A Homozygous CARD9 Mutation in a Family with Susceptibility to Fungal Infections (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 29.
Published in final edited form as: N Engl J Med. 2009 Oct 29;361(18):1727–1735. doi: 10.1056/NEJMoa0810719
Abstract
BACKGROUND
Chronic mucocutaneous candidiasis may be manifested as a primary immunodeficiency characterized by persistent or recurrent infections of the mucosa or the skin with candida species. Most cases are sporadic, but both autosomal dominant inheritance and autosomal recessive inheritance have been described.
METHODS
We performed genetic studies in 36 members of a large, consanguineous five-generation family, in which 4 members had recurrent fungal infections and an additional 3 members died during adolescence, 2 after invasive infection of the brain with candida species. All 36 family members were enrolled in the study, and 22 had blood samples taken for DNA analysis. Homozygosity mapping was used to locate the mutated gene. In the 4 affected family members (patients) and the 18 unaffected members we sequenced CARD9, the gene encoding the caspase recruitment domain-containing protein 9_,_ carried out T-cell phenotyping, and performed functional studies, with the use of either leukocytes from the patients or a reconstituted murine model of the genetic defect.
RESULTS
We found linkage (lod score, 3.6) to a genomic interval on chromosome 9q, including CARD9. All four patients had a homozygous point mutation in CARD9, resulting in a premature termination codon (Q295X). Healthy family members had wild-type expression of the CARD9 protein; the four patients lacked wild-type expression, which was associated with low numbers of Th17 cells (helper T cells producing interleukin-17). Functional studies based on genetic reconstitution of myeloid cells from _Card9_−/− mice showed that the Q295X mutation impairs innate signaling from the antifungal pattern-recognition receptor dectin-1.
CONCLUSIONS
An autosomal recessive form of susceptibility to chronic mucocutaneous candidiasis is associated with homozygous mutations in CARD9.
Chronic mucocutaneous candidiasis is characterized by impaired clearance of fungal infections and results in colonization and infections of the mucosa or skin, predominantly with Candida albicans.1,2 A variety of clinical conditions, such as infection with the human immunodeficiency virus or the use of corticosteroids, favor the development of chronic mucocutaneous candidiasis, but the disease may also be a primary immunodeficiency arising from unknown genetic defects.1,3 In chronic mucocutaneous candidiasis, the most common infections are due to C. albicans; however, patients may also have an increased susceptibility to dermatophytes.1,3 Severe complications rarely develop in patients with chronic mucocutaneous candidiasis, although reports on invasive infections with candida species — Cryptococcus neoformans or Histoplasma capsulatum — have been published.4–6
Research conducted in the eight decades since the first report on primary chronic mucocutaneous candidiasis appeared7 has shown that it is a heterogeneous syndrome that may be accompanied by endocrine and inflammatory disorders, including hypothyroidism and adrenocortical failure. 2 Most cases of chronic mucocutaneous candidiasis are sporadic, but multiplex families with dominant8–12 and recessive13,14 inheritance have been described.
Recurrent and severe candidiasis can have a defining role in primary immunodeficiencies. The autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) syndrome is caused by biallelic mutations in AIRE, the autoimmune regulator gene. Heterozygous mutations in the signal transducer and activator of transcription 3 gene (STAT3) cause the hyper-IgE syndrome, another multisystem disorder in which candidiasis is a common clinical feature.17–20 Genetic linkage of an autosomal dominant candidiasis–thyroiditis syndrome to chromosome 2 has been reported,12 and candidiasis associated with a low expression of intercellular adhesion molecule 1 (ICAM-1) has been traced to chromosome 11,21 but in both cases the causative genes remain unknown.
We undertook genetic studies of a large, consanguineous Iranian family with multiple cases of chronic mucocutaneous candidiasis to determine whether a mutated gene was associated with this form of nonsyndromic candidiasis. Recent work has shown that innate antifungal immunity in mice is controlled by a signaling pathway that does not involve toll-like receptors. Mice lacking either the C-type lectin receptor dectin-1 (encoded by the Clec7a gene) or the intracellular adapter molecule Card9, which is essential for dectin-1 signaling, have impaired antifungal immunity.22,23
We identified a homozygous mutation in CARD9 that results in a loss-of-function mutation due to a premature stop codon in the coding sequence. Experiments in the murine _Card9_−/− model showed that only wild-type CARD9 — not the mutated human CARD9 gene found in patients — could restore cytokine production in response to the triggering of dectin-1, a pattern-recognition receptor for fungal cell-wall antigens.
METHODS
STUDY PARTICIPANTS
We enrolled 36 members of a large, consanguineous Iranian family in the study. Blood samples were obtained for DNA analysis, and the participants were classified as likely to be affected or likely to be unaffected according to the results of physical examination and microbiologic culture. Laboratory personnel were unaware of the classification of the samples. The participants provided written informed consent on forms approved by local ethics committees. DNA samples from 50 healthy Iranian donors and 180 healthy white donors of other nationalities were used as controls.
GENOTYPING AND ANALYSIS
DNA samples from five family members deemed likely to be affected and eight deemed likely to be unaffected were genotyped with use of the Affymetrix 250k _Nsp_I single-nucleotide-polymorphism (SNP) mapping array, as described previously.24 Genotypes of the SNP arrays were assigned with the use of the Bayesian Robust Linear Modeling and Mahalanobis (BRLMM) distance method, which was implemented as described in the Genotyping Console software and introduced as an improvement over the RLMM method.25
To further evaluate one region on chromosome 9 that was suggestive of linkage, four microsatellite markers were genotyped on 13 available samples, as described previously.26 Single-marker lod scores were computed with Fastlink27–29; multipoint lod scores were computed with Superlink.30,31 (For details on the use of the BRLMM method, Fastlink, and Superlink, see the Supplementary Appendix, available with the full text of this article at NEJM.org.)
MOLECULAR GENETICS AND T-CELL PHENOTYPING
DNA from the study participants was isolated, and coding regions of CARD9 (Ensembl number, ENSG00000187796) were amplified and sequenced. Total RNA was isolated and transcribed into complementary DNA (cDNA). SYBR green-based real-time polymerase-chain-reaction quantification of CARD9 was performed with the use of standard curves. Screening for the Q295X mutation with the use of restriction-enzyme digestion was carried out in heterozygous and homozygous healthy family members as well as in healthy controls.
T-cell phenotyping, regulatory T-cell staining, and detection of Th17 cells (helper T cells producing interleukin-17) were performed in accordance with protocols published previously.32–36 Extracts of peripheral-blood mononuclear cells from patients with a deficiency of the CARD9 protein were stained with a polyclonal goat anti-CARD9 antibody. (Details of these procedures are available in the Supplementary Appendix.)
RETROVIRAL TRANSDUCTION OF HUMAN CARD9 VARIANTS
CARD9 cDNA was generated from human peripheral-blood mononuclear cells and cloned into a retroviral expression vector (based on murine stem-cell virus) expressing green fluorescent protein.37 The Q295X mutation was introduced with the use of site-directed mutagenesis. Retroviruses were generated by transfecting the Phoenix ecotropic-packaging cell line and were used to infect bone marrow cells as described previously.37 Bone marrow cells were differentiated into macrophages and stimulated with either curdlan, a selective dectin-1 agonist, or ultrapure lipopolysaccharide. Concentrations of tumor necrosis factor α (TNF-α) in the supernatants were analyzed with the use of an enzyme-linked immunosorbent assay.22,23,38
RESULTS
PATIENTS’ MEDICAL HISTORY
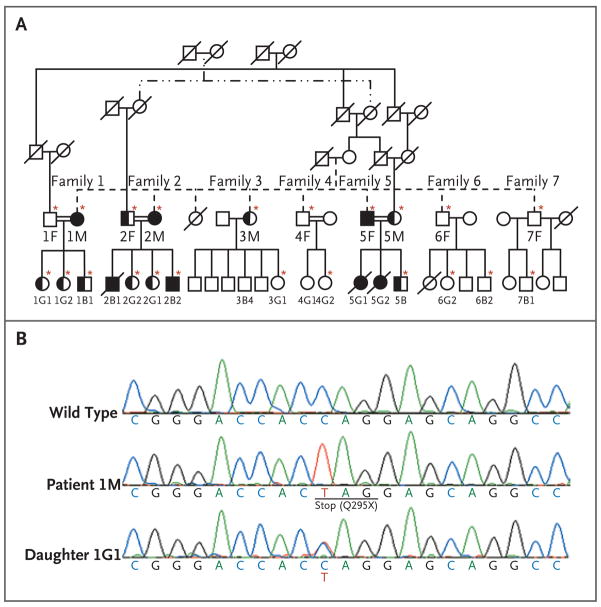
The pedigree (Fig. 1A) for the consanguineous Iranian family shows that multiple members were affected by chronic mucocutaneous candidiasis through a presumably autosomal recessive mode of inheritance. Recurrent fungal infections were diagnosed clinically in eight family members, three of whom died in early adolescence — two with proven and one with presumed invasive candida infection of the brain. None of the eight infected patients had unusual bacterial or viral infections, suggesting that the host defense against these pathogens was normal.
Figure 1. Pedigree of an Iranian Family with Chronic Mucocutaneous Candidiasis.
In Panel A, circles denote female family members; squares male family members; solid circles and squares patients with chronic mucocutaneous candidiasis, who were homozygous for the Q295X mutation; half-solid circles and squares members who were heterozygous for the Q295X mutation; open circles and squares healthy members with wild-type CARD9, and double horizontal lines consanguinity in a married couple. A slash denotes a deceased family member. Asterisks indicate family members whose samples were sequenced. In Panel B, the sequence at the top is for a healthy family member with wild-type CARD9. The middle sequence, obtained from Patient 1M, is characteristic of a person with a homozygous CARD9 mutation, in which a single-nucleotide exchange (C→T) in exon 6 of CARD9 results in a premature stop codon (Q295X). The bottom sequence, from Patient 1G1, is characteristic of a healthy heterozygous person.
The index patient (Patient 2B2) was a 19-year-old man who had had oral candidiasis (thrush) since the age of 3 years. Candida infection was confirmed with the use of microbiologic testing, and prophylaxis with ketoconazole was ongoing. He was otherwise healthy.
Patient 2B1, a sibling of Patient 2B2, had had intermittent thrush since early childhood. Seizures developed suddenly, with loss of consciousness, when he was 18 years old. Hydrocephalus developed, and he died of candida meningitis at the age of 19 years.
Patient 2M, the mother of Patients 2B1 and 2B2, was 50 years old at the time of our study and had had vaginal candidiasis since the age of 42 years. In addition to being infected with C. albicans, she had a 5-year history of dermatophytosis of her hands and neck. She also had intermittent aphthous lesions, type 2 diabetes mellitus, and nephrolithiasis.
Patient 1M, the sister of Patient 2M, had had oral and vaginal candidiasis since early childhood. She also had tinea corporis on her chest and neck. She was otherwise healthy.
Patient 5F, the affected brother of Patients 1M and 2M, had had dermatophytosis since childhood, with little improvement in response to local treatment. Both of his daughters were given a postmortem diagnosis of invasive candida infection.
The older daughter (Patient 5G1) had a ventricular septal defect during infancy and had a geographic tongue, which is suggestive of chronic candidiasis. A unilateral paresthesia developed when she was 13 years old, and she died from what was vaguely defined as a brain tumor, with severe skull destruction, at the age of 15 years.
Her sister (Patient 5G2) had recurrent, severe, refractory thrush starting in early childhood. At 15 years of age, a severe headache and fevers developed, followed by diplopia. A brain tumor was suspected, but candida meningoencephalitis was identified during surgery. She died 6 months after the onset of symptoms.
None of the patients with invasive fungal infections had a condition or took any medication that predisposed them to infection. The three deceased patients could not be enrolled in the study because of the lack of available tissue samples.
Patient 1B1, a child of Patient 1M and a cousin of the index patient (Patient 2B2), had had one mild episode of candida infection in adulthood. In our genetic analysis, we considered Patient 1B1 to be affected. We found that the episode of infection in Patient 1B1 was the result of a phenocopy, which is consistent with the fact that it was clinically much milder than the infections in all the other affected family members (see the Supplementary Appendix for the definition of phenocopy).
CELL COUNTS AND T-CELL PHENOTYPING
In Patients 1M, 2M, and 5F, complete blood counts were within the normal range, as were total counts of CD3+ T cells, CD4+ T cells, CD8+ T cells, memory T cells, follicular helper T cells, effector memory T cells, regulatory T cells, B cells, and natural killer cells (for results see Table S1 in the Supplementary Appendix). Basal levels of serum immunoglobulin were also normal. In Patient 2B2, the results of a delayed-type hypersensitivity skin test were negative for tuberculin but positive for candida.
AIRE was shown to be of wild-type sequence in Patient 2B2. The absence of a specific immunologic disorder led us to use a positional cloning approach to identify the patients’ underlying genetic defect.
GENETIC LINKAGE ANALYSIS
Analysis of the SNP genotypes showed a region of perfect segregation on chromosome 9 (137.5 to 138.8 Mbp in human genome build 36), provided that Patient 1B1’s episode of candida infection was the result of a phenocopy. This finding was confirmed by genotyping four microsatellite markers, yielding a peak multipoint lod score of 3.6 (see the Supplementary Appendix).
There were 121 genes in the maximal linkage interval defined by the microsatellite markers D9S2157 (135.0 Mbp) and D9S1838 (139.8 Mbp). Among these 121 genes, 41 are located in the perfectly segregating 1.3-Mbp subinterval suggested by the SNP data. After exploring the literature, we identified CARD9 — which is among the 41 ideally located genes (see Table S2 in the Supplementary Appendix) — as a functional candidate because _Card9_−/− mice are susceptible to fungal infections (for details, see the Supplementary Appendix). 22,39
HOMOZYGOUS MUTATIONS IN CARD9
We sequenced CARD9 in the four affected patients and 18 other relatives and identified in all affected persons a single homozygous point mutation from C to T in exon 6 at codon 295, resulting in a premature termination codon (Q295X). Patient 1B1 and his healthy relatives had either a heterozygous Q295X mutation or wild-type alleles only (Fig. 1B).
To assess the frequency of this previously unknown genetic abnormality in CARD9 and to exclude the possibility of a genetic variation, we examined the affected site in 50 unrelated healthy Iranians and 180 unrelated healthy white subjects by means of a sequencing assay or a restriction-enzyme assay. None of the 230 controls had the Q295X mutation in CARD9.
CARD9 mRNA LEVELS AND PROTEIN EXPRESSION
Among CARD9 wild-type cells, average levels of CARD9 mRNA were highest in monocytes, followed by granulocytes, B cells and T cells, and the colon-cell line HT-29. The peripheral-blood mononuclear cells from our patients still had substantial levels of mutated CARD9 mRNA, thereby escaping nonsense-mediated RNA decay (see the Supplementary Appendix).
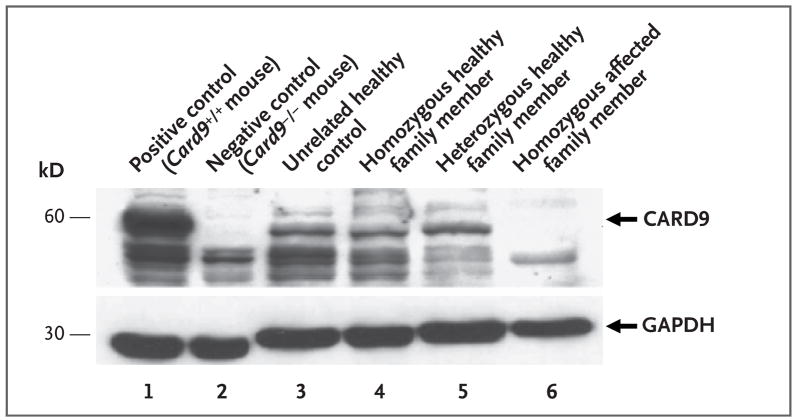
To examine the effect of the CARD9 Q295X mutation at the protein level, we assessed the expression of CARD9 in peripheral-blood mononuclear cells from the patients, using Western blotting. As compared with unrelated healthy controls and homozygous or heterozygous healthy family members, patients with the homozygous Q295X mutation completely lacked expression of the wild-type CARD9 protein (Fig. 2), indicating the detrimental consequences of the mutation. However, expression of the truncated CARD9 (amino acid positions 1 through 294) protein cannot be ruled out, since the polyclonal antibody used is directed against the C-terminal part of CARD9.
Figure 2. Western Blot Detection of CARD9 in Peripheral-Blood Mononuclear Cells.
Macrophages from Card9 wild-type mice (Card9+/+) and Card9 knockout mice (Card9−/−) were used as positive and negative controls (lanes 1 and 2), respectively. The blot in lane 4 is from Relative 4F, that in lane 5 from Relative 2G2, and that in lane 6 from Patient 2M. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control.
EFFECT OF CARD9 Q295X ON SIGNAL TRANSDUCTION
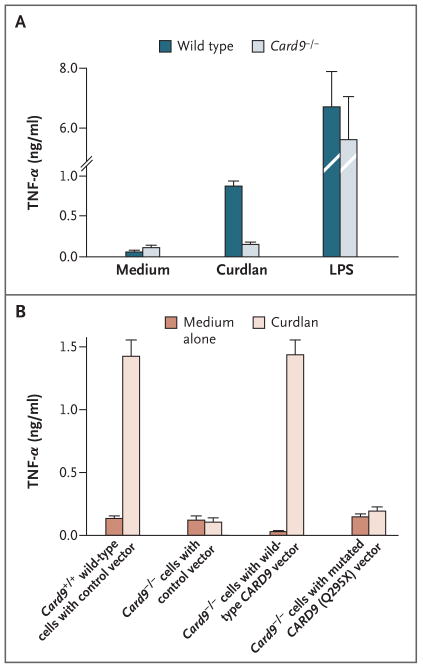
Primary bone marrow cells from Card9-deficient mice were retrovirally transduced with human wild-type and mutated (Q295X) CARD9, differentiated into macrophages in vitro, and analyzed with the use of flow cytometry (see the Supplementary Appendix). We then stimulated the transduced or nontransduced cells with the _β_-glucan preparation curdlan as a specific and selective agonist for dectin-1 or with the TLR4 ligand lipopolysaccharide and measured TNF-α production to test innate immune cell activation.38 As is consistent with previous data, Card9−/− cells showed severe defects in dectin-1–triggered TNF-α production, although they responded normally to stimulation with lipopolysaccharide (Fig. 3A).22 Expression of human full-length CARD9 corrected the dectin-1 signaling defect in Card9−/− cells, indicating that the human protein can complement the murine mutation. Expression of the human mutant CARD9 Q295X did not increase TNF-α production, on stimulation with dectin-1, above the level in uninfected cells or those transduced with green fluorescent protein only, showing that CARD9 Q295X is a loss-of-function mutation (Fig. 3B).
Figure 3. Analysis of the Functional Defect and Genetic Reconstitution of the Card9 Q295X Mutation.
Bone-marrow–derived macrophages from wild-type mice and _Card9_−/− mice were stimulated for 16 hours with both the dectin-1 agonist curdlan (300 _μ_g per milliliter) and the toll-like-receptor agonist lipopolysaccharide (LPS, 100 ng per milliliter) (Panel A). The concentration of secreted tumor necrosis factor (TNF-α) was determined in the cell supernatant with the use of an enzyme-linked immunosorbent assay. To examine the effect of mutated CARD9 Q295X on signal transduction (Panel B), we cloned human wild-type CARD9 complementary DNA (cDNA) and mutated CARD9 Q295X cDNA into a retroviral expression vector. Using these constructs, we retrovirally transduced primary bone marrow cells from _Card9_−/− mice with either wild-type CARD9 or the mutated CARD9 Q295X. To establish a control, we transduced bone marrow cells from Card9+/+ wild-type mice and _Card9_−/− mice with a control vector only. The transduced bone marrow cells were then differentiated into macrophages in vitro. After stimulation of the macrophages with curdlan for 16 hours, the concentration of secreted TNF-α was determined. The macrophages of _Card9_−/− mice transduced with human wild-type CARD9 secreted as much TNF-α upon stimulation with curdlan as did the control macrophages of wild-type Card9+/+ mice with the control vector. In contrast with these cells, the macrophages of _Card9_−/− mice transduced with mutated human CARD9 Q295X or with a control vector only did not respond with increased secretion of TNF-α upon stimulation with curdlan, a finding showing that CARD9 Q295X is a loss-of-function mutation that cannot correct the dectin-1/Card9 signaling pathway in the cells of _Card9_−/− mice. T bars indicate standard deviations in three independent experiments.
HOMOZYGOUS Q295X MUTATIONS AND TH17 CELLS
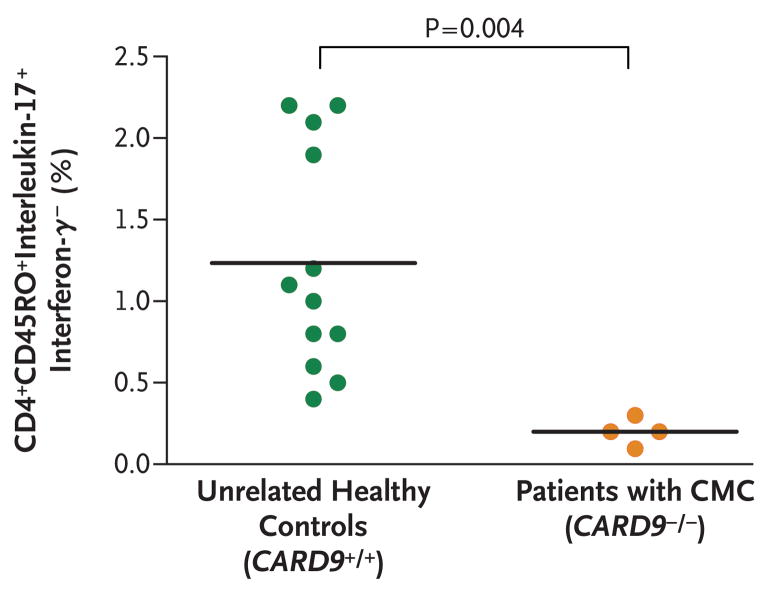
Since Th17 cells40 are important for antifungal immunity and _Card9_−/− mice have an impairment in Th17 polarization,22,38 we compared Th17 cells in our four patients with those in family members with wild-type CARD9 and nine healthy controls. The mean proportion of Th17 cells in the four affected patients was significantly lower than that in healthy controls (mean, 0.2% of CD4+CD45RO+ interleukin-17+interferon-_γ_− cells; P = 0.004) (Fig. 4). Healthy controls and family members with wild-type CARD9 had an average of 1.2% Th17 cells ex vivo.
Figure 4. Proportion of Interleukin-17–Producing CD4+CD45RO+ Cells That Were Negative for Interferon-γ Production in Four Patients with a Homozygous Mutation in CARD9 as Compared with Unrelated Healthy Controls.
Cells were surface-stained for CD4 and CD45RO and then subjected to intracellular staining for interleukin-17 and interferon-γ. CMC denotes chronic mucocutaneous candidiasis.
DISCUSSION
Pattern-recognition receptors of the innate immune system bind components of microbes and initiate intracellular signal cascades that result in the activation of transcription factors, up-regulation of defense-associated target genes, and release of cytokines. Dectin-1 is a transmembrane pattern-recognition receptor that senses the _β_-glucan component of fungal cell walls.23,41–43 On ligand binding, dectin-1 sends signals through an immunoreceptor tyrosine-based activation motif (ITAM), which becomes phosphorylated by Src family kinases (proto-oncogenic tyrosine kinases), leading to the recruitment and activation of the spleen tyrosine kinase (Syk).44,45
Dectin-1–Syk engages CARD9, which together with B-cell leukemia–lymphoma 10 (BCL10) and mucosa-associated lymphoid tissue 1 (MALT1) forms an intracellular signaling complex that in cells recognizing fungi leads to the activation of the transcription factor nuclear factor κ_B and mitogen-activated protein kinases.22,46–48 This signaling pathway is operative in myeloid cells and promotes the production of key cytokines, including interleukin-1_β, interleukin-6, and interleukin-23, which are required to control antifungal immune responses.24,38,49–52 Apart from dectin-1, the C-type lectins dectin-2 and macrophage-inducible C-type lectin (MINCLE) may also recognize fungi, engage the ITAM adapter FcR_γ_ for Syk activation, and transmit signals through the CARD9 pathway. 23,53–55 Thus, CARD9 plays a central role in antifungal defense by receiving signals from several antifungal pattern-recognition receptors and stimulating proinflammatory responses. Since murine Card9 deficiency results in susceptibility to fungal infections,22,39 this signaling pathway seems to be conserved between mice and humans.
Our study shows that a homozygous point mutation in CARD9, resulting in a premature termination codon and a loss of function in the adapter protein CARD9, is associated with a susceptibility to fungal infections, as evidenced by a chronic mucocutaneous candidiasis phenotype. In the family in our study, two members died from a fungal infection and a third presumably died from a similar cause. Further studies may clarify whether human CARD9 deficiency accounts only for recurrent mucosal infections or also accounts for an increased susceptibility to severe invasive fungal infections. In this consanguineous family, we cannot exclude the possibility that a second genetic defect may have contributed to a more severe phenotype in the deceased family members.
Unfortunately, we were unable to study viable cells from the family members in vitro because of logistical constraints. However, to understand the function of the human mutated CARD9 gene, we used an in vivo model with cells from _Card9_−/− mice and showed that the truncated human CARD9 protein fails to correct the dectin-1 signaling defect. In contrast, the human wild-type CARD9 protein restores the dectin-1–Card9 pathway in murine Card9−/− macrophages.
In _Card9_−/− mice, stimulation of dendritic cells with the cell-wall component zymosan or whole C. albicans cells results in a considerable reduction in the release of cytokines, including interleukin-2, interleukin-6, interleukin-10, and TNF-α, and decreased numbers of Th17 cells, which are implicated in adaptive antifungal immunity.22,38 All _CARD9_−/− patients had significantly reduced numbers of Th17 cells, further supporting the notion that CARD9-mediated signaling contributes to Th17-cell differentiation. Th17 cells and their production of interleukin-17 have been shown to play a pivotal role in mucosal host defense against candidiasis in mice.56,57 Moreover, Eyerich et al. reported decreased numbers of Th17 cells in two sporadic cases of chronic mucocutaneous candidiasis, 58 but the role of these cells in human anti-fungal immunity remains elusive. If the lack of Th17 cells and their cytokines were critical for the pathogenesis of mucosal candidiasis, one could speculate that in patients with a low total CD4 count, such as in the low-CD4 syndrome, another rare primary immunodeficiency, or in patients with the acquired immunodeficiency syndrome, the lack of CD4 differentiation into Th17 cells is critical for maintaining the mucosal host defense against candida. Patients with the hyper-IgE syndrome, who lack Th17 cells because of heterozygous mutations in STAT3, also have recurrent candidiasis.18,20 Whether Th17 cells are also implicated in the pathogenesis of candidiasis in APECED is currently being studied. The phenotype of susceptibility to fungal infections in human CARD9 deficiency serves as another example of a rare primary immunodeficiency that gives insight into the signaling pathways involved in immune regulation.
Supplementary Material
Supplementary Appendix
Supplementary Figures and Legends
Acknowledgments
Supported in part by a European Commission Marie Curie Excellence Grant (MEXT-CT-2006-042316); the young investigator award of 2008 from the European Society for Immunodeficiencies; the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine; a Max Eder Program Grant from Deutsche Krebshilfe (to Dr. Ruland); and Sonderforschungsbereich grants from the Deutsche Forschungs-gemeinschaft (to Dr. Ruland).
We thank Judy Levin and Charlotte Holden of the NIH for administrative support and encouragement and Dr. Hans Stauss for his critical reading of an earlier version of this article.
APPENDIX
The authors’ affiliations are as follows: the Department of Immunology and Molecular Pathology, Royal Free Hospital and University College London, London (E.-O.G., C.W., F.T., S.J., A.M., B.G.); the Departments of Rheumatology and Clinical Immunology (A.H., U.S., K.W.) and Hematology and Oncology (D.P., H.V.), University Hospital Freiburg, Freiburg, Germany; Semnan University of Medical Science, Semnan, Iran (M.N.); the National Center for Biotechnology Information, National Institutes of Health, Department of Health and Human Services, Bethesda, MD (A.A.S.); the Growth and Development Research Center, Center of Excellence for Pediatrics, Children’s Medical Center (N.R.), and the Immunogenetic Laboratory, Department of Immunology (A.A.A.), School of Medicine, Tehran University of Medical Sciences, Tehran, Iran; Clinica Pediatrica, Università di Brescia and Istituto Medicina Molecolare Angelo Nocivelli, Spedali Civili, Brescia, Italy (A.P.); Medizinische Klinik 3, Klinikum rechts der Isar, Technische Universität München, Munich, Germany (N.H., O.G., J.R.); and the Laboratory of Signaling in the Immune System, Helmholtz Zentrum München — German Research Center for Environmental Health, Neuherberg, Germany (J.R.)
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Kirkpatrick CH. Chronic mucocutaneous candidiasis. Pediatr Infect Dis J. 2001;20:197–206. doi: 10.1097/00006454-200102000-00017. [DOI] [PubMed] [Google Scholar]
- 2.Lilic D. New perspectives on the immunology of chronic mucocutaneous candidiasis. Curr Opin Infect Dis. 2002;15:143–7. doi: 10.1097/00001432-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/s1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 4.Drouhet E, Dupont B. Chronic mucocutaneous candidosis and other superficial and systemic mycoses successfully treated with ketoconazole. Rev Infect Dis. 1980;2:606–19. doi: 10.1093/clinids/2.4.606. [DOI] [PubMed] [Google Scholar]
- 5.Kauffman CA, Shea MJ, Frame PT. Invasive fungal infections in patients with chronic mucocutaneous candidiasis. Arch Intern Med. 1981;141:1076–9. [PubMed] [Google Scholar]
- 6.van ’t Wout JW, de Graeff-Meeder ER, Paul LC, Kuis W, van Furth R. Treatment of two cases of cryptococcal meningitis with fluconazole. Scand J Infect Dis. 1988;20:193–8. doi: 10.3109/00365548809032437. [DOI] [PubMed] [Google Scholar]
- 7.Thorpe ES, Handley HE. Chronic tetany and chronic mycelial stomatitis in a child aged four and one-half years. Am J Dis Child. 1929;38:328–38. [Google Scholar]
- 8.Canales L, Middlemas RO, III, Louro JM, South MA. Immunological observations in chronic mucocutaneous candidiasis. Lancet. 1969;2:567–71. doi: 10.1016/s0140-6736(69)90264-5. [DOI] [PubMed] [Google Scholar]
- 9.Kroll JJ, Einbinder JM, Merz WG. Mucocutaneous candidiasis in a mother and son. Arch Dermatol. 1973;108:259–62. [PubMed] [Google Scholar]
- 10.Sams WM, Jr, Jorizzo JL, Snyderman R, et al. Chronic mucocutaneous candidiasis: immunologic studies of three generations of a single family. Am J Med. 1979;67:948–59. doi: 10.1016/0002-9343(79)90635-1. [DOI] [PubMed] [Google Scholar]
- 11.Loeys BL, Van Coster RN, Defreyne LR, Leroy JG. Fungal intracranial aneurysm in a child with familial chronic mucocutaneous candidiasis. Eur J Pediatr. 1999;158:650–2. doi: 10.1007/s004310051169. [DOI] [PubMed] [Google Scholar]
- 12.Atkinson TP, Schäffer AA, Grimbacher B, et al. An immune defect causing dominant chronic mucocutaneous candidiasis and thyroid disease maps to chromosome 2p in a single family. Am J Hum Genet. 2001;69:791–803. doi: 10.1086/323611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells RS, Higgs JM, Macdonald A, Valdimarsson H, Holt PJ. Familial chronic muco-cutaneous candidiasis. J Med Genet. 1972;9:302–10. doi: 10.1136/jmg.9.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahonen P, Myllarniemi S, Sipila I, et al. Clinical variation of autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322:1829–36. doi: 10.1056/NEJM199006283222601. [DOI] [PubMed] [Google Scholar]
- 15.The Finnish-German APECED Consortium. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 16.Nagamine K, Peterson P, Scott HS, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–8. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 17.Minegishi Y, Saito M, Tsuchiya S, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 18.Holland SM, DeLeo FR, Elloumi HZ, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 19.Van Scoy RE, Hill HR, Ritts RE, Quie PG. Familial neutrophil chemotaxis defect, recurrent bacterial infections, mucocutaneous candidiasis, and hyperimmunoglobulinemia E. Ann Intern Med. 1975;82:766–71. doi: 10.7326/0003-4819-82-6-766. [DOI] [PubMed] [Google Scholar]
- 20.Grimbacher B, Holland SM, Gallin JI, et al. Hyper-IgE syndrome with recurrent infections — an autosomal dominant multisystem disorder. N Engl J Med. 1999;340:692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 21.Mangino M, Salpietro DC, Zuccarello D, et al. A gene for familial isolated chronic nail candidiasis maps to chromosome 11p12–q12.1. Eur J Hum Genet. 2003;11:433–6. doi: 10.1038/sj.ejhg.5200985. [DOI] [PubMed] [Google Scholar]
- 22.Gross O, Gewies A, Finger K, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–6. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 23.Taylor PR, Tsoni SV, Willment JA, et al. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–8. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeifer D, Woellner C, Petersen A, et al. The hyper-IgE syndrome is not caused by a microdeletion syndrome. Immunogenetics. 2007;59:913–26. doi: 10.1007/s00251-007-0257-z. [DOI] [PubMed] [Google Scholar]
- 25.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 26.Finck A, Van der Meer JWM, Schäffer AA, et al. Linkage of autosomal-dominant common variable immunodeficiency to chromosome 4q. Eur J Hum Genet. 2006;14:867–75. doi: 10.1038/sj.ejhg.5201634. [DOI] [PubMed] [Google Scholar]
- 27.Lathrop GM, Lalouel J-M, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984;81:3443–6. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cottingham RW, Jr, Idury RM, Schäffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–63. [PMC free article] [PubMed] [Google Scholar]
- 29.Schäffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44:225–37. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 30.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18(Suppl 1):S189–S198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 31.Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D. Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. Am J Hum Genet. 2006;78:922–35. doi: 10.1086/504158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giovannetti A, Mazzetta F, Caprini E, et al. Skewed T-cell receptor repertoire, decreased thymic output, and predominance of terminally differentiated T cells in ataxia telangiectasia. Blood. 2002;100:4082–9. doi: 10.1182/blood-2002-03-0976. [DOI] [PubMed] [Google Scholar]
- 33.Giovannetti A, Pierdominici M, Mazzetta F, et al. Unravelling the complexity of T cell abnormalities in common variable immunodeficiency. J Immunol. 2007;178:3932–43. doi: 10.4049/jimmunol.178.6.3932. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Putnam AL, Xu-Yu Z, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–11. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seddiki N, Santner-Nanan B, Martinson J, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203:1693–700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milner JD, Brenchley JM, Laurence A, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–6. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miething C, Grundler R, Fend F, et al. The oncogenic fusion protein nucleophosmin- anaplastic lymphoma kinase (NPM-ALK) induces two distinct malignant phenotypes in a murine retroviral transplantation model. Oncogene. 2003;22:4642–7. doi: 10.1038/sj.onc.1206575. [DOI] [PubMed] [Google Scholar]
- 38.LeibundGut-Landmann S, Gross O, Robinson MJ, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–8. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 39.Hsu Y-MS, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune response to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 40.Romagnani S. Human Th17 cells. Arthritis Res Ther. 2008;10:206. doi: 10.1186/ar2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herre J, Marshall ASJ, Caron E, et al. Dectin-1 uses novel mechanisms for yeast phagocytosis in macrophages. Blood. 2004;104:4038–45. doi: 10.1182/blood-2004-03-1140. [DOI] [PubMed] [Google Scholar]
- 42.Rogers NC, Slack EC, Edwards AD, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–17. doi: 10.1016/j.immuni.2005.03.004. [Erratum, Immunity 2005;22:773–4.] [DOI] [PubMed] [Google Scholar]
- 43.Willment JA, Brown GD. C-type lectin receptors in antifungal immunity. Trends Microbiol. 2008;16:27–32. doi: 10.1016/j.tim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Underhill DM, Goodridge HS. The many faces of ITAMs. Trends Immunol. 2007;28:66–73. doi: 10.1016/j.it.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 46.Colonna M. All roads lead to CARD9. Nat Immunol. 2007;8:554–5. doi: 10.1038/ni0607-554. [DOI] [PubMed] [Google Scholar]
- 47.Hruz P, Eckmann L. Caspase recruitment domain-containing sensors and adaptors in intestinal innate immunity. Curr Opin Gastroenterol. 2008;24:108–14. doi: 10.1097/MOG.0b013e3282f50fdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertin J, Guo Y, Wang L, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-κB. J Biol Chem. 2000;275:41082–6. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- 49.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–17. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178:3107–15. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- 51.Dillon S, Agrawal S, Banerjee K, et al. Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J Clin Invest. 2006;116:916–28. doi: 10.1172/JCI27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma CS, Chew GYJ, Simpson N, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205:1551–7. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato K, Yang X-L, Yudate T, et al. Dectin- 2 is a pattern recognition receptor for fungi that couples with the Fc receptor γ chain to induce innate immune responses. J Biol Chem. 2006;281:38854–66. doi: 10.1074/jbc.M606542200. [DOI] [PubMed] [Google Scholar]
- 54.Bugarcic A, Hitchens K, Beckhouse AG, Wells CA, Ashman RB, Blanchard H. Human and mouse macrophage-inducible C-type lectin (Mincle) bind Candida albicans. Glycobiology. 2008;18:679–85. doi: 10.1093/glycob/cwn046. [DOI] [PubMed] [Google Scholar]
- 55.Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol. 2008;9:1179–88. doi: 10.1038/ni.1651. [DOI] [PubMed] [Google Scholar]
- 56.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190:624–31. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 57.Conti HR, Shen F, Nayyar N, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eyerich K, Foerster S, Rombold S, et al. Patients with chronic mucocutaneous candidiasis exhibit reduced production of Th17-associated cytokines IL-17 and IL-22. J Invest Dermatol. 2008;128:2640–5. doi: 10.1038/jid.2008.139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Appendix
Supplementary Figures and Legends