Timing of acquisition of deletion 13 in plasma cell dyscrasias is dependent on genetic context (original) (raw)
Monoclonal gammopathy of undetermined significance (MGUS) and smoldering myeloma (SMM) are characterized by an expansion of monoclonal plasma cells and can progress to symptomatic multiple myeloma. This study assessed the incidence and the association of monosomy 13 with IgH translocations, ploidy status and deletions of 16q23 and TP53 in a large series of MGUS and SMM patients.
Keywords: deletion 13, plasma cell dyscrasias, genetic context
Abstract
Background
Multiple myeloma, monoclonal gammopathy of undetermined significance and smoldering multiple myeloma harbor common chromosomal abnormalities but the prevalence and relative association of aberrations in these diagnostic groups remains controversial. We investigated these aspects in a large series of patients.
Design and Methods
Chromosome 13 deletion (Δ13), deletion of TP53, ploidy status and immunoglobulin heavy chain (IgH) translocations were evaluated by fluorescence in situ hybridization in patients with monoclonal gammopathy of undetermined significance (n=189), smoldering multiple myeloma (n=127) and multiple myeloma (n=400).
Results
Overall, Δ13 (25%, 34% and 47%), 16q23 deletions (6%, 8% and 21%) and 17p13 deletions (3%, 1% and 10%) were less frequent in patients with monoclonal gammopathy of undetermined significance and smoldering multiple myeloma than in those with multiple myeloma. When distinct genetic groups were considered, no differences in the prevalence of Δ13 were found with t(4;14)(p16;q32) and t(14;16)(q32;q23) among the three diagnostic groups; in contrast Δ13 was rarer in t(11;14)(q13;q32) in patients with monoclonal gammopathy (1/28) and smoldering myeloma (2/13) than in those with multiple myeloma (40%). Similar results were seen for the few t(6;14)(p21;q32) cases: 0/3 patients with monoclonal gammopathy or smoldering myeloma had the Δ13, whereas 4/6 (67%) patients with multiple myeloma and this translocation also had the deletion. In multiple myeloma patients with both an IgH translocation and Δ13, the proportions of cells affected by the two abnormalities were similar, as was the case for t(4;14) and t(14;16) monoclonal gammopathy patients positive for Δ13. In contrast, in monoclonal gammopathy patients with t(14;20)(q32;q11), the translocation was present in almost all cells, while the Δ13 was present in only a sub-population.
Conclusions
These results indicate that the presence and time of occurrence of Δ13 depends on the presence of specific concurrent abnormalities. The observation that Δ13 was extremely rare in monoclonal gammopathy of undetermined significance and smoldering multiple myeloma with translocations directly involving cyclin D genes (CCND1 and CCND3) suggest a possible role of Δ13 in the progression of the disease specifically in these genetic sub-groups. (clinicaltrials.gov identifier: ISRCTN 68454111; UKCRN ID 1176).
Introduction
Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM) are characterized by an expansion of monoclonal plasma cells and both can progress to symptomatic multiple myeloma (MM) or other related conditions.1–3 MGUS is the most frequent plasma cell disorder and its incidence increases markedly with age, reaching approximately 3% in subjects over 70 years old.1,3–5 MGUS is defined by a serum M-protein concentration of less than 30 g/L and fewer than 10% of plasma cells in the bone marrow, while patients with SMM meet the diagnostic criteria for MM but are asymptomatic. SMM resembles MGUS in that end-organ damage is absent, but clinically it is far more likely to progress to active MM or amyloidosis.3
The paucity of plasma cells within the bone marrow of patients with MGUS, together with the low proliferative capacity of these cells, has precluded significant karyotypic studies in these patients. Interphase fluorescence in situ hybridization (FISH) provides an alternative approach to investigate chromosomal aberrations in tumor cells from which metaphases are difficult to obtain. Using interphase FISH, chromosomal aberrations were consistently detected in a high proportion of MGUS patients, with roughly 50% of them carrying one of the primary IgH translocations and the remaining patients displaying a hyperdiploid karyotype. These findings suggested that ploidy status and_IgH_ rearrangements were early events delineating different pathogenic pathways.6–9
Conflicting results have been reported on the prevalence of deletion/monosomy 13 (Δ13) in MGUS. Avet-Loiseau et al. found a substantially lower frequency of this abnormality in MGUS (~25%) than in MM (~50%),10,11 while others reported a similar incidence in both conditions.7,12 Fonseca et al. also indicated that when Δ13 was detected in MGUS it occurred in the majority of clonal plasma cells,7 consistent with that normally observed in MM,13,14 while others reported a greater heterogeneity in MGUS.13 There has also been controversy regarding the prognostic significance of Δ13 in MM. This chromosomal aberration, detected by interphase FISH, was one of the first established genetic prognostic factors, independent of the mode of treatment.15 However, it now appears that the dismal prognosis previously thought to be conferred by the deletion is actually due to its association with other poor-risk genetic markers, such as specific primary IgH translocations or TP53 deletions,16,17 and that Δ13 on its own probably does not affect prognosis.
In this study, we used interphase FISH to assess the incidence and the association of Δ13 with IgH translocations, ploidy status and deletions of 16q23 and TP53 in a large series of MGUS and SMM patients. We compared the results with the frequencies found in a group of newly diagnosed MM patients in order to determine whether the patterns of chromosomal aberrations differ within the different diagnostic groups. We also explored the clonal heterogeneity of MGUS by comparing the frequencies of the different chromosomal aberrations detected in individual patients.10
Design and Methods
Patients
We evaluated a consecutive series of 716 patients with plasma cell disorders reported to the LRF UK Myeloma Forum Cytogenetic Database between November 2000 and January 2007. Samples were received from different centers throughout the UK with informed consent for cytogenetic/FISH analysis. The cohort consisted of 189 patients with MGUS (median age, 69 years; range, 36–92 years) and 127 with SMM (median age, 69 years; range, 31–89 years) not requiring therapy, and 400 newly diagnosed MM patients entered into the MRC Myeloma IX Trial (median age, 64 years; range, 30–89 years). Patients with an IgM heavy chain subtype were not eligible for this study, given the different biology of this disease.15 Patients were classified as having MGUS or SMM according to standard criteria18 and were required to have no evidence of organ damage indicative of MM. All but four SMM and eight MGUS patients were studied at diagnosis.
Cytogenetic testing
Density gradient centrifugation of bone marrow aspirates over Lymphoprep was performed to separate mononuclear cells by standard protocols. CD138+ plasma cells were isolated by magnetic-activated cell sorting using anti-CD138 immunobeads (Miltenyi Biotec Ltd., Bisley, UK). Interphase FISH was performed on plasma cells using a panel of commercial and in-house probes, as previously described.19,20 Results were available for Δ13, IgH rearrangements, t(4;14)(p16;q32), CCND3 break-apart (6p21), t(11;14)(q13;q32), t(14;16)(q32;q23), MAFB break-apart (20q11), deletion of TP53 (17p13) together with 17 centromere, and ploidy status. The interphase FISH method used to estimate ploidy and classify patients into groups with and without hyperdiploidy had been previously designed and assessed in MM patients,20 using a modification of the method described by Wuilleme et al.21 Break-apart patterns for the CCND3 or the MAFB probes in cases with a concomitant break-apart of the IgH probe were suggestive of t(6;14) and t(14;20), respectively. Cases with a suspected t(14;20) were tested using a fusion probe approach to confirm the presence of the translocation.
The cut-off levels for interphase FISH scoring recommended by the European Myeloma Network (EMN) FISH workshop (10% for fusion/break-apart probes and 20% for numerical abnormalities) were followed.
Statistical analysis
The frequencies of chromosomal aberrations in the groups of patients were compared by Fisher’s exact test or the Kruskall-Wallis test, as appropriate.
Results
Frequencies of chromosomal abnormalities
FISH analysis was performed according to the availability of patients’ material: a minimum of seven loci were tested (4p16, 5p15, CEP 9, 11q13, 13q14, 14q32 and CEP 15). In more than 80% of patients, 12 different loci were analyzed. We observed copy number changes or structural alterations for at least one of the chromosomal regions tested in 169/189 (89%) MGUS, 124/127 (98%) SMM and 396/400 (99%) MM.
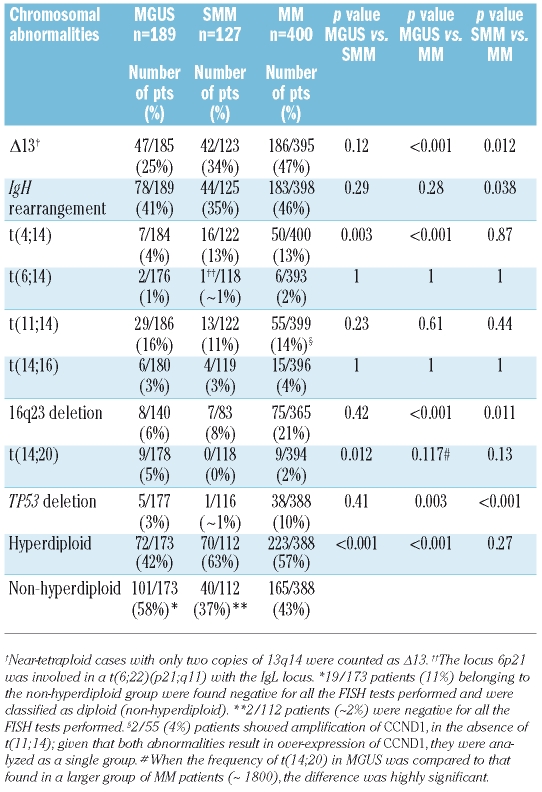
Table 1 summarizes the frequencies of the specific chromosomal aberrations within each diagnostic group. The incidence of Δ13 was substantially lower in MGUS (25%) and SMM (34%) than in MM (47%); the difference across the three groups was statistically highly significant (Kruskall-Wallis test: MGUS versus SMM versus MM,p<0.001).
Table 1.
Incidence of specific chromosomal abnormalities in the three groups of patients with MGUS, SMM and MM.
Rearrangements involving the IgH heavy chain locus located at 14q32 were detected with similar frequencies in MGUS and MM (41% and 46%, respectively); the incidence in SMM (35%) was lower, but the difference was not statistically significant. When the individual chromosomal partners of the primary translocations were considered, similar frequencies of t(6;14), t(11;14) and t(14;16) were observed among the three groups; t(4;14) was rare in MGUS (4%), but was observed at the same incidence in SMM and MM (13%) (MGUS versus MM,p<0.001). The frequency of t(14;20) was higher in MGUS (5%) than in either SMM (0%) or MM (2%). The difference in the incidence of this translocation was not statistically significant in this analysis, but became so when the incidence in the same MGUS group was compared to that in a larger population of MM patients (unpublished data). Deletion of 16q23 was found to be less common in MGUS (6%) and SMM (8%) than in MM (21%) (MGUS versus MM,p<0.001). Deletion of TP53 was also very rare in MGUS (3%) and SMM (~1%).
Patients with MGUS and SMM were classified as being hyperdiploid or not in the same way as previously described for MM.20 The distribution into the two ploidy classes differed between the diagnostic groups. A hyperdiploid karyotype was indicated in 63% of SMM and 57% of MM patients, while only 42% of MGUS cases were assigned to this category. The non-hyperdiploid MGUS group also included those patients found to be negative for all the interphase FISH tests performed; in MGUS these patients represented 11% of the total group and accounted for most of the difference between the groups.
IgH rearrangements involving the five recurrent loci (4p16, 6p21, 11q13, 16q23 and 20q11) were highly associated with a non-hyperdiploid karyotype in all three diagnostic groups: 94% of MGUS cases, 82% of SMM cases and 73% of MM cases with one of these_IgH_ translocations were found in the context of non-hyperdiploidy (p<0.001 for all diagnostic groups). In contrast 35 of 49 MM cases with an IgH rearrangement not involving one of these loci were found in association with hyperdiploidy (p_=0.043). In the SMM group the six unidentified IgH rearrangements were equally distributed between the two ploidy groups. In MGUS, 12 of 16 unidentified_IgH rearrangements were found in the context of a non-hyperdiploid karyotype but the association was not statistically significant (_p_=0.18).
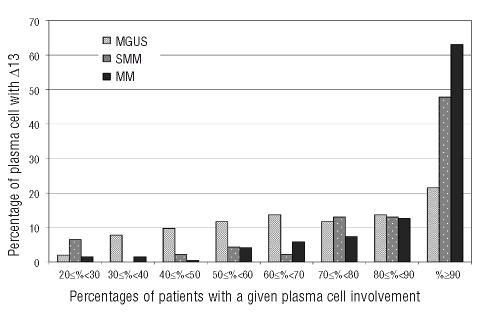
Percentage of plasma cells in patients with chromosome 13 deletion
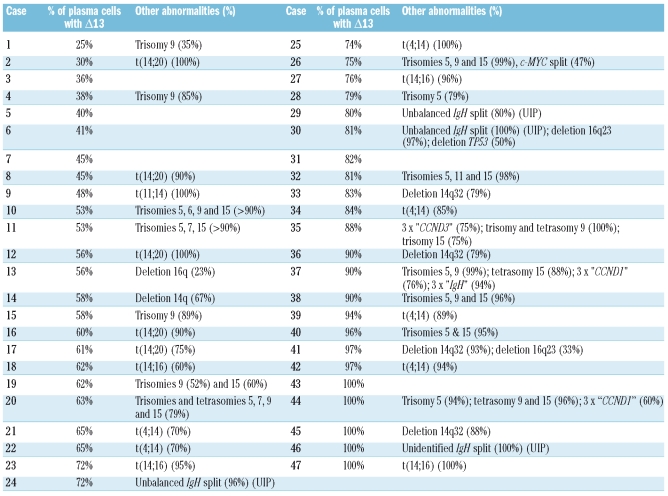
When present, Δ13 involved a variable proportion of plasma cells (Figure 1). The median percentage of plasma cells carrying the abnormality was 65% in MGUS, 88.5% in SMM and 95% in MM. No variation was seen for illegitimate IgH rearrangements: in patients with MGUS the median percentage of cells displaying a 14q32 translocation was 91.5% (range, 24%–100%). The level of plasma cell involvement by different chromosomal aberrations was compared for all MGUS patients who exhibited Δ13 with at least one other abnormality (Table 2). Because of the differences in false positive rates between probes, unequivocal evidence of heterogeneity within the neoplastic clone was only accepted when the difference in the proportions of cells affected by distinct chromosomal aberrations was 30% or more. In 16 of 47 patients with Δ13, the abnormality was present in 60% or less of plasma cells (cases 1–16 in Table 2); of these 16 cases, 12 had other chromosomal aberrations for comparison. In all but three of these 12, the plasma cell involvement by the non-Δ13 abnormality was at least 30% greater than that shown by Δ13.
Figure 1.
Distribution of the percentages of abnormal plasma cells with Δ13 in patients found positive for the abnormality, among the three groups of patients.
Table 2.
List of 47 MGUS patients with Δ13 ordered on the basis of the percentage of plasma cell involvement by the abnormality. For each case, concomitant numerical or structural abnormalities are specified with the percentage of plasma cell involvement (UIP, unidentified partner).
Interestingly five of six (83%) MGUS cases positive for both t(4;14) and Δ13 showed the same proportion (±5%) of plasma cells with the two abnormalities. In contrast, four of five (80%) cases of t(14;20) MGUS with Δ13 showed at least 30% fewer Δ13-positive plasma cells (median 51%) than t(14;20)-positive ones (median 95%). In MM, seven of nine (78%) cases of t(14;20) were associated with Δ13 and the median difference in plasma cell involvement by Δ13 and the translocation was 10% (range, 0–27%). In MGUS, 16q23 deletions were often present in a sub-clone of plasma cells (median, 63%; range, 23–100%), while in MM the median percentage of plasma cells with the abnormality was 87% (range, 21–100%). Those MGUS cases in which the proportion of plasma cells carrying the 16q deletion was small displayed at least one of the other chromosomal aberrations in the majority of clonal plasma cells (deletion 16q23 versus other chromosomal aberrations: 23%versus 82% IgH split; 33% versus 93% deletion 14q32; 44% versus 92% IgH split; 47% versus 99% IgH split).
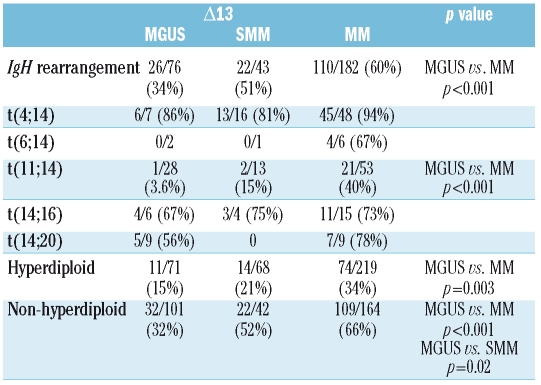
Association of chromosome 13 deletion with other abnormalities
Table 3 shows the association between Δ13 and other chromosomal aberrations. The Δ13 was less frequently associated with any of the IgH rearrangements in MGUS than in MM (MGUS versus MM, 34% versus 60%;p<0.001). However, when the individual translocations were examined, no differences were found in the frequencies of association of t(4;14), t(14;16), or t(14;20) with Δ13 among the three diagnostic groups.
Table 3.
Association between Δ13 and the different chromosomal abnormalties.
In MM, Δ13 was found in 40% of t(11;14) cases and involved a large proportion of plasma cells (>85%) while only one of 28 (3.6%) MGUS cases with t(11;14) had Δ13 (p<0.001). Furthermore, in this case, only 45% of the plasma cells had Δ13, while the translocation was present in all cells. In SMM only two of t(11;14) cases had Δ13 (15%), but this was not significantly different from the percentage found in MM. Both SMM patients had Δ13 in 70% of their plasma cells and the translocation in 100%. In this study, the presence of t(6;14) was detected in only 1–2% of patients, in agreement with other reported series. Despite the small number of cases, it was notable that Δ13 was present in four of the six (67%) MM cases, while in MGUS the two t(6;14) were both negative for Δ13 (in one of these two cases, 12% of the plasma cells carried the deletion).
Discussion
Our analysis of chromosomal aberrations in MGUS and SMM revealed a significantly lower frequency of Δ13 in the pre-malignant conditions than in MM. This is in accordance with findings reported by Avet-Loiseau et al.10 The frequency of Δ13 progressively increased from MGUS to SMM to MM, suggesting a possible role of this abnormality in disease progression. The incidence of deletion of 16q23 and_TP53_ also increased progressively from MGUS to MM (p<0.001 and _p_=0.003, respectively). In contrast, a similar frequency of IgH rearrangements was observed in the three groups. When the individual incidences of the specific translocations were compared, only t(4;14) was significantly less frequent in MGUS, in agreement with other reports.7,10
Interphase FISH was used to classify patients according to their ploidy status into those with hyperdiploidy and those without. Δ13 was found more frequently in patients with a non-hyperdiploid karyotype than in those with hyperdiploidy in all three groups of patients (MGUS, 15%versus 32%; SMM, 21%versus 52%; MM, 34% versus 66%), suggesting that the specific association between Δ13 and non-hyperdiploidy, extensively reported in MM,15,17 is already established at the stage of MGUS. These findings differ from those reported by Brousseau et al.22 In MGUS, they found Δ13 more frequently in patients with hyperdiploidy (11/29, 38%) than in those without (3/27, 11%), although the reverse association was seen in MM. They defined ploidy by measuring the plasma cell DNA content using the Feulgen reaction and image cytometry. We are unable to explain this discrepancy although the fact that ploidy was evaluated by two different methods may be partially responsible. Pseudodiploidy and low chromosome count hyperdiploidy (48–49 chromosomes) are potentially difficult to identify by interphase FISH, compared to true hypodiploidy or high chromosome count hyperdiploidy. However, the comparison of our interphase FISH ploidy results with actual karyotypes for those MGUS (n=8) and SMM (n=24) patients with abnormal cytogenetics showed that all cases but one were accurately classified. Thus interphase FISH misclassification of ploidy is unlikely to account for the significant difference in results between the two series.
Abnormalities of 14q32 were observed in the majority of clonal plasma cells, independently of the stage of the disease, whereas the percentages of plasma cells carrying Δ13 or the 16q23 deletion varied significantly between MGUS, SMM and MM, with MGUS patients showing the greatest heterogeneity. In MGUS, Δ13 was often present in a sub-clone of the abnormal plasma cells. Although low level clones in MGUS may be due to only a small proportion of the CD138-positive plasma cells being part of the neoplastic clone, our results indicate that, in these cases, the Δ13 is a later change following_IgH_ translocations or multiple trisomies. Similar findings were observed for most low level 16q23 deletions in which the cells were found to be 100% positive for other chromosomal aberrations.
Our results clearly show that the time of occurrence of specific abnormalities is crucially dependent on genetic context. The t(4;14), t(14;16) and t(14;20) are highly associated with Δ13 in MM.15,17,20 The same association was observed in MGUS and SMM patients. Moreover, in cases with t(4;14) and t(14;16),IgH rearrangement and Δ13 were found in a similar proportion of abnormal cells in the three diagnostic groups, suggesting that Δ13 occurred early in disease pathogenesis. However, a different time of occurrence of Δ13 was observed in relation to t(14;20). In MGUS, Δ13 appeared to originate later than t(14;20). A striking difference between MGUS and MM was seen regarding the association of Δ13 with t(11;14). While in MM 21/53 of t(11;14) cases also showed Δ13, in MGUS only 1/28 of cases with the translocation was associated with Δ13 (p<0.001). In MM the median percentage of plasma cells carrying the Δ13 in patients with t(11;14) was 98% while in the only case of MGUS with both t(11;14) and Δ13, all the plasma cells were positive for t(11;14) but only 48% of the plasma cells had Δ13. The translocation t(11;14) has been related to t(6;14) on the basis of a similar biological and clinical behavior.23 Both translocations directly activate a_cyclin_ D family member (CCND1 and_CCND3_, respectively) and gene expression profiling studies demonstrated that cases carrying either one or the other translocation exhibited dysregulation of similar transcriptional programs showing overlapping gene expression profiles.23,24 As for t(11;14), Δ13 was not found in the MGUS cases with t(6;14), while it was present in the majority of MM cases with this translocation.
These findings are consistent with those reported by Bochtler et al.,25 who applied an oncogenic tree model to study patients with amyloid light chain amyloidosis, MGUS and MM, in order to detect clustering of chromosomal abnormalities. Patients with amyloidosis and MGUS showed the t(11;14) branch independent of Δ13, while t(4;14), and gain of 1q21 were grouped together with Δ13. In our study, Δ13 occurred less frequently in SMM cases with t(11;14), but not to a degree that was statistically significantly different from the MM group.
Conclusions
In this study, we examined a range of numerical and structural chromosomal changes in MGUS, SMM and MM patients. None of the chromosomal aberrations tested was exclusive to a single diagnostic group, confirming the extensive overlap between the different conditions from a genetic point of view. However, statistically significant differences were observed in the incidence of specific abnormalities between the three conditions, in particular for Δ13, 16q23 deletion and TP53 deletion. In MGUS, the greatest variation in the proportion of abnormal plasma cells carrying the abnormality was seen for Δ13 and 16q23 deletion. In particular, the temporal appearance of Δ13 was related to the presence of specific concomitant abnormalities: early when t(4;14) or t(14;16) was present, later with t(14;20), and even later with t(11;14) or t(6;14). These data suggest a possible role of Δ13 in the transition from MGUS to MM specifically in cases with t(11;14) or t(6;14).
Acknowledgments
the authors would like to thank Paul Strike of the Salisbury District Hospital Research and Development Support Unit for help with the statistical analysis.
Footnotes
Funding: this work was supported by the Leukaemia Research Fund.
Authorship and Disclosures
LC designed and performed research, analyzed data and wrote the first draft of the paper; GPD, AHI, EDC, RKMP and DMS performed research; KHO, NCPC and CJH contributed to the analysis of the data; FMR designed and performed research, and analyzed data. All the authors contributed to the final draft of the paper. The authors reported no potential conflicts of interest.
References
- 1.Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med. 2002;346:564–9. doi: 10.1056/NEJMoa01133202. [DOI] [PubMed] [Google Scholar]
- 2.Kyle RA, Rajkumar SV. Monoclonal gammopathy of undetermined significance. Br J Haematol. 2006;134:573–89. doi: 10.1111/j.1365-2141.2006.06235.x. [DOI] [PubMed] [Google Scholar]
- 3.Kyle RA, Remstein ED, Therneau TM, Dispenzieri A, Kurtin PJ, Hodnefield JM, et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N Engl J Med. 2007;356:2582–90. doi: 10.1056/NEJMoa070389. [DOI] [PubMed] [Google Scholar]
- 4.Kyle RA. "Benign" monoclonal gammopathy–after 20 to 35 years of follow-up. Mayo Clin Proc. 1993;68:26–36. doi: 10.1016/s0025-6196(12)60015-9. [DOI] [PubMed] [Google Scholar]
- 5.Kyle RA, Rajkumar SV. Monoclonal gammopathies of undetermined significance. Hematol Oncol Clin North Am. 1999;13:1181–202. doi: 10.1016/s0889-8588(05)70120-9. [DOI] [PubMed] [Google Scholar]
- 6.Avet-Loiseau H, Li JY, Facon T, Brigaudeau C, Morineau N, Maloisel F, et al. High incidence of translocations t(11;14)(q13;q32) and t(4;14) (p16;q32) in patients with plasma cell malignancies. Cancer Res. 1998;58:5640–5. [PubMed] [Google Scholar]
- 7.Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA, et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood. 2002;100:1417–24. [PubMed] [Google Scholar]
- 8.Harrison CJ, Mazzullo H, Ross FM, Cheung KL, Gerrard G, Harewood L, et al. Translocations of 14q32 and deletions of 13q14 are common chromosomal abnormalities in systemic amyloidosis. Br J Haematol. 2002;117:427–35. doi: 10.1046/j.1365-2141.2002.03438.x. [DOI] [PubMed] [Google Scholar]
- 9.Chng WJ, Van Wier SA, Ahmann GJ, Winkler JM, Jalal SM, Bergsagel PL, et al. A validated FISH trisomy index demonstrates the hyperdiploid and nonhyperdiploid dichotomy in MGUS. Blood. 2005;106:2156–61. doi: 10.1182/blood-2005-02-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avet-Loiseau H, Facon T, Daviet A, Godon C, Rapp MJ, Harousseau JL, et al. 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Intergroupe Francophone du Myelome. Cancer Res. 1999;59:4546–50. [PubMed] [Google Scholar]
- 11.Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL, et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99:2185–91. doi: 10.1182/blood.v99.6.2185. [DOI] [PubMed] [Google Scholar]
- 12.Konigsberg R, Ackermann J, Kaufmann H, Zojer N, Urbauer E, Kromer E, et al. Deletions of chromosome 13q in monoclonal gammopathy of undetermined significance. Leukemia. 2000;14:1975–9. doi: 10.1038/sj.leu.2401909. [DOI] [PubMed] [Google Scholar]
- 13.Avet-Louseau H, Daviet A, Sauner S, Bataille R. Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br J Haematol. 2000;111:1116–7. doi: 10.1046/j.1365-2141.2000.02488.x. [DOI] [PubMed] [Google Scholar]
- 14.Fonseca R, Oken MM, Harrington D, Bailey RJ, Van Wier SA, Henderson KJ, et al. Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia. 2001;15:981–6. doi: 10.1038/sj.leu.2402125. [DOI] [PubMed] [Google Scholar]
- 15.Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546–58. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 16.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–95. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 17.Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007;20:571–96. doi: 10.1016/j.beha.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durie BG. Staging and kinetics of multiple myeloma. Semin Oncol. 1986;13:300–9. [PubMed] [Google Scholar]
- 19.Ross FM, Ibrahim AH, Vilain-Holmes A, Winfield MO, Chiecchio L, Protheroe RK, et al. Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia. 2005;19:1634–42. doi: 10.1038/sj.leu.2403857. [DOI] [PubMed] [Google Scholar]
- 20.Chiecchio L, Protheroe RK, Ibrahim AH, Cheung KL, Rudduck C, Dagrada GP, et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia. 2006;20:1610–7. doi: 10.1038/sj.leu.2404304. [DOI] [PubMed] [Google Scholar]
- 21.Wuilleme S, Robillard N, Lode L, Magrangeas F, Beris H, Harousseau JL, et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19:275–8. doi: 10.1038/sj.leu.2403586. [DOI] [PubMed] [Google Scholar]
- 22.Brousseau M, Leleu X, Gerard J, Gastinne T, Godon A, Genevieve F, et al. Hyperdiploidy is a common finding in monoclonal gammopathy of undetermined significance and monosomy 13 is restricted to these hyperdiploid patients. Clin Cancer Res. 2007;13:6026–31. doi: 10.1158/1078-0432.CCR-07-0031. [DOI] [PubMed] [Google Scholar]
- 23.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–8. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 24.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochtler T, Hegenbart U, Cremer FW, Heiss C, Benner A, Hose D, et al. Evaluation of the cytogenetic aberration pattern in amyloid light chain amyloidosis as compared with monoclonal gammopathy of undetermined significance reveals common pathways of karyotypic instability. Blood. 2008;111:4700–5. doi: 10.1182/blood-2007-11-122101. [DOI] [PubMed] [Google Scholar]