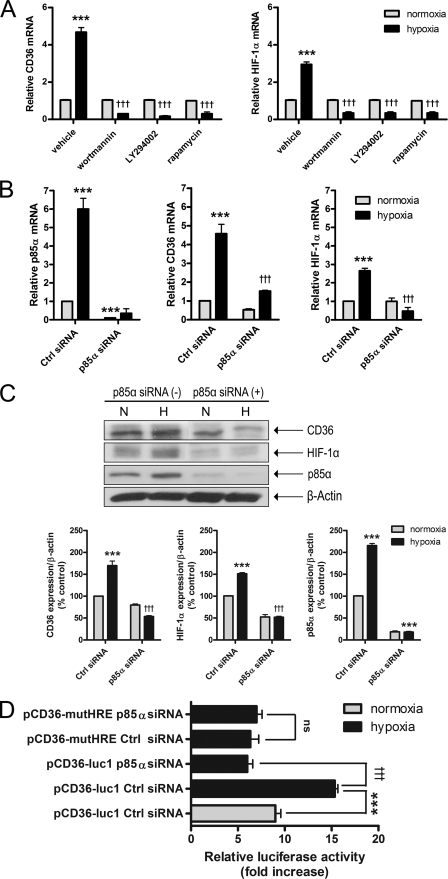
Our findings thus far suggested that ongoing translation (Fig. 3), ROS production (Fig. 4), and HIF-1 activation (Fig. 5) were essential for the CD36 hypoxic response. Because ROS-dependent activation of the PI3K pathway has been linked to the translational regulation of several hypoxia-inducible genes (49, 50), including HIF-1, we evaluated the contribution of this cascade. Pre-treatment of ARPEs with two structurally distinct PI3K inhibitors, wortmannin (200 nm) and LY294002 (20 μm), and an inhibitor of the downstream PI3K effector, mTOR (rapamycin, 100 nm), significantly ablated both CD36 and HIF-1α hypoxic responses (Fig. 7A). Similarly, knock-down of the _p85_α regulatory subunit of PI3K via siRNA (30 nm) prevented hypoxic accumulation of both CD36 and HIF-1α mRNA (Fig. 7B) and protein (Fig. 7C). Interestingly, hypoxia-dependent stimulation of pCD36-luc1 luciferase activity was blunted by _PI3K p85_α siRNA (Fig. 7D, p < 0.001).
FIGURE 7.
Implication of the PI3K pathway in the hypoxic accumulation of CD36. ARPEs were incubated with or without PI3K (A) (wortmannin, 200 nm; LY294002, 20 μm) and mTOR (rapamycin, 100 nm) inhibitors, or PI3K p85α siRNA (B and C) (30 nm) prior to 24 h hypoxic exposure and quantitative real time PCR analysis (A) or Western blot analysis for CD36 and HIF-1α expression (B). For each experiment, results are expressed as mean ± S.E. of three to six independent experiments performed in quadruplicate. In D, ARPEs were co-transfected with or without PI3K p85α siRNA and the indicated reporter constructs containing the wild-type CD36 promoter were fused to the pGL3-basic (pCD36-luc1) vector; pcDNA3.1 LacZ served as transfection control (Ctrl). Transfected cells were exposed to normoxia or hypoxia for 1 h before measurement of luciferase activity. Promoter activity was expressed as the fold-increase in relative luciferase activity (mean ± S.E. of three independent experiments performed in quadruplicate). ***, p < 0.001 versus normoxia vehicle or normoxia Ctrl siRNA; †††, p < 0.001 versus hypoxia vehicle or hypoxia Ctrl siRNA; ns, nonsignificant.
Functional Consequences of CD36 Hypoxic Up-regulation
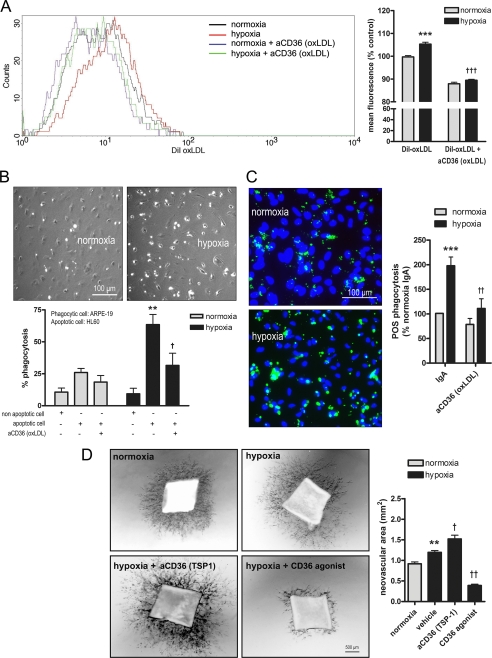
Although we provide abundant data corroborating the hypoxic up-regulation of CD36, we considered it pertinent to correlate these findings with a functional significance. Of relevance, studies have implicated CD36 in the recognition and uptake of oxLDLs, apoptotic cells, and POS (16, 24, 32, 35, 51), whereas we and others have demonstrated that hypoxia promotes lipid peroxidation (Fig. 4B) (52, 53) and apoptosis (54). We pre-exposed ARPEs to 2% O2 for 16 h followed by incubation with DiI-oxLDL (20 μg/ml, 6 h) and FACS analysis. Our results show that hypoxia caused a significant increase in DiI mean fluorescence (Fig. 8A, p < 0.001) that was blocked by an anti-oxLDL binding site CD36 mAb (aCD36 (oxLDL)) (Fig. 8A, p < 0.001).
FIGURE 8.
Effect of hypoxia on CD36-mediated phagocytosis and angiostasis. A–C, representative images and histograms of ARPEs pre-treated with vehicle (anti-human IgA) or an anti-oxLDL binding site CD36 monoclonal antibody (aCD36 (oxLDL), 10 μg/ml) for 2 h and exposed to normoxia (21% O2) or hypoxia (2% O2) for 16 h. ARPEs were subsequently incubated for 3 h with DiI-labeled oxLDL (A) (20 μg/ml) followed by flow cytometry analysis, viable or CellTracker Green 5-fluoromethylfluorescein diacetate (B) fluorescently labeled apoptotic (camptotecin, 5 μm, for 4 h; as monitored by a DNA laddering assay and annexin V staining) HL60 cells followed by immunofluorescence, or FITC-labeled POS and immunofluorescence (C). The rate of phagocytosis was determined as the ratio of bound apoptotic cells (B) or POS (C) relative to the total number of phagocytic ARPEs. D, mouse aortas were seeded on Matrigel and on day 4 incubated in the presence of vehicle (anti-mouse IgG1 or anti-mouse IgM), an anti-TSP-1 binding site CD36 mAb (aCD36 (TSP-1), 10 μg/ml), or a CD36 agonist (SMO, 5 μg/ml) prior to 24 h hypoxia. Photographs were taken on day 5 and the neovascular area was quantified in the histogram. In all cases, results are representative of three to four independent experiments performed in quadruplicate. **, p < 0.01; ***, p < 0.001 versus normoxia control; †, p < 0.05; ††, p < 0.01; †††, p < 0.001 versus hypoxia control.
To explore the influence of hypoxia on CD36-mediated engulfment of apoptotic cells and POS, confluent monolayers of normoxia- and hypoxia-exposed ARPEs were co-cultured with: 1) viable versus apoptotic (5 μm camptotecin) fluorescently labeled HL60 cells, or 2) FITC-labeled POS. After incubation, non-adherent/non-phagocytosed cells were removed by vigorous washing, and the rate of phagocytosis was determined as the ratio of bound cells to the total number of adherent ARPEs. Relative to normoxia, hypoxia significantly increased both apoptotic cell (Fig. 8B, p < 0.01) and POS (Fig. 8C, p < 0.001) phagocytosis; these events were abrogated by aCD36 (oxLDL) (Fig. 8, B and C).
It is well established that CD36 transduces signals leading to apoptosis-dependent inhibition of angiogenesis (55). We therefore tested the outcome of blocking or activating CD36 in a hypoxia-driven model of aortic ring angiogenesis. As expected, compared with normoxia (0.91 ± 0.04 mm2), hypoxia (1.2 ± 0.05 mm2) increased microvessel sprouting. Conversely, incubation of hypoxia-exposed aortic rings with CD36 functionally blocking (aCD36 (TSP-1)) or activating (SMO) mAbs, respectively, exacerbated (1.5 ± 0.1 mm2) or suppressed (0.4 ± 0.04 mm2) the neovascular area (Fig. 8D).
DISCUSSION
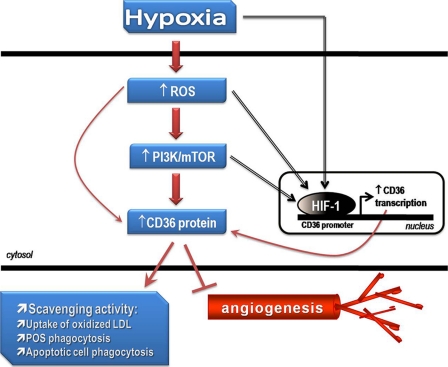
The CD36 scavenger receptor has emerged as a pivotal player in diverse homeostatic and pathological settings particularly those affecting ocular tissue (16, 18, 19, 21–24). Although hypoxia is considered a major stimulus of numerous disease states including ischemic retinopathies and ocular neovascularization, the molecular mechanisms and pathways governing the regulation of CD36 by hypoxia have yet to be elucidated. Herein we report that CD36 is activated at low oxygen tension through mechanisms that involve ROS, HIF-1, and the PI3K/mTOR pathway, as depicted by the schematic model in Fig. 9.
FIGURE 9.
Schematic model depicting the putative mechanisms involved in the hypoxic regulation of CD36. In the proposed model, generation of ROS by hypoxia stimulates the PI3K/mTOR pathway, leading to accumulation of the CD36 protein with consequent increases in CD36 scavenging and anti-angiogenic activities. Involvement of HIF-1 is proposed via direct or indirect activation by hypoxia, ROS, or the PI3K/mTOR cascade. Once activated, HIF-1 binds to the CD36 promoter to induce CD36 transcription and ultimately protein synthesis. Hypoxia-related lipid peroxidation further generates ligands, such as oxidized lipids, which serve to enhance receptor activation and function.
Hypoxic injury to ocular tissue elicits lipid peroxidation thus generating oxidized lipids that are prominent CD36 ligands (16, 53). The cornea and retina express abundant surface CD36 (18, 19, 23) and are rich in polyunsaturated phospholipids (32, 56, 57), which renders them particularly susceptible to oxidant stress. It was therefore intriguing to observe a substantial rise in CD36 corneal and retinal mRNA expression following hypoxia (Fig. 1), a finding that was confirmed by comparable augmentations in CD36 expression in microvascular endothelial cells, RPE cells, and smooth muscle cells (Fig. 2A). Based on this data, we surmise that hypoxic induction of CD36 may represent a universal adaptive response aimed at protecting susceptible cellular or tissue milieus from the deleterious consequences of hypoxia.
Ample studies have documented that oxidative stress stimulates CD36 expression and that antioxidants reduce its expression and function (29–31). Indeed our results link the CD36 hypoxic response to the cellular redox state given that lipid peroxidation and ROS production were increased during hypoxia and in this process CD36 mRNA and ROS levels were positively correlated (Fig. 4, A–C). Experimental data has also revealed that the NADPH oxidase (58, 59) and mitochondrial electron transport complexes I and III are major cellular sources of ROS, principally in the form of the superoxide anion and hydrogen peroxide (14, 37, 60). Our observations concur with this data because inhibition of the aforesaid ROS sources abrogated the CD36 hypoxic signal (Fig. 4, D and E) and is substantiated by a study reporting that induction of CD36 expression in human monocytes involves redox signaling via the NADPH oxidase (59).
The current data provide presumptive evidence of translational modulation of CD36. First, we demonstrate that cycloheximide, a protein synthesis inhibitor, ablated the CD36 hypoxic response (Fig. 3B). Second, both CD36 mRNA and protein were not stabilized by hypoxia (Fig. 3, A and C), suggesting that CD36 accumulation may be dependent on new protein synthesis. Third, hypoxic up-regulation of CD36 was sensitive to inhibition of the PI3K/mTOR pathway, which is known to regulate the translation rate of particular mRNAs (Fig. 7, A–C) (61–64). We therefore hypothesize that overproduction of ROS during hypoxia activates the PI3K/mTOR pathway thus initiating a translation stimulation signal leading to enhanced CD36 protein expression. Indeed, similar models depicting translational modulation of HIF-1α through the PI3K/mTOR cascade have been proposed (9, 12, 64).
Our results provide strong evidence of transcriptional modulation of CD36 by HIF-1, the master regulator of oxygen homeostasis (3–5, 7). In line, we attained an increase in CD36 mRNA levels following administration of CoCl2, a pharmacological inducer of HIF-1 (Fig. 5A), and significant attenuation when the HIF-1 inhibitor YC-1 (3, 43–47) and HIF-1α siRNA were employed (Fig. 5, B–E).
HIFs commonly interact with HREs with the core sequence 5′-RCGTG-3′. Accordingly, analysis of the 5′ region of the human CD36 promoter revealed at least one putative HIF-1 binding site with the consensus sequence 5′-CGTG-3′ (Fig. 6, A and B). When this region was cloned into luciferase reporter vectors and transfected into ARPEs, luciferase activity was markedly induced by both hypoxia and CoCl2 (Fig. 6C); mutation of this potential binding site abrogated the hypoxia-induced promoter activation (Fig. 6C). Additionally, DNA binding activity of HIF-1 to this putative HRE was demonstrated by chromatin immunoprecipitation assay in hypoxia- and CoCl2-treated cells (Fig. 6D). Together, our data strongly support a role for HIF-1 in the transcriptional modulation of CD36, although it is conceivable that other transcription factors are involved that may act synergistically with HIF-1 to regulate hypoxic/ischemic CD36 expression.
Numerous studies have identified the PI3K/mTOR pathway as an important element in hypoxic induction of HIF-1α protein and activity (11–13). Interestingly, YC-1 has been shown to limit HIF-1 accumulation during hypoxia by antagonizing the PI3K/mTOR pathway (12). In the present study, PI3K inhibition by specific inhibitors LY294002 and wortmannin, and down-regulation via _PI3K p85_α siRNA, significantly blunted both CD36 and HIF-1α accumulation upon hypoxia (Fig. 7, A–C). Suppression of the PI3K pathway also prevented hypoxia-induced CD36 promoter activity (Fig. 7D). Collectively, these findings suggest that activation of CD36 by hypoxia is mediated by HIF-1 and PI3K cascade.
Recent efforts have demonstrated that a variety of stimuli including hypoxia trigger translocation of CD36 from intracellular pools to the plasma membrane with consequent elevations in fatty acid transport (36). Although our data reveal analogous hypoxia-elicited increases in CD36 surface expression (Fig. 2C), it was not accompanied by subcellular redistribution of the receptor (data not shown), but was nonetheless associated with enhanced functional activity (Fig. 8). It is thus intriguing to speculate on the functional consequences of CD36 hypoxic up-regulation. Consistent with the role of CD36 as a scavenger receptor (16, 24, 32, 35, 51), we show that oxLDL uptake and apoptotic cell and POS phagocytosis were markedly enhanced during hypoxia in a CD36-specific manner (Fig. 8, A–C). It is likely that these events occur via similar mechanisms because CD36 recognizes oxidized phospholipid species in the apoptotic cell membrane that mimic the oxLDL structure (35). Of pathophysiological relevance, our data propose that the hypoxic activation of CD36 in the retina (Fig. 1B), particularly the RPE (Fig. 2), results in increased clearance of POS, a process that is critical for normal visual function (23, 24, 32, 51). Studies have also affirmed that CD36 signaling may be indispensable for RPE POS phagocytosis under conditions of heightened oxidative stress (32), such as occurs during hypoxia.
Present dogma states that angiogenesis, a critical physiological response to hypoxia, is controlled by a dynamic balance between pro- and anti-angiogenic factors (65–67). Angiogenesis stimulators such as vascular endothelial growth factor have also been shown to induce expression of anti-angiogenic factors (68). Moreover, we previously reported up-regulation of CD36 during corneal neovascularization wherein CD36 activation suppressed angiogenesis by antagonizing the vascular endothelial growth factor pathway (18). Consequently, our findings that hypoxia augmented CD36 expression in microvascular endothelial cells (Fig. 2A) and modulated hypoxia-driven microvessel sprouting (Fig. 8D) are intriguing given the angioinhibitory nature of this receptor (16, 18, 55). We speculate that CD36 may act as a negative feedback regulator of pathological angiogenesis, a premise that has been proposed for other anti-angiogenic factors (68, 69). Indeed, studies have reported overexpression of the CD36 ligand thrombospondin-1, in hypoxia-exposed endothelial cells (69) and leg ischemia, where such an overexpression is thought to exert an anti-angiogenic effect aimed at counteracting the hypoxia-induced pro-angiogenic drive (70). Similar negative-feedback mechanisms between CD36 and HIF-1 may thus be operative under hypoxic stress.
In conclusion, our studies provide the first characterization of CD36 as a hypoxia responsive gene that is rapidly activated at the transcriptional and translational levels via ROS-, HIF-1-, and PI3K-dependent mechanisms, culminating in enhanced CD36 functional activity, as summarized in Fig. 9. Given the importance of CD36 in cellular biology and pathophysiology, as well as the central role of hypoxia in many disease states, delineating the mechanisms underlying CD36 hypoxic regulation may provide a valuable framework for future studies investigating the regulatory aspects of this multifunctional receptor. This may have notable implications for the development of therapeutic strategies against eye diseases involving abnormal neovascularization or retinal degeneration.
Acknowledgment
We thank Carmen Gagnon for invaluable technical assistance.
*
This work was supported in part by Fight for Sight Foundation Grant GA05003, Canadian Institutes of Health Research Grant MOP-79332, Hospital for Sick Children Grant XG 03-105, and the Canadian National Institute for the Blind (CNIB_E.A. Baker Foundation).
4
The abbreviations used are:
HIF-1
hypoxia-inducible factor-1
HRE
hypoxia response element
ROS
reactive oxygen species
PI3K
phosphatidylinositol 3-kinase
mTOR
mammalian target of rapamycin
POS
photoreceptor outer segments
ActD
actinomycin D
FITC
fluorescein isothiocyanate
CHX
cycloheximide
DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
oxLDL
oxidized lipoprotein lipase
qRT
quantitative real time
ARPE
human retinal pigment epithelial cells
siRNA
silencing RNA
mAb
monoclonal antibody
FACS
fluorescence-activated cell sorting.
REFERENCES
- 1.Chandel N. S., Budinger G. R. (2007) Free Radic. Biol. Med. 42, 165–174 [DOI] [PubMed] [Google Scholar]
- 2.Semenza G. L. (2007) Science 318, 62–64 [DOI] [PubMed] [Google Scholar]
- 3.Jiang B. H., Zheng J. Z., Leung S. W., Roe R., Semenza G. L. (1997) J. Biol. Chem. 272, 19253–19260 [DOI] [PubMed] [Google Scholar]
- 4.Semenza G. L. (2000) J. Appl. Physiol. 88, 1474–1480 [DOI] [PubMed] [Google Scholar]
- 5.Semenza G. L. (2001) Cell 107, 1–3 [DOI] [PubMed] [Google Scholar]
- 6.Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 7.Wang G. L., Semenza G. L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4304–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treins C., Giorgetti-Peraldi S., Murdaca J., Monthouël-Kartmann M. N., Van Obberghen E. (2005) Mol. Endocrinol. 19, 1304–1317 [DOI] [PubMed] [Google Scholar]
- 9.Pagé E. L., Robitaille G. A., Pouysségur J., Richard D. E. (2002) J. Biol. Chem. 277, 48403–48409 [DOI] [PubMed] [Google Scholar]
- 10.Lin X., David C. A., Donnelly J. B., Michaelides M., Chandel N. S., Huang X., Warrior U., Weinberg F., Tormos K. V., Fesik S. W., Shen Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belaiba R. S., Bonello S., Zähringer C., Schmidt S., Hess J., Kietzmann T., Görlach A. (2007) Mol. Biol. Cell 18, 4691–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H. L., Liu Y. N., Huang Y. T., Pan S. L., Huang D. Y., Guh J. H., Lee F. Y., Kuo S. C., Teng C. M. (2007) Oncogene 26, 3941–3951 [DOI] [PubMed] [Google Scholar]
- 13.Mottet D., Dumont V., Deccache Y., Demazy C., Ninane N., Raes M., Michiels C. (2003) J. Biol. Chem. 278, 31277–31285 [DOI] [PubMed] [Google Scholar]
- 14.Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., Schumacker P. T. (2000) J. Biol. Chem. 275, 25130–25138 [DOI] [PubMed] [Google Scholar]
- 15.Bonello S., Zähringer C., BelAiba R. S., Djordjevic T., Hess J., Michiels C., Kietzmann T., Görlach A. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 755–761 [DOI] [PubMed] [Google Scholar]
- 16.Febbraio M., Hajjar D. P., Silverstein R. L. (2001) J. Clin. Invest. 108, 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh U., Devaraj S., Jialal I. (2005) Annu. Rev. Nutr. 25, 151–174 [DOI] [PubMed] [Google Scholar]
- 18.Mwaikambo B. R., Sennlaub F., Ong H., Chemtob S., Hardy P. (2006) Invest. Ophthalmol. Vis. Sci. 47, 4356–4364 [DOI] [PubMed] [Google Scholar]
- 19.Sennlaub F., Valamanesh F., Vazquez-Tello A., El-Asrar A. M., Checchin D., Brault S., Gobeil F., Beauchamp M. H., Mwaikambo B., Courtois Y., Geboes K., Varma D. R., Lachapelle P., Ong H., Behar-Cohen F., Chemtob S. (2003) Circulation 108, 198–204 [DOI] [PubMed] [Google Scholar]
- 20.Yang C., Mwaikambo B. R., Zhu T., Gagnon C., Lafleur J., Seshadri S., Lachapelle P., Lavoie J. C., Chemtob S., Hardy P. (2008) Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R467–R476 [DOI] [PubMed] [Google Scholar]
- 21.Houssier M., Raoul W., Lavalette S., Keller N., Guillonneau X., Baragatti B., Jonet L., Jeanny J. C., Behar-Cohen F., Coceani F., Scherman D., Lachapelle P., Ong H., Chemtob S., Sennlaub F. (2008) PLoS Med. 5, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mwaikambo B. R., Sennlaub F., Ong H., Chemtob S., Hardy P. (2008) Cornea 27, 1037–1041 [DOI] [PubMed] [Google Scholar]
- 23.Ryeom S., Sparrow J., Silverstein R. (1996) J. Cell Sci. 108, 387–395 [DOI] [PubMed] [Google Scholar]
- 24.Finnemann S. C., Silverstein R. L. (2001) J. Exp. Med. 194, 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abumrad N. A., el-Maghrabi M. R., Amri E. Z., Lopez E., Grimaldi P. A. (1993) J. Biol. Chem. 268, 17665–17668 [PubMed] [Google Scholar]
- 26.Ibrahimi A., Bonen A., Blinn W. D., Hajri T., Li X., Zhong K., Cameron R., Abumrad N. A. (1999) J. Biol. Chem. 274, 26761–26766 [DOI] [PubMed] [Google Scholar]
- 27.Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. (1993) J. Biol. Chem. 268, 11811–11816 [PubMed] [Google Scholar]
- 28.Kwapiszewska G., Wilhelm J., Wolff S., Laumanns I., Koenig I. R., Ziegler A., Seeger W., Bohle R. M., Weissmann N., Fink L. (2005) Respir. Res. 6, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho S., Park E., Febbraio M., Anrather J., Park L., Racchumi G., Silverstein R., Iadecola C. (2005) J. Neuro. 25, 2504–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricciarelli R., Zingg J., Azzi A. (2000) Circulation 102, 82–87 [DOI] [PubMed] [Google Scholar]
- 31.Cho S., Szeto H. H., Kim E., Kim H., Tolhurst A. T., Pinto J. T. (2007) J. Biol. Chem. 282, 4634–4642 [DOI] [PubMed] [Google Scholar]
- 32.Sun M., Finnemann S. C., Febbraio M., Shan L., Annangudi S. P., Podrez E. A., Hoppe G., Darrow R., Organisciak D. T., Salomon R. G., Silverstein R. L., Hazen S. L. (2006) J. Biol. Chem. 281, 4222–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molday R. S., Hicks D., Molday L. (1987) Invest. Ophthalmol. Vis. Sci. 28, 50–61 [PubMed] [Google Scholar]
- 34.Sun K., Cai H., Tezel T. H., Paik D., Gaillard E. R., Del Priore L. V. (2007) Mol. Vis. 13, 2310–2319 [PubMed] [Google Scholar]
- 35.Greenberg M. E., Sun M., Zhang R., Febbraio M., Silverstein R., Hazen S. L. (2006) J. Exp. Med. 203, 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chabowski A., Górski J., Calles-Escandon J., Tandon N. N., Bonen A. (2006) FEBS Lett. 580, 3617–3623 [DOI] [PubMed] [Google Scholar]
- 37.Guzy R. D., Schumacker P. T. (2006) Exp. Physiol. 91, 807–819 [DOI] [PubMed] [Google Scholar]
- 38.Ward J. P. (2006) J. Appl. Physiol. 101, 993–995 [DOI] [PubMed] [Google Scholar]
- 39.Milne G., Musiek E., Morrow J. (2005) Biomarkers 10, (Suppl. 1) S10–S23 [DOI] [PubMed] [Google Scholar]
- 40.Morrow J. D., Awad J. A., Kato T., Takahashi K., Badr K. F., Roberts L. J., 2nd, Burk R. F. (1992) J. Clin. Invest. 90, 2502–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montuschi P., Barnes P. J., Roberts L. J., 2nd (2004) FASEB J. 18, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 42.Becker L., van den Hoek T., Shao Z., Li C., Schumacker P. (1999) Am. J. Physiol. 277, H2240–H2246 [DOI] [PubMed] [Google Scholar]
- 43.Goldberg M. A., Dunning S. P., Bunn H. F. (1988) Science 242, 1412–1415 [DOI] [PubMed] [Google Scholar]
- 44.An W. G., Kanekal M., Simon M. C., Maltepe E., Blagosklonny M. V., Neckers L. M. (1998) Nature 392, 405–408 [DOI] [PubMed] [Google Scholar]
- 45.Yeh W. L., Lu D. Y., Lin C. J., Liou H. C., Fu W. M. (2007) Mol. Pharmacol. 72, 440–449 [DOI] [PubMed] [Google Scholar]
- 46.Yeo E. J., Chun Y. S., Cho Y. S., Kim J., Lee J. C., Kim M. S., Park J. W. (2003) J. Natl. Cancer Inst. 95, 516–525 [DOI] [PubMed] [Google Scholar]
- 47.Yeo E. J., Chun Y. S., Park J. W. (2004) Biochem. Pharmacol. 68, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 48.Semenza G. L., Nejfelt M. K., Chi S. M., Antonarakis S. E. (1991) Proc. Natl. Acad. Sci. 88, 5680–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zampetaki A., Mitsialis S. A., Pfeilschifter J., Kourembanas S. (2004) FASEB J. 18, 1090–1092 [DOI] [PubMed] [Google Scholar]
- 50.Gerasimovskaya E. V., Tucker D. A., Stenmark K. R. (2005) J. Appl. Physiol. 98, 722–731 [DOI] [PubMed] [Google Scholar]
- 51.Finnemann S. C., Bonilha V. L., Marmorstein A. D., Rodriguez-Boulan E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Shabrawey M., Bartoli M., El-Remessy A. B., Platt D. H., Matragoon S., Behzadian M. A., Caldwell R. W., Caldwell R. B. (2005) Am. J. Pathol. 167, 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behn C., Araneda O. F., Llanos A. J., Celedón G., González G. (2007) Respir. Physiol. Neurobiol. 158, 143–150 [DOI] [PubMed] [Google Scholar]
- 54.Harris A. L. (2002) Nat. Rev. Cancer 2, 38–47 [DOI] [PubMed] [Google Scholar]
- 55.Jiménez B., Volpert O. V., Crawford S. E., Febbraio M., Silverstein R. L., Bouck N. (2000) Nat. Med. 6, 41–48 [DOI] [PubMed] [Google Scholar]
- 56.Strauss O. (2005) Physiol. Rev. 85, 845–881 [DOI] [PubMed] [Google Scholar]
- 57.Bazan H. E. (2005) Exp. Eye Res. 80, 453–463 [DOI] [PubMed] [Google Scholar]
- 58.Bedard K., Krause K. H. (2007) Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]
- 59.Boyer J. F., Balard P., Authier H., Faucon B., Bernad J., Mazières B., Davignon J. L., Cantagrel A., Pipy B., Constantin A. (2007) Arthritis Res. Ther. 9, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Q., Vazquez E. J., Moghaddas S., Hoppel C. L., Lesnefsky E. J. (2003) J. Biol. Chem. 278, 36027–36031 [DOI] [PubMed] [Google Scholar]
- 61.Gingras A. C., Kennedy S. G., O'Leary M. A., Sonenberg N., Hay N. (1998) Genes Dev. 12, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gingras A. C., Raught B., Sonenberg N. (1999) Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 63.Hay N., Sonenberg N. (2004) Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 64.Chachami G., Simos G., Hatziefthimiou A., Bonanou S., Molyvdas P. A., Paraskeva E. (2004) Am. J. Respir. Cell Mol. Biol. 31, 544–551 [DOI] [PubMed] [Google Scholar]
- 65.Carmeliet P., Jain R. K. (2000) Nature 407, 249–257 [DOI] [PubMed] [Google Scholar]
- 66.Folkman J. (1971) N. Engl. J. Med. 285, 1182–1186 [DOI] [PubMed] [Google Scholar]
- 67.Hanahan D., Folkman J. (1996) Cell 86, 353–364 [DOI] [PubMed] [Google Scholar]
- 68.Lejmi E., Leconte L., Pédron-Mazoyer S., Ropert S., Raoul W., Lavalette S., Bouras I., Feron J. G., Maitre-Boube M., Assayag F., Feumi C., Alemany M., Jie T. X., Merkulova T., Poupon M. F., Ruchoux M. M., Tobelem G., Sennlaub F., Plouët J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12491–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phelan M. W., Forman L. W., Perrine S. P., Faller D. V. (1998) J. Lab. Clin. Med. 132, 519–529 [DOI] [PubMed] [Google Scholar]
- 70.Favier J., Germain S., Emmerich J., Corvol P., Gasc J. M. (2005) J. Pathol. 207, 358–366 [DOI] [PubMed] [Google Scholar]