Reengineering Escherichia coli for Succinate Production in Mineral Salts Medium (original) (raw)
Abstract
The fermentative metabolism of glucose was redirected to succinate as the primary product without mutating any genes encoding the native mixed-acid fermentation pathway or redox reactions. Two changes in peripheral pathways were together found to increase succinate yield fivefold: (i) increased expression of phosphoenolpyruvate carboxykinase and (ii) inactivation of the glucose phosphoenolpyruvate-dependent phosphotransferase system. These two changes increased net ATP production, increased the pool of phosphoenolpyruvate available for carboxylation, and increased succinate production. Modest further improvements in succinate yield were made by inactivating the pflB gene, encoding pyruvate formate lyase, resulting in an E scherichia coli pathway that is functionally similar to the native pathway in Actinobacillus succinogenes and other succinate-producing rumen bacteria.
Succinic acid is used as a specialty chemical in the agricultural, food, and pharmaceutical industries (17, 32). It has also been identified by the U.S. Department of Energy as one of the top 12 building block chemicals (30), because it can be converted into a variety of products, including green solvents, pharmaceutical products, and biodegradable plastics (17, 32). Although succinic acid is currently produced from petroleum-derived maleic anhydride, considerable interest in the fermentative production of succinate from sugars has emerged during the past decade (9, 10, 17).
Several natural succinate-producing rumen bacteria that have high rates of succinate production and high succinate yields, such as Anaerobiospirillum succiniciproducens (22), Actinobacillus succinogenes (13, 28), and “_Mannheimia succiniciproducens_” (15, 16), have been isolated. However, these strains require complex organic nutrients that increase the costs associated with production, purification, and waste disposal (15, 22, 28). Low levels of succinate are produced by native strains of Escherichia coli in complex and mineral salts media (1, 4). Most mutant strains of E. coli that have been described previously as succinate producers also require complex organic nutrients (18, 23-26, 29, 31). Many involve two-step aerobic and anaerobic processes (3, 23-25, 29) and the addition of foreign genes (5, 6, 23-26, 29, 31).
Novel E. coli biocatalysts (KJ060, KJ071, and KJ073) for the anaerobic production of succinate in mineral salts medium have been developed recently without the use of foreign genes or resident plasmids (9, 10). These biocatalysts were developed by combining constructed mutations to eliminate alternative routes of NADH oxidation in the mixed-acid pathway with growth-based selection (metabolic evolution). In subsequent studies (33), these strains were found to have recruited the glucose-repressed (7), gluconeogenic pck gene (11, 12, 19, 21, 27), encoding phosphoenolpyruvate carboxykinase (PCK) (derepressed via a point mutation in the promoter region), to replace the native phosphoenolpyruvate carboxylase (ppc) and serve as the primary route for CO2 fixation (Fig. 1). A second acquired mutation was also identified as a frameshift mutation in the carboxy terminus of ptsI, inactivating the phosphoenolpyruvate-dependent phosphotransferase system (33). Glucose uptake by the phosphotransferase system was functionally replaced by galactose permease (galP) and glucokinase (glk).
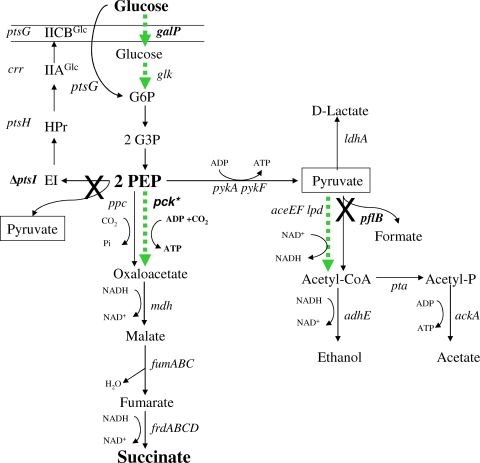
FIG. 1.
Anaerobic metabolism of E. coli using the mixed-acid fermentation pathway (data from reference 1). The native phosphotransferase system pathway for glucose uptake and the mixed-acid pathway for fermentation are shown with black arrows. Peripheral reactions for glucose uptake, carboxylation, and acetyl-CoA synthesis are shown as dotted green arrows and represent new metabolic functions that have been recruited for succinate production from glucose. Reactions that have been blocked by gene deletions or point mutations are marked with an X. pck* indicates a novel mutation that derepressed pck, allowing the enzyme to serve as the primary route for oxaloacetate production. Pyruvate (boxed) appears at two sites but is presumed to exist as a single intracellular pool.
Based on these previous studies, we have now determined the core mutations needed to direct carbon flow from glucose to succinate in E. coli and have constructed new succinate-producing strains with a minimum of genetic change.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The strains used in this study are listed in Table 1. During strain construction, cultures were grown aerobically at 30, 37, or 39°C in Luria broth (10 g liter−1 Difco tryptone, 5 g liter−1 Difco yeast extract, and 5 g liter−1 NaCl) containing 2% (wt/vol) glucose or 5% (wt/vol) arabinose. Ampicillin (50 mg liter−1), kanamycin (50 mg liter−1), or chloramphenicol (40 mg liter−1) was added as needed.
TABLE 1.
Sources and characteristics of E. coli strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 8739 | Wild type | Lab collection |
| KJ012 | Δ_ldhA_ Δ_adhE_ Δ_ackA_ | 10 |
| XZ02 | Δ_ldhA_ | This study |
| XZ04 | Δ_ldhA_ Δ_pflB_ | This study |
| XZ14 | Δ_ldhA_ Δ_adhE_ | This study |
| XZ15 | Δ_adhE_ | This study |
| XZ17 | Δ_pflB_ | This study |
| XZ468b | pck* Δ_ptsH_ | This study |
| XZ469b | pck* Δ_ptsI_W | This study |
| XZ470 | pck* Δ_ptsG_ | This study |
| XZ629 | Δ_ldhA_ Δ_adhE_ Δ_ackA pck_* | This study |
| XZ632 | pck* | This study |
| XZ635 | Δ_ldhA pck_* | This study |
| XZ638 | Δ_ldhA_ Δ_adhE pck_* | This study |
| XZ639 | Δ_adhE pck_* | This study |
| XZ640 | Δ_pflB pck_* | This study |
| XZ641 | Δ_ldhA_ Δ_pflB pck_* | This study |
| XZ647 | pck* Δ_ptsI_ | This study |
| XZ650 | Δ_ptsI_ | This study |
| XZ721 | pck* Δ_ptsI_ Δ_pflB_ | This study |
| XZ723 | pck* Δ_ptsI_ Δ_adhE_ | This study |
| Plasmids | ||
| pck overexpression (pLOI4677) | bla kan; pck (including ribosomal binding site, coding region, and terminator fragment) from E. coli ATCC 8739 cloned into the pCR2.1-TOPO vector | This study |
| ldhA deletion | ||
| pLOI4162 | bla; cat-sacB cassette | 10 |
| pLOI4652 | bla kan; ldhA (PCR) from E. coli cloned into the pCR2.1-TOPO vector | This study |
| pLOI4653 | cat-sacB cassette cloned into ldhA of pLOI4652 | This study |
| pLOI4655 | PacI digestion of pLOI4653 and self-ligation | This study |
| pflB deletion | ||
| pLOI4667 | bla kan; pflB (PCR) from E. coli cloned into the pCR2.1-TOPO vector | This study |
| pLOI4668 | cat-sacB cassette cloned into pflB of pLOI4667 | This study |
| pLOI4669 | PacI digestion of pLOI4668 and self-ligation | This study |
| adhE deletion | ||
| pLOI4707 | bla kan; adhE (PCR) from E. coli cloned into the pCR2.1-TOPO vector | This study |
| pLOI4708 | cat-sacB cassette cloned into adhE of pLOI4707 | This study |
| pLOI4709 | PacI digestion of pLOI4707 and self-ligation | This study |
| pck promoter change | ||
| pLOI4736 | bla kan; pck -p from E. coli cloned into the pCR2.1-TOPO vector | This study |
| pLOI4737 | cat-sacB cassette cloned into pck -p of pLOI4736 | This study |
| pLOI4752 | trc promoter cloned into pck -p of pLOI4736 | This study |
| ptsI deletion | ||
| pLOI4734 | bla kan; ptsI from E. coli cloned into the pCR2.1-TOPO vector | This study |
| pLOI4735 | cat-sacB cassette cloned into ptsI of pLOI4734 | This study |
| pLOI4735B | PacI digestion of pLOI4735 and self-ligation | This study |
| ptsG deletion | ||
| pLOI4683 | bla kan; ptsG from E. coli cloned into the pCR2.1-TOPO vector | This study |
| pLOI4684 | cat-sacB cassette cloned into ptsG of pLOI4683 | This study |
| pLOI4685 | PacI digestion of pLOI4684 and self-ligation | This study |
| Primers | ||
| ldhA deletion | ||
| XZ-ldhA-up | GATAACGGAGATCGGGAATG | This study |
| XZ-ldhA-down | CTTTGGCTGTCAGTTCACCA | |
| XZ-ldhA-1 | TCTGGAAAAAGGCGAAACCT | |
| XZ-ldhA-2 | TTTGTGCTATAAACGGCGAGT | |
| pflB deletion | ||
| PflB-up2 | TGTCCGAGCTTAATGAAAAGTT | This study |
| PflB-down2 | CGAGTAATAACGTCCTGCTGCT | |
| PflB-5 | AAACGGGTAACACCCCAGAC | |
| PflB-6 | CGGAGTGTAAACGTCGAACA | |
| adhE deletion | ||
| adhE-up | CATGCTAATGTAGCCACCAAA | This study |
| adhE-down | TTGCACCACCATCCAGATAA | |
| adhE-1 | TCCGGCTAAAGCTGAGAAAA | |
| adhE-2 | GTGCGTTAAGTTCAGCGACA | |
| pck mutation | ||
| pck-Pro-up | CACGGTAGCAACAACATTGC | This study |
| pck-Pro-down | AGAAAGCGTCGACAACGAAC | |
| pck-Pro-1 | ATGCGCGTTA ACAATGGTTT | |
| pck-Pro-2 | ATGGATAACG TTGAACTTTC | |
| pck-P-2 | TTCACTGCTC CTTAGCCAAT | |
| ptsI truncation | ||
| ptsI-up | CGCATTATGTTCCCGATGAT | This study |
| ptsI-down | GCCTTTCAGTTCAACGGTGT | |
| ptsI-1 | CGGCCCAATTTACTGCTTAG | |
| ptsI-2 | ATCCCCAGCAACAGAAGTGT | |
| ptsI deletion | ||
| ptsI-D-up | ATGATTTCAGGCATTTTAGCATCCCCGGGTATCGCTTTCGGTAAA GTGTAGGCTGGAGCTGCTTC | This study |
| ptsI-D-down | TTAGCAGATTGTTTTTTCTTCAATGAACTTGTTAACCAGCGTCAT CATATGAATATCCTCCTTAG | |
| ptsG deletion | ||
| ptsG-up | GAAGAACTGGCGCAGGTAAC | This study |
| ptsG-down | AAGGAAACGCCGTTAATCCT | |
| ptsG-1 | CCTGAAAACCGAGATGGATG | |
| ptsG-2 | CATCAGCGATTTACCGACCT | |
| ptsH deletion | ||
| ptsH-D-up | ATGTTCCAGCAAGAAGTTACCATTACCGCTCCGAACGGTCTGCAC GTGTAGGCTGGAGCTGCTTC | This study |
| ptsH-D-down | TTACTCGAGTTCCGCCATCAGTTTAACCAGATGTTCAACCGCTTT CATATGAATATCCTCCTTAG |
Genetic methods.
Chromosomal genes were deleted seamlessly without leaving segments of foreign DNA, as described previously (10, 34). Red recombinase technology (Gene Bridges GmbH, Dresden, Germany) was used to facilitate chromosomal integration. The plasmids and primers used during construction are listed in Table 1.
Fermentation.
Strains were grown without antibiotics at 37°C in Luria broth medium or NBS mineral salts medium (2) supplemented with 5% (wt/vol) glucose and 100 mM potassium bicarbonate unless stated otherwise. Preinocula for fermentations were grown by transferring fresh colonies to a 250-ml flask (100 ml Luria broth or NBS medium, 2% glucose). After 16 h (37°C, 120 rpm), this culture was diluted into a small fermentation vessel containing 300 ml Luria broth or NBS medium (5% glucose, 100 mM potassium bicarbonate) to provide an inoculum of 0.033 g (dry weight) of cells liter−1. Fermentations were maintained at pH 7.0 by the automatic addition of a base containing additional CO2 (2.4 M potassium carbonate containing 1.2 M potassium hydroxide).
PCK assay.
The activity of PCK was determined as described previously (33). Activity is reported in micromoles per milligram of protein per minute.
Analysis.
The dry weight of cells was estimated by measuring the optical density at 550 nm. Organic acids and glucose were measured by high-performance liquid chromatography (34).
RESULTS
Eliminating competing fermentation routes for NADH.
Examination of the mixed-acid pathway in E. coli indicates three primary routes for NADH oxidation, leading to succinate, lactate, and ethanol (Fig. 1). Deletion of genes that block these competing routes for NADH oxidation was insufficient to direct carbon flow to succinate (Table 2). Deletion of individual genes had little effect on cell growth during fermentation in NBS mineral salts medium relative to that of the parent (ATCC 8739). The succinate yield increased only after deletion of adhE (12%) (P < 0.05; _n_ = 3 replicates) during fermentation in NBS mineral salts medium, while it decreased slightly after _ldhA_ deletion (6%) (_P_ > 0.05; n = 3) and modestly after pflB deletion (44%) (P < 0.05; n = 3). Deletion of both alternative NADH oxidation pathways (ldhA with adhE or ldhA with pflB) substantially reduced both cell growth and succinate production in NBS mineral salts medium.
TABLE 2.
Effects of inactivating alternative NADH oxidizing pathways on succinate production in NBS mineral salts medium and complex medium (Luria broth)
| Strain | Genetic modification | Medium | Growth | Glu concn used (mM) | Suc yielda | Fermentation product concn (mM)b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Cell mass (g/liter) | Suc | Ace | Pyr | Lac | For | EtOH | |||||
| ATCC 8739 | Wild type | NBS | 3 | 3.0 | 278 | 0.16 ± 0.01 | 46 ± 2 | 151 | 110 | 318 | 133 | |
| XZ02 | Δ_ldhA_ | NBS | 3 | 2.9 | 278 | 0.15 ± 0.01 (−6) | 43 ± 2 | 165 | 11 | 353 | 199 | |
| XZ15 | Δ_adhE_ | NBS | 3 | 2.7 | 278 | 0.18 ± 0 (+12) | 50 ± 1 | 84 | 352 | 102 | ||
| XZ17 | Δ_pflB_ | NBS | 3 | 2.2 | 278 | 0.09 ± 0 (−44) | 26 ± 1 | 11 | 460 | |||
| XZ14 | Δ_ldhA_ Δ_adhE_ | NBS | 6 | 0.5 | 57 | 0.12 ± 0.02 (−25) | 6 ± 1 | 48 | 44 | |||
| XZ04 | Δ_ldhA_ Δ_pflB_ | NBS | 6 | 0.5 | 39 | 0.13 ± 0.03 (−19) | 5 ± 1 | 22 | 22 | 12 | ||
| KJ012c | Δ_ldhA_ Δ_adhE_ Δ_ackA_ | NBS | 6 | 0.3 | 46 | 0.13 (−19) | 6 | 26 | ||||
| ATCC 8739 | Wild type | LB | 2 | 2.0 | 257 | 0.13 ± 0 | 34 ± 1 | 138 | 214 | 161 | 127 | |
| XZ02 | Δ_ldhA_ | LB | 2 | 1.8 | 257 | 0.21 ± 0.01 (+61) | 55 ± 2 | 224 | 17 | 152 | 206 | |
| XZ15 | Δ_adhE_ | LB | 2 | 1.9 | 268 | 0.11 ± 0.02 (−15) | 30 ± 5 | 43 | 459 | |||
| XZ17 | Δ_pflB_ | LB | 2 | 2.0 | 261 | 0.09 ± 0 (−31) | 24 ± 1 | 8 | 469 | |||
| XZ14 | Δ_ldhA_ Δ_adhE_ | LB | 6 | 0.3 | 31 | 0.23 ± 0.03 (+77) | 7 ± 1 | 45 | 17 | |||
| XZ04 | Δ_ldhA_ Δ_pflB_ | LB | 6 | 1.2 | 150 | 0.33 ± 0.08 (+154) | 48 ± 12 | 9 | 86 | 35 | 23 | 89 |
| KJ012c | Δ_ldhA_ Δ_adhE_ Δ_ackA_ | LB | 6 | 1.5 | 154 | 0.70 (+438) | 108 | 61 | 14 |
Quite different results were observed in Luria broth medium (Table 2). The succinate yield increased to different degrees in all strains that contained a deletion in ldhA (alone or in combination) (P < 0.05; _n_ = 3). The yield increased 61% with a single _ldhA_ deletion, while it increased 77% with _ldhA_ and _adhE_ double deletion and 154% with _ldhA_ and _pflB_ double deletion. However, the succinate yield showed a small decrease after _adhE_ deletion (15%) (_P_ > 0.05; n = 3) and a modest decrease after pflB deletion (31%) (P < 0.05; n = 3). Again, growth was reduced when both alternative NADH oxidation pathways were inactivated. Almost none of these deletions were sufficient to redirect substantial amounts of glucose carbon to succinate in either mineral salts medium or complex medium. A significant increase was obtained only with ldhA, adhE, and ackA triple deletion (438%) (P < 0.05; n = 3). Overall, deletion strains constructed for succinate production based on observational analysis of the native mixed-acid pathway in E. coli were less productive than would be expected in complex medium and were unsuccessful in NBS mineral salts medium.
PCK.
Our previous investigations (9, 10, 33) discovered a promoter mutation (G to A at position −64 relative to the ATG start codon; designated pck*) that increased PCK activity and increased succinate production from glucose in the presence of many additional mutations. This was surprising, because pck is typically repressed by glucose (7) and has been shown to function primarily in gluconeogenesis during the aerobic metabolism of organic acids (11, 12, 19, 21, 27, 31). To examine this further, the pck gene (ribosomal binding site, coding region, and terminator region) was amplified and cloned into pCR2.1-TOPO to produce pLOI4677 with pck expression under the control of the lac promoter. This plasmid was transformed into ATCC 8739. Also, the native pck gene in ATCC 8739 was replaced with the mutant pck* gene to construct XZ632. Glucose fermentation was examined in both strains using NBS mineral salts medium.
PCK activity was similar for XZ632 and the isopropyl-β-d-thiogalactopyranoside (IPTG)-induced plasmid strain, more than eightfold that of the parent. Upon chromosomal integration, the single-copy-number pck* gene was almost as effective for PCK production as the IPTG-induced multicopy plasmid. The succinate yield increased 69% (P < 0.05; _n_ = 3) with the plasmid overexpressing _pck_ and 38% (_P_ > 0.05; n = 3) with pck* over the yield from the parent, ATCC 8739 (Table 3). Increased PCK activity alone was insufficient to redirect glucose carbon to succinate.
TABLE 3.
Effects of increasing PCK activity on succinate production and yield in NBS mineral salts medium
| Strain | Genetic modificationa | Growth | PCK activity [μmol min−1 (mg of protein)−1] | Glu concn used (mM) | Suc yieldb | Fermentation product concn (mM)c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Cell mass (g/liter) | Suc | Ace | Pyr | Lac | For | EtOH | |||||
| ATCC 8739 | Wild type | 3 | 3.0 | 0.2 ± 0 | 278 | 0.16 ± 0.01 | 46 ± 2 | 151 | 110 | 318 | 133 | |
| ATCC 8739 | pLOI4677d | 3 | 2.8 | 1.8 ± 0.3 | 278 | 0.27 ± 0.01 (+69) | 73 ± 4 | 141 | 24 | 112 | 266 | 116 |
| XZ632 | pck* | 3 | 2.9 | 1.6 ± 0.2 | 278 | 0.22 ± 0.02 (+38) | 61 ± 6 | 161 | 88 | 302 | 152 | |
| XZ635 | Δ_ldhA pck_* | 3 | 2.7 | 1.7 ± 0.2 | 278 | 0.22 ± 0.01 (+38) | 60 ± 2 | 151 | 13 | 327 | 178 | |
| XZ639 | Δ_adhE pck_* | 3 | 2.7 | 1.6 ± 0.1 | 278 | 0.25 ± 0 (+56) | 70 ± 1 | 80 | 329 | 99 | ||
| XZ640 | Δ_pflB pck_* | 3 | 2.5 | 1.6 ± 0.2 | 278 | 0.14 ± 0 (−12) | 39 ± 1 | 12 | 448 | |||
| XZ638 | Δ_ldhA_ Δ_adhE pck_* | 6 | 0.8 | 0.9 ± 0.1 | 66 | 0.24 ± 0.02 (+50) | 16 ± 1 | 71 | 79 | |||
| XZ641 | Δ_ldhA_ Δ_pflB pck_* | 6 | 0.8 | 0.9 ± 0.1 | 61 | 0.39 ± 0.05 (+144) | 24 ± 3 | 9 | 2 | 15 |
The pck* mutation was tested in combination with other mutations that eliminated pathways for NADH oxidation (Table 3). The succinate yield increased in nearly all strains with pck*, from 38% to 144% (P < 0.05; n = 3), over that of isogenic strains containing the native pck gene, except for a small decrease (12%) (P < 0.05; n = 3) from that of the strain with pflB deletion (Table 3). The combined action of mutations to eliminate competing routes for NADH oxidation and increased PCK activity was also insufficient to substantially redirect glucose carbon to succinate.
The phosphotransferase system.
The phosphoenolpyruvate-dependent phosphotransferase system is the primary mechanism for glucose uptake in E. coli and an integral part of the glucose catabolite repression system (12, 20). A frameshift mutation within the carboxyl end of ptsI (a single-base-pair deletion at bp 1673) was discovered in the succinate-producing strains KJ060, KJ071, and KJ073 as a result of metabolic evolution (32). This mutation would be expected to disrupt the phosphotransferase system (20). In this mutant, glucose uptake was functionally replaced by galP (galactose permease) and glk (glucokinase) (8, 12, 33). To investigate the effect of the ptsI disruption on succinate production, the carboxy-terminal 175 bp of ptsI was deleted from wild-type E. coli ATCC 8739 to produce strain XZ650. Surprisingly, succinate production and yield showed small decreases in this mutant from those of the parent (12%) (P > 0.05; n = 3) (Table 4) .
TABLE 4.
Effects of combining a mutation in ptsI, mutations that disrupted the ethanol pathway, and increased expression of pck on succinate production in NBS mineral salts medium
| Strain | Genetic modificationa | Growth | Glu concn used (mM) | Suc yieldb | Fermentation product concn (mM)c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Cell mass (g/liter) | Suc | Ace | Pyr | Lac | For | EtOH | ||||
| ATCC 8739 | Wild-type ATCC 8739 | 3 | 3.0 | 278 | 0.16 ± 0.01 | 46 ± 2 | 151 | 110 | 318 | 133 | |
| XZ632 | pck* | 3 | 2.9 | 278 | 0.22 ± 0.02 (+38) | 61 ± 6 | 161 | 88 | 302 | 152 | |
| XZ650 | Δ_ptsI_ | 3 | 2.6 | 259 | 0.14 ± 0.01 (−12) | 35 ± 2 | 131 | 28 | 130 | 273 | 119 |
| XZ647 | pck* Δ_ptsI_ | 3 | 2.5 | 242 | 0.89 ± 0.09 (+456) | 216 ± 22 | 174 | 5 | 199 | 22 | |
| XZ650 | Δ_ptsI_; pLOI4677d | 3 | 2.4 | 224 | 0.75 ± 0.14 (+369) | 167 ± 32 | 167 | 2 | 8 | 224 | 52 |
| XZ469 | pck* Δ_ptsI_W | 3 | 3.0 | 245 | 0.84 ± 0.06 (+425) | 206 ± 14 | 143 | 3 | 164 | 31 | |
| XZ468 | pck* Δ_ptsH_ | 3 | 2.6 | 266 | 0.67 ± 0.10 (+319) | 179 ± 26 | 151 | 17 | 242 | 69 | |
| XZ470 | pck* Δ_ptsG_ | 3 | 2.5 | 269 | 0.63 ± 0.05 (+294) | 170 ± 13 | 175 | 32 | 245 | 63 | |
| XZ721 | pck* Δ_ptsI_ Δ_pflB_ | 3 | 2.3 | 262 | 1.25 ± 0.11 (+681) | 327 ± 28 | 70 | 29 | |||
| XZ723 | pck* Δ_ptsI_ Δ_adhE_ | 3 | 2.4 | 259 | 0.86 ± 0.02 (+438) | 222 ± 6 | 175 | 55 | 171 |
Two approaches were used to investigate the effect of combining the ptsI truncation with high levels of PCK using the pck* mutation or the native pck gene overexpressed from the lac promoter (plasmid pLOI4677). In combination with the ptsI truncation, both approaches resulted in dramatic increases in succinate production (456% and 369%) (P < 0.05; n = 3) (Table 4). Strain XZ647 (pck*; ptsI truncation) produced 216 mmol succinate in NBS mineral salts medium with 5% glucose, with a yield of 0.89 mol succinate per mol glucose, 4.7-fold higher than that of wild-type E. coli ATCC 8739 (Table 2). For both XZ647 and XZ650(pLOI4677), formate, ethanol, and small amounts of lactate remained as minor side products. As expected, a similar increase in succinate production was observed following deletion of any other gene essential for the glucose phosphotransferase system (ptsH, ptsG) (319% and 294%, respectively) (P < 0.05; n = 3) (Table 4). Based on these results, we conclude that two changes, inactivation of the phosphoenolpyruvate-dependent phosphotransferase system and elevated levels of PCK activity, represent the core modifications required to effectively redirect glucose metabolism to pyruvate in NBS mineral salts medium. These core modifications do not involve genes directly concerned with fermentative redox balance.
Further genetic improvements in succinate production.
Although strain XZ647 (pck*; ptsI truncation) produced succinate as the dominant fermentation product, significant levels of unwanted coproducts (lactate, ethanol, formate, and acetate) were also produced (Table 4). Deletion of either adhE (XZ723) or pflB (XZ721) eliminated the production of ethanol. The production of acetate and formate was substantially reduced or eliminated only by the deletion of pflB. The resulting strain (XZ721) produced high levels of succinate with a molar yield of >1.2 mol succinate per mol glucose.
DISCUSSION
Succinate typically represents a minor product of glucose fermentation in E. coli. Most of the glucose carbon is converted to ethanol and lactate via alternative NADH-oxidizing pathways (Fig. 1). Derivatives of E. coli have been constructed in efforts to improve succinate production for more than a decade, with variable success (3, 5, 6, 18, 23-26, 29, 31). The strategy used to construct these strains has typically focused on the elimination of competing pathways for NADH oxidation (3, 6, 23-25, 29, 31). Target genes for deletion were selected primarily based on inspection of the pathway (Fig. 1). However, successes with this strategy have been limited to complex media, heterologous genes, and two-step (an aerobic growth phase followed by an anaerobic production phase) processes (3, 6, 18, 23-25, 29, 31).
In a complex medium such as Luria broth, most of the building block molecules required for biosynthesis and growth are supplied directly as intermediates in nutrients such as yeast extract or tryptone. In this medium, deletion of genes based on observation of the fermentation pathway itself generally improved succinate production (Fig. 1; Table 2). Deletion of ldhA and all combinations of target genes including ldhA resulted in increased succinate production and an increased succinate yield per mole of glucose during fermentation in Luria broth (Table 2).
With a mineral salts medium such as NBS broth, however, deletion of the same target genes in the mixed-acid fermentation pathway was not helpful. Deletions of most of these genes were detrimental for both succinate production and succinate yield. With the exception of a small increase after the deletion of adhE, deletions of single genes and combinations of genes concerned directly with the mixed-acid fermentation pathway all reduced succinate production and yields in NBS mineral salts medium (Table 2). KJ012 (ATCC 8739 Δ_ldhA_ Δ_adhE_ Δ_ackA_), the genetic equivalent of a strain patented for two-step (an aerobic growth phase followed by an anaerobic production phase) succinate production in a complex medium (23), produced even less succinate than the wild-type parent in NBS mineral salts medium. These results demonstrate that the rational selection of target genes for deletion based solely on observations of pathways is an unreliable predictor of success for improvements in succinate production when one is using mineral salts medium. In mineral salts medium, carbon must be precisely partitioned between energy generation, fermentation, and the biosynthesis of building block molecules needed for cell growth.
A new strategy was developed to construct strains for succinate production in mineral salts medium. No mutations were required in genes encoding the E. coli mixed-acid fermentation pathway (Fig. 1) to substantially redirect glucose carbon to succinate. The combination of two core changes in peripheral pathways resulted in a fivefold increase in succinate yield: (i) increased expression of the energy-conserving (gluconeogenic) enzyme PCK to replace the native fermentative phosphoenolpyruvate carboxylase (energy wasting) and (ii) replacement of the glucose phosphoenolpyruvate-dependent phosphotransferase system with an alternative permease such as galP and ATP-dependent phosphorylation (glk). Together, these changes increased net ATP production for growth, increased the pool of phosphoenolpyruvate available for carboxylation, and increased succinate production.
Additional mutations in mixed-acid fermentation genes, such as the deletion of pflB and others, represent opportunities for modest further increases in succinate yield and production. Note that acetyl coenzyme A (acetyl-CoA) is an essential metabolite for biosynthesis that is produced primarily by pflB during fermentative growth. This function is presumed to be replaced in pflB mutants by native expression of the pyruvate dehydrogenase complex (aceEF, lpd), an enzyme that typically serves as the dominant route for acetyl-CoA production during oxidative metabolism (14). A low pyruvate dehydrogenase activity was found in E. coli ATCC 8739 and its derivatives under our fermentation conditions (data not shown). The resulting succinate pathway in E. coli strains optimally engineered for succinate production in mineral salts medium (Fig. 1) is functionally similar to the native pathway that evolved in succinate-producing rumen bacteria (13, 15, 16, 22, 28).
Acknowledgments
This research was supported by grants from the U.S. Department of Energy (FG02-96ER20222), the Florida High Technology Corridor Council, and Myriant Technologies, LLC.
Footnotes
▿
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Bock, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 2.Causey, T. B., K. T. Shanmugam, L. P. Yomano, and L. O. Ingram. 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc. Natl. Acad. Sci. USA 101**:**2235-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnelly, M. I., C. S. Millard, and L. Stols. June 1998. Mutant E. coli strain with increased succinic acid production. U.S. patent 5,770,435.
- 4.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 5.Gokarn, R. R., M. A. Eiteman, and E. Altman. 2000. Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl. Environ. Microbiol. 66**:**1844-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gokarn, R. R., M. A. Eiteman, and E. Altman. September 2002. Metabolically engineered E. coli for enhanced production of oxaloacetate-derived biochemicals. U.S. patent 6,455,284.
- 7.Goldie, H. 1984. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J. Bacteriol. 159**:**832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández-Montalvo, V., A. Martinez, G. Hernandez-Chavez, F. Bolivar, F. Valle, and G. Gosset. 2003. Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnol. Bioeng. 83**:**687-694. [DOI] [PubMed] [Google Scholar]
- 9.Jantama, K., M. J. Haupt, S. A. Svoronos, X. Zhang, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99**:**1140-1153. [DOI] [PubMed] [Google Scholar]
- 10.Jantama, K., X. Zhang, J. C. Moore, K. T. Shanmugam, S. A. Svoronos, and L. O. Ingram. 2008. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol. Bioeng. 101**:**881-893. [DOI] [PubMed] [Google Scholar]
- 11.Kao, K. C., L. M. Tran, and J. C. Liao. 2005. A global regulatory role of gluconeogenic genes in Escherichia coli revealed by transcriptome network analysis. J. Biol. Chem. 280**:**36079-36087. [DOI] [PubMed] [Google Scholar]
- 12.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33**:**D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, P., M. Laivenieks, C. Vieille, and J. G. Zeikus. 2004. Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl. Environ. Microbiol. 70**:**1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, Y., L. O. Ingram, and K. T. Shanmugam. 2007. Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Appl. Environ. Microbiol. 73**:**1766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, P. C., S. Y. Lee, S. H. Hong, and H. N. Chang. 2002. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl. Microbiol. Biotechnol. 58**:**663-668. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. J., H. Song, and S. Y. Lee. 2006. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl. Environ. Microbiol. 72**:**1939-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinlay, J. B., C. Vieille, and J. G. Zeikus. 2007. Prospects for a bio-based succinate industry. Appl. Microbiol. Biotechnol. 76**:**727-740. [DOI] [PubMed] [Google Scholar]
- 18.Millard, C. S., Y. P. Chao, J. C. Liao, and M. I. Donnelly. 1996. Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl. Environ. Microbiol. 62**:**1808-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh, M. K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277**:**13175-13183. [DOI] [PubMed] [Google Scholar]
- 20.Postma, P. W., J. W. Lengela, and G. R. Jacobson. 1996. Phosphoenolpyruvate: carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 21.Ramseier, T. M., S. Bledig, V. Michotey, R. Feghali, and M. H. Saier, Jr. 1995. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 16**:**1157-1169. [DOI] [PubMed] [Google Scholar]
- 22.Samuelov, N. S., R. Lamed, S. Lowe, and J. G. Zeikus. 1991. Influence of CO2-HCO3 levels and pH on growth, succinate production, and enzyme-activities of Anaerobiospirillum succiniciproducens. Appl. Environ. Microbiol. 57**:**3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San, K. Y., G. N. Bennett, and A. M. Sanchez. May 2007. Mutant E. coli strain with increased succinic acid production. U.S. patent 7,223,567.
- 24.Sánchez, A. M., G. N. Bennett, and K. Y. San. 2005. Efficient succinic acid production from glucose through overexpression of pyruvate carboxylase in an Escherichia coli alcohol dehydrogenase and lactate dehydrogenase mutant. Biotechnol. Prog. 21**:**358-365. [DOI] [PubMed] [Google Scholar]
- 25.Sánchez, A. M., G. N. Bennett, and K. Y. San. 2005. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab. Eng. 7**:**229-239. [DOI] [PubMed] [Google Scholar]
- 26.Stols, L., and M. I. Donnelly. 1997. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63**:**2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unden, G., and A. Kleefeld. 30 July 2004, posting date. Chapter 3.4.5, C4-dicarboxylate degradation in aerobic and anaerobic growth. In A. Bock, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nystrom, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org/.
- 28.Van der Werf, M. J., M. V. Guettler, M. K. Jain, and J. G. Zeikus. 1997. Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch. Microbiol. 167**:**332-342. [DOI] [PubMed] [Google Scholar]
- 29.Vemuri, G. N., M. A. Eiteman, and E. Altman. 2002. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl. Environ. Microbiol. 68**:**1715-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werpy, T., and G. Petersen (ed.). 2004. Top value added chemicals from biomass. U.S. Department of Energy, Washington, DC. http://www1.eere.energy.gov/biomass/pdfs/35523.pdf.
- 31.Wu, H., Z. Li, L. Zhou, and Q. Ye. 2007. Improved succinic acid production in the anaerobic culture of an Escherichia coli pflB ldhA double mutant as a result of enhanced anaplerotic activities in the preceding aerobic culture. Appl. Environ. Microbiol. 73**:**7837-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeikus, J. G., M. K. Jain, and P. Elankovan. 1999. Biotechnology of succinic acid production and markets for derived industrial products. Appl. Microbiol. Biotechnol. 51**:**545-552. [Google Scholar]
- 33.Zhang, X., K. Jantama, J. C. Moore, L. R. Jarboe, K. T. Shanmugam, and L. O. Ingram. 16 November 2009. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc. Natl. Acad. Sci. USA. doi: 10.1073/pnas.0905396106. [DOI] [PMC free article] [PubMed]
- 34.Zhang, X., K. Jantama, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2007. Production of l-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77**:**355-366. [DOI] [PubMed] [Google Scholar]