Analysis of VHL gene alterations and their relationship to clinical parameters in sporadic conventional renal cell carcinoma (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 15.
Abstract
Purpose:
To carry out a comprehensive analysis of genetic and epigenetic changes of the von Hippel Lindau (VHL) gene in patients with conventional (clear cell) renal cell carcinoma (RCC) and to determine their significance relative to clinicopathological characteristics and outcome.
Experimental design:
The VHL status in 86 conventional RCCs was determined by mutation detection, loss of heterozygosity (LOH) and promoter methylation analysis, extending our original cohort to a total of 177 patients. Data was analysed to investigate potential relationships between VHL changes, clinical parameters and outcome.
Results:
LOH was found in 89.2%, mutation in 74.6% and methylation in 31.3% of evaluable tumours; evidence of biallelic inactivation (LOH and mutation or methylation alone) was found in 86.0% whilst no involvement of VHL was found in only 3.4% of samples. Several associations were suggested including between LOH and grade, nodal status and necrosis, between mutation and sex and between methylation and grade. Biallelic inactivation may be associated with better overall survival compared to patients with no VHL involvement although small sample numbers in the latter group severely limit this analysis which requires independent confirmation.
Conclusions:
This study reports one of the highest proportions of conventional RCC with VHL changes, and suggests possible relationships between VHL status and clinical variables. The data suggests that VHL defects may define conventional RCCs but the clinical significance of specific VHL alterations will only be clarified by the determination of their biological effect at the protein level rather than through genetic or epigenetic analysis alone.
Keywords: VHL, Renal cell carcinoma (RCC), mutation, methylation, prognosis
Introduction1
The incidence of renal cancer is increasing and currently accounts for ~2-3% of cancers worldwide, with around 210000 new cases diagnosed annually (GLOBOCAN 2002)1. Patients often present late with advanced or unresectable disease and up to 30% of patients relapse after potentially curative surgery. Metastatic renal cancer is associated with a 5-year survival rate of ~10% (1, 2). Traditional treatments, such as interferon and interleukin, only have response rates of 15-20% but more recently, therapies such as the tyrosine kinase inhibitors sunitinib and sorafenib (3) have shown response rates and survival benefits.
The von Hippel Lindau (VHL) tumour suppressor gene on chromosome 3p plays a central role in the development of the conventional (clear cell) histological subtype of renal cell carcinoma (RCC). Identified in patients with familial VHL disease, an autosomal dominant cancer syndrome associated with the development of a number of tumours including conventional RCC (4), VHL has also been shown to be important in sporadic conventional RCCs, with a large number of studies reporting potential loss of VHL function as a result of allele loss, mutation and promoter methylation (5-47). In one of the most comprehensive studies, 91% of histologically confirmed tumours were recently found to have mutation or methylation of VHL although loss of heterozygosity (LOH) and relationship to outcome was not examined (34).
The most well characterised function of VHL is as the substrate recognition subunit of an E3 ubiquitin ligase complex that targets hypoxia inducible factor (HIF-α) for ubiquitination and degradation by the proteasome (48). Stabilisation of HIF-α due to loss of VHL function, resulting in constitutive activation of a hypoxic response, has been shown to be central to development of conventional RCC. VHL has been implicated in many cellular processes including extracellular matrix assembly, organisation of actin and tubulin cytoskeletons and cell growth and apoptosis, and there is evidence that it has HIF-independent activities. It is possible that particular VHL defects may result in loss of different VHL functions which may alter their specific clinical impact.
In sporadic RCC, VHL mutation or methylation has been associated with improved prognosis in patients with stage I to III disease but not stage IV patients (46). Other studies have reported poor prognosis to be associated with loss of function (frameshift and nonsense) mutations of the VHL gene (28, 39). Despite increasing numbers of studies examining sporadic RCC and VHL status, many involve small numbers of samples, inconsistent methodology and varying stringency and further larger studies are needed to consolidate results. We have extended our previous study (9), analysing samples from a total of 177 patients with sporadic conventional RCC to look for somatic allele loss, mutation and promoter methylation and evaluated their significance in relationship to clinical and pathological variables and patient outcomes.
Materials and Methods
Patient samples
Renal tissue samples from 86 previously untreated patients undergoing nephrectomy for sporadic conventional RCC between December 2001 and December 2006 were collected with informed consent and approval of the Local Research Ethics Committee. All tumours included in the study were reviewed by a single consultant pathologist to confirm their histological classification. Tumour tissue was dissected, rinsed in ice cold PBS and snap frozen and stored in liquid nitrogen. Venous blood samples (EDTA) were centrifuged at 2000g at 20°C for 10 min and the buffy coats were removed and stored at −80°C.
DNA was extracted from tissue samples and buffy coats using the QIAamp DNA mini kit (Qiagen, Crawley, United Kingdom) and quantified using the PicoGreen dsDNA assay (Molecular Probes, Leiden, Netherlands). Samples of patients which had no VHL involvement, LOH only or mutation only on analysis of frozen tissues were reviewed and in 27 cases, these samples contained <50% tumour cells. In 21 of these cases, sections from corresponding FFPE tissue were cut, subjected to gross macrodissection to select tumour-rich areas and DNA extracted using the Ambion recoverAll total nucleic acid isolation kit (Applied Biosystems, Warrington, UK) for subsequent mutation and LOH analysis only. This included 8 tumours from the original study (9). In the remaining 6 cases, this was not possible due either to unavailability of blocks or failure of DNA extraction/amplification.
Loss of heterozygosity (LOH) analysis
Six highly polymorphic microsatellite markers flanking the VHL gene (D3S1038, D3S1435, D3S1317, D3S1597, D3S1537 and D3S3691) were used for LOH analysis and scored as previously described (9). Normal DNA samples were screened to identify informative markers, initially using D3S1597 and DS31435 then further markers as required; in 20 cases it was not possible to confirm LOH with markers on both sides of the VHL gene.
Promotor methylation analysis
The methylation status of the VHL promoter was examined by sodium bisulfite modification and methylation-specific PCR as previously described (9).
Mutation detection
Primers were designed to amplify the entire coding sequence plus flanking splice sites of VHL using Primer3 software and a genomic reference sequence retrieved from Ensembl. Primer sequences were exon 1F AGCGCGTTCCATCCTCTAC; exon 1R AGTTCCCCGTCTGCAAAAT; exon 2F GGACGGTCTTGATCTCCTGA; exon 2R CAGGCAAAAATTGAGAACTGG; exon 3F GTTGGCAAAGCCTCTTGTTC; exon 3R GCAATGGTGCCTATTTTACTCTG. PCR (15μL final volume) was carried out using HotStarTaq Mastermix (Qiagen), 11.25 pmol of each primer and 20 ng of DNA. Reaction products were checked for size and purity by agarose electrophoresis and then used for DNA sequencing. Sequencing primers were identical to the PCR primers except 2F (GGGATTACAGGTGTGGGCC) and 3R (TCATCAGTACCATCAAAAGC). An alternative set of PCR/sequencing primers which amplify the coding region in a series of short, overlapping fragments were designed for use with degraded DNA extracted from FFPE material (Supplementary Table 1).
Processing of PCR products and DNA sequencing using the BigDye (v1.1) Terminator kit (Applied Biosystems) was carried out as previously described (9) using an ABI Prism 3730 Genetic Analyzer (Applied Biosystems). Data was processed using Sequencing Analysis software (Applied Biosystems) and analysed using Mutation Surveyor software (SoftGenetics, State College, USA) and by visual inspection of electropherograms. All samples were initially sequenced in a single direction. Samples which contained a mutation and samples in which the first trace was equivocal were additionally sequenced in the opposite direction. Mutations were verified by confirming their absence in matched normal DNA.
Multiplex ligation–dependent probe amplification (MLPA)
MLPA analysis to detect copy number changes in the VHL gene was carried out using the SALSA P016B VHL probe kit (MRC-Holland, Amsterdam, Netherlands) as previously described (9). Data was generated using PeakScanner software (Applied Biosystems) and analysed by calculating a dosage quotient for every possible pairwise combination of the relative peak heights of test and control probes in the test sample compared to a wild type reference sample. Where the mean dosage quotient for any test peak fell below 0.7, it was considered as potentially deleted; where it rose above 1.2, it was considered as potentially duplicated.
Genotyping of SNP rs779805
The common polymorphism rs779805 located in the 5′UTR of VHL 195 bases upstream of the ATG, which was previously suggested to correlate with methylation (9), was analysed by pyrosequencing. Primers for amplification of a 108bp region containing SNP rs779805 (forward primer: TACAGTAACGAGTTGGCCTAGCCT and reverse primer: biotin-ACGCGCTCGCGGAAATAG) and pyrosequencing analysis of the SNP (TCGCCTCCGTTACAA) were designed using proprietary pyrosequencing assay design software.
PCR reactions (final volume 25 μL) contained 12.5 μL of Qiagen HotStarTaq Master Mix (Qiagen, Crawley, UK), additional magnesium chloride to give a final concentration of 2 mM, 200 nM each of forward and reverse primers and 20 ng of genomic DNA. PCR products were amplified using 94°C for 12 min, 40 cycles of 94°C for 10 sec, 55°C for 20 sec and 72°C for 20 sec. PCR products were sequenced by pyrosequencing on a PyroMark ID system (Biotage AB, Sweden) following the manufacturer's instructions. Data analysis was performed by the PyroMark ID software set to the SNP allele quantification (AQ) mode.
Statistical analysis
The data analysis for this study was undertaken using SAS software, Version 9.1 (SAS Institute Inc., Cary, NC, USA). Summary statistics were calculated using contingency table analysis and evaluated using Fisher's exact test. Due to the large number of tests performed, the outcomes are regarded as exploratory and only _p_-values of 0.01 or lower suggest significance. Patients with grade 1 or grade 2 tumours were grouped together due to the small numbers in each group. Disease-free (defined as date of operation to date of relapse or death due to cancer), cancer-specific and overall survivals were calculated and analysed using Kaplan-Meier estimates of the survivor function, the log-rank test (with trend tests for variables with ordered categories) and Cox's Multivariate Proportional Hazards analysis to account for the clinical parameters grade, stage and microvascular invasion (MVI) as well as the VHL alterations mutation, methylation and LOH.
Results
Genetic and epigenetic analysis of VHL in sporadic conventional RCC
Our previous study (9) was extended by analysis of further banked samples and additionally incorporated more stringent review in terms of histological classification of tumours as conventional RCC and estimation of tumour cell numbers in tissue samples used for DNA extraction. Seven tumours originally diagnosed as conventional RCC were reclassified as chromophobe RCC and removed from the analysis; this included three patients from the original study (73, 108 and 153) for which no VHL involvement had been identified. After review, 177 patients with sporadic conventional RCC were included (Table 1).
Table 1.
Clinical characteristics of the 177 patients with conventional (clear cell) RCC. Fuhrman's grading system and UICC TMN staging system were used for pathological diagnosis.
| Variables | Number of patients |
|---|---|
| Sex | 110 male / 67 female |
| Age | Range 35 – 86; median 63 |
| Grade Unknown 1 2 3 4 | 1 6 46 77 47 |
| Tumour 1a 1b 2 3a 3b 3c 4 | 31 35 12 41 (3 at least 3a) 57 (9 at least 3b) 0 1 |
| Node X 0 1/2 | 11 138 28 |
| Metastasis X 0 1 | 1 127 49 |
| Stage I II III IV | 59 9 60 (5 at least stage III) 49 |
The common single nucleotide polymorphism rs779805 was determined in 85 of the 87 additional patients added to this study and showed a genotype distribution of A/A 31, A/G 39 and G/G 15, therefore 63.5% were heterozygous or homozygous for the polymorphism. Other polymorphisms identified were rs34661876 IVS2+43 A>G (c.463+43 A>G) in 7 patients, rs61758376 IVS1+5 G>C (c.340+5 G>C) in one patient and rs35460768 c.74 C>T P25L in one patient.
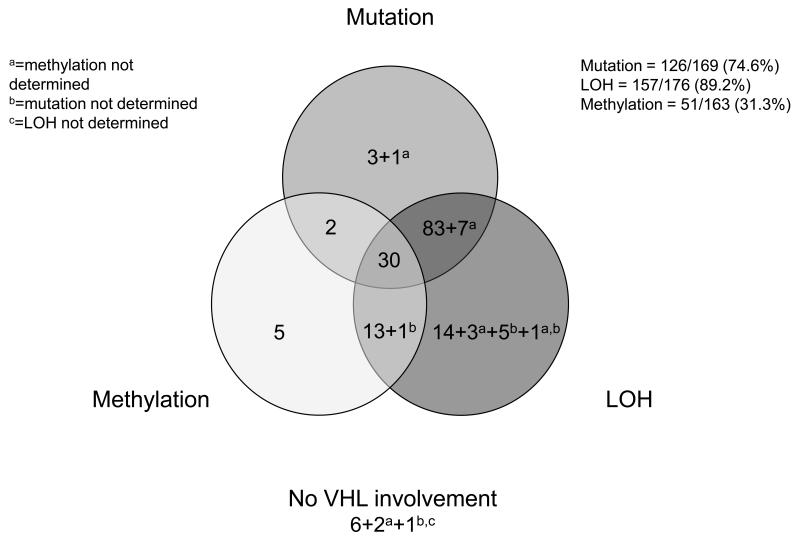
In the combined data set, LOH, mutation and methylation data were all collected for 156/177 tumours. Data were incomplete as reanalysis of samples with low tumour cell number using FFPE material was not possible for all, particularly for methylation (Figure 1, Table 2). LOH was found in 157/176 (89.2%) tumours; in 26 cases this was confirmed by MLPA. Only one tumour showed evidence of altered gene dosage by MLPA in the absence of loss of flanking markers by LOH analysis. Mutation in the VHL coding region or within the region of introns close to intron-exon boundaries was found in 126/169 (74.6%) cases of which 120 (95.2%) also had LOH. One further tumour was found to have a mutation in the 5'UTR (sample 36 (9)); this mutation may have an effect on the efficiency of transcription and/or translation. Methylation was found in 51/163 (31.3%) tumours; of these 30 were accompanied by LOH and mutation, 14 by LOH and 2 by mutation. Evidence of biallelic inactivation, defined as LOH and mutation or methylation alone, was found in 141/164 (86.0%) cases. Mutation or methylation was found in 145/165 (87.9%) cases, similar to that reported by Nickerson et al (34). There was no apparent VHL involvement (i.e. no LOH, methylation or mutation) in only 6/174 (3.4%) of samples.
Figure 1.
Summary of VHL alterations identified in 177 patients with sporadic conventional RCC.
Table 2.
Summary of the alterations in the VHL gene in patients with sporadic conventional RCC. The table includes 86 new tumours (IDs 157 onwards) and updated results for 16 samples (IDs 4 to 151) from our initial study. Three patients from the initial study (73, 108 and 153) were reclassified as chromophobe whilst patients 47 and 79 had low tumour cell numbers and FFPE material was not available for re-analysis so these were removed from the analysis. Results for the remaining 75 cases in the initial study were unchanged (9). Mutation nomenclature is in accordance with guidelines at http://www.hgvs.org/mutnomen/. Ensembl was used for both nucleotide numbering and mRNA (ENST00000256474) with A of the first initiator ATG being 1.
| ID | Histology | Grade & pTNM | LOH? | Methyln? | Mutation? | Mutation? | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomic change | mRNA change | Exon | Codon/ amino acid change | Mutation type | Previously found? | ||||||
| 4 | RCC Conv | G1 T1b,N0,M0 | + | nd | + | 10166569_10166570 del CT | 562_563 delCT | 3 | L188fs | FS | N* |
| 7 | RCC Conv | G2 T3a,N0,M0 | + | − | + | 10158781_10158784 del4 | 250_253 del4 | 1 | V84fs | FS | N |
| 24 | RCC Conv | G2 T1a,N0,M0 | + | − | + | 10166603 dup A | 596 dup A | 3 | R200fs | FS | N |
| 34 | RCC Conv | G3 T3b,N0,M0 | + | − | + | nd | nd | nd | nd | nd | − |
| 52 | RCC Conv | G3 T3b,N0,M0 | + | − | nd | nd | nd | nd | nd | nd | − |
| 58 | RCC Conv + sc | G4 T3b,N1,M0 | + | nd | − | − | − | − | − | − | − |
| 64 | RCC Conv | G2 T1a,NX,M1 | + | − | nd | nd | nd | nd | nd | nd | − |
| 81 | RCC Conv | G2 T3a,N0,M0 | − | − | − | − | − | − | − | − | − |
| 88 | RCC Conv | G4 T2,N0,M0 | + | nd | + | 10166551 del A | 544 delA | 3 | R182fs | FS | N |
| 112 | RCC Conv | G3 T3a,N0,M0 | − | nd | + | 10166486_10166500 del 15 | 479_493 del15 | 3 | E160_Q164 del | IFD | N* |
| 132 | RCC Conv | G3 T1b,N0,M0 | + | nd | + | 10158862 A>G | 331 A>G | 1 | S111G | MS | Y |
| 137 | RCC Conv | G3 T3b,N0,M0 | + | + | nd | nd | nd | nd | nd | nd | − |
| 143 | RCC Conv + sc | G4 T3b,N1,M0 | − | − | − | − | − | − | − | − | − |
| 144 | RCC Conv | G3 T1b,N0,M0 | + | − | nd | nd | nd | nd | nd | nd | − |
| 150 | RCC Conv | G3 T3a,NX,M0 | + | nd | − | − | − | − | − | − | − |
| 151 | RCC Conv + sc | G4 T1b,N0,M0 | + | − | + | 10166480 T>A | 473 T>A | 3 | L158Q | MS | Y |
| 157 | RCC Conv | G4 T1a,N0,M1 | + | − | + | 10158787 C>G | 256 C>G | 1 | P86A | MS | N* |
| 160 | RCC Conv | G2 T1a,N0,M0 | + | − | + | 10158851 G>C +10158852C>A | 320_321 delinsCA | 1 | R107P | MS | Y |
| 163 | RCC Conv | G3 T2,N0,M0 | + | − | − | − | − | − | − | − | − |
| 164 | RCC Conv | G3 T3a,N0,M0 | + | − | + | 10166532 C>A | 525 C>A | 3 | Y175X | N | Y |
| 165 | RCC Conv | G3 T2,N1,M0 | + | − | − | − | − | − | − | − | − |
| 166 | RCC Conv | G4 T3b,N0,M0 | + | − | + | 10163279 dup A | 422 dup A | 2 | N141fs | FS | Y |
| 167 | RCC Conv | G3 T1b,N0,M0 | + | + | + | 10158873 del T | 340+2 del T (IVS1+2del) | 1 | Spl | − | |
| 171 | RCC Conv | G3 T3b,N0,M0 | + | + | + | 10163226 del G | 369 del G | 2 | T124fs (G123fs) | FS | Y |
| 172 | RCC Conv | G4 T1b,N1,M1 | + | + | + | 10163301 dup T | 444 dup T | 2 | A149fs (F148fs) | FS | Y |
| 173 | RCC Conv | G3 T1a,N0,M0 | − | + | − | − | − | − | − | − | − |
| 177 | RCC Conv | G3 T3a,N0,M0 | + | + | + | 10166484 delA | 477 delA | 3 | E160fs (K159fs) | FS | N* |
| 179 | RCC Conv | G3 T1b,N0,M0 | + | + | − | − | − | − | − | − | − |
| 180 | RCC Conv | GX T1b,N0,M1 | nd | − | nd | nd | nd | nd | nd | nd | − |
| 181 | RCC Conv | G4 T3b,N1,M0 | + | − | + | 10163300-10163301 del TT | 443_444 del TT | 2 | F148fs | FS | Y |
| 182 | RCC Conv | G3 T1a,N0,M0 | + | − | + | 10166474 dup A | 467 dup A | 3 | Y156X | N | N* |
| 183 | RCC Conv | G3 T3b,N0,M0 | + | + | + | 10163218 G>T | 361 G>T | 2 | D121Y | MS | N* |
| 186 | RCC Conv | G4 T1b,N0,M0 | + | − | + | 10166565 del A | 558 del A | 3 | D187fs | FS | N* |
| 187 | RCC Conv | G4 T3a,N0,M0 | − | + | − | − | − | − | − | − | − |
| 189 | RCC Conv | G2 T1b,N0,M0 | + | − | + | 10158757-10158766 del 10 | 226_235 del 10 | 1 | F76fs | FS | N* |
| 194 | RCC Conv | G2 T1b,N0,M0 | + | nd | nd | nd | nd | nd | nd | nd | − |
| 195 | RCC Conv | G3 T1b,N0,M1 | + | + | − | − | − | − | − | − | − |
| 197 | RCC Conv | G4 T3b,N1,M1 | − | + | + | 10158714 del C | 183 del C | 1 | V62fs (P61fs) | FS | Y |
| 198 | RCC Conv | G4 T3b,N1,M1 | + | − | + | 10158862-10158867 del 6 | 331_336 del 6 | 1 | S111_Y112 del | IFD | N |
| 199 | RCC Conv | G3 T3a,N0,M1 | + | − | + | 10163307 del T | 450 del T | 2 | N150fs | FS | Y |
| 200 | RCC Conv | G3 T1a,N1,M1 | + | + | + | 10166558 T>C | 551 T>C | 3 | L184P | MS | Y |
| 201 | RCC Conv | G2 T1a,N0,M0 | + | + | − | − | − | − | − | − | − |
| 202 | RCC Conv | G3 T3a,N0,M1 | + | − | + | 10166570 dup T | 563 dup T | 3 | E189fs (L188fs) | FS | N* |
| 203 | RCC Conv | G4 T3a,N1,M1 | − | − | − | − | − | − | − | − | − |
| 205 | RCC Conv | G2 T1a,N0,M0 | + | − | + | 10166549-10166552 del 4 | 542_545 del 4 | 3 | V181fs | FS | N |
| 207 | RCC Conv | G3 T1a,N0,M0 | + | nd | − | − | − | − | − | − | − |
| 209 | RCC Conv | G4 T3a,Nx,M1 | + | nd | + | 10158863 G>T | 332 G>T | 1 | S111I | MS | N* |
| 212 | RCC Conv | G2 T1b,N0,M0 | + | − | + | 10166495 T>A | 488 T>A | 3 | L163H | MS | N* |
| 214 | RCC Conv | G3 T1b,N0,M0 | − | − | + | 10158783 del G | 252 del G | 1 | L85fs (V84fs) | FS | N* |
| 215 | RCC Conv | G4 T2,Nx,M0 | + | − | + | 10158702 del G | 171 del G | 1 | R58fs (G57fs) | FS | Y |
| 217 | RCC Conv | G3 T1b,N0,M0 | + | − | − | − | − | − | − | − | − |
| 218 | RCC Conv | G3 T3b,N0,M1 | + | + | + | 10166504 T>G | 497 T>G | 3 | V166G | MS | Y |
| 220 | RCC Conv | G4 T3b,N0,M1 | − | − | − | − | − | − | − | − | − |
| 221 | RCC Conv | G2 T3a,N0,M0 | + | − | + | 10158827 del C | 296 del C | 1 | P99fs | FS | Y |
| 222 | RCC Conv | G2 T3a,N0,M1 | + | − | + | 10166480-10166505 dup 26 | 473_489 dup 26 | 3 | R167X | N | N |
| 224 | RCC Conv | G3 T1b,N0,M0 | + | + | + | 10163261 dup T | 404 dup T | 2 | L135fs | FS | N* |
| 225 | RCC Conv | G3 T1b,N1,M0 | − | nd | − | − | − | − | − | − | − |
| 226 | RCC Conv | G4 T3b,N2,M1 | + | − | − | − | − | − | − | − | − |
| 227 | RCC Conv | G4 T3b,N2,M1 | + | − | + | 10166485 delins TC | 478 delins TC | 3 | E160fs | FS | N |
| 228 | RCC Conv | G2 T1a,N0,M0 | + | − | − | − | − | − | − | − | − |
| 230 | RCC Conv | G3 T3b,N0,M0 | + | + | − | − | − | − | − | − | − |
| 231 | RCC Conv | G4 T3a,N2,M1 | + | + | − | − | − | − | − | − | − |
| 232 | RCC Conv | G3 T3b,N0,M1 | + | − | + | 10158740 del A | 209 del A | 1 | E70fs | FS | N* |
| 233 | RCC Conv | G4 T3b,N1,M1 | + | + | − | − | − | − | − | − | − |
| 234 | RCC Conv | G2 T3a,N0,Mx | + | nd | + | 10158702_10158711 del 10 | 171_180 del 10 | 1 | R58fs | FS | N* |
| 235 | RCC Conv | G3 T3b,N0,M1 | + | − | + | 10158731 del A | 200 del A | 1 | N67fs | FS | N* |
| 236 | RCC Conv | G3 T3a,N0,M0 | + | − | + | 10158805-10158809 del5 | 274_278 del5 | 1 | D92fs | FS | N* |
| 237 | RCC Conv | G3 T3a,N0,M0 | + | + | + | 10158744 del C | 213 del C | 1 | S72fs (P71fs) | FS | Y |
| 239 | RCC Conv | G3 T3b,N0,M0 | + | − | + | 10163288 del G | 431 del G | 2 | G144fs | FS | Y |
| 242 | RCC Conv | G3 T1b,N0,M0 | + | + | − | − | − | − | − | − | − |
| 243 | RCC Conv | G3 T3b,N0,M0 | + | − | + | 10158868 C>T | 337 C>T | 1 | R113X | N | N |
| 244 | RCC Conv | G4 T3b,N0,M1 | + | − | + | 10158779-10158786 del 8 | 248-255 del 8 | 1 | V83fs | FS | N* |
| 245 | RCC Conv | G3 T3a,N0,M1 | + | − | + | 10158759-10158767 delins GA | 228-236 delins GA | 1 | F76fs | FS | N* |
| 247 | RCC Conv | G3 T3a,N0,M0 | + | + | + | 10158793 T>A | 262 T>A | 1 | W88R | MS | Y |
| 248 | RCC Conv | G3 T1a,N0,M0 | + | + | − | − | − | − | − | − | − |
| 252 | RCC Conv + sc | G4 T3b,N2,M1 | + | − | + | 10166627-10166642 dup16 | 620_635 dup16 | 3 | D213fs (G212fs) | FS | N* |
| 253 | RCC Conv | G2 T2,N0,M0 | + | − | − | − | − | − | − | − | − |
| 254 | RCC Conv | G3 T3b,N0,M0 | + | − | nd | nd | nd | nd | nd | nd | − |
| 255 | RCC Conv | G3 T2,N0,M0 | + | + | + | 10163217-10163224 del 8 | 360-367 del 8 | 2 | A122fs | FS | N* |
| 256 | RCC Conv | G4 T3b,N0,M1 | + | + | − | − | − | − | − | − | − |
| 261 | RCC Conv | G3 T3a,N0,M0 | + | − | + | 10158800 A>T | 269 A>T | 1 | N90I | MS | Y |
| 267 | RCC Conv | G3 T3b,N1,M0 | + | + | + | 10163245 G>C | 388 G>C | 2 | V130L | MS | Y |
| 269 | RCC Conv + sc | G4 T3b,N0,M1 | + | − | + | 10163283 del T | 426 del T | 2 | D143fs (V142fs) | FS | Y |
| 270 | RCC Conv | G2 T3a,N0,M0 | + | − | + | 10158749 del A | 218 del A | 1 | Q73fs | FS | N* |
| 271 | RCC Conv | G3 T1b,N0,M0 | + | + | + | 10158725 C>A | 194 C>A | 1 | S65X | N | Y |
| 272 | RCC Conv | G4 T3b,N1,M1 | + | nd | + | 10158703_10158704 delins TA | 172_173 delins TA | 1 | R58X | N | − |
| 273 | RCC Conv | G3 T3b,N0,M0 | + | − | + | 10163206 T>C | 349 T>C | 2 | W117R | MS | N* |
| 274 | RCC Conv | G2 T2,N0,M0 | − | nd | − | − | − | − | − | − | − |
| 276 | RCC Conv | G4 T3b,N0,M0 | + | − | − | − | − | − | − | − | − |
| 278 | RCC Conv | G2 T1b,N0,M0 | + | − | + | 10163281 delG | 424 delG | 2 | V142fs | FS | N* |
| 279 | RCC Conv | G3 T3b,N0,M0 | + | + | + | 10166492 G>A | 485 G>A | 3 | C162Y | MS | Y |
| 280 | RCC Conv | G3 T3b,N0,M1 | + | − | − | − | − | − | − | − | − |
| 281 | RCC Conv | G3 T3b,N0,M0 | + | + | + | 10163187 T>A | 341-11 T>A | 2 | − | Spl? | |
| 282 | RCC Conv | G3 T3b,N0,M0 | − | + | − | − | − | − | − | − | − |
| 283 | RCC Conv + sc | G4 T3b,N1,M0 | + | + | + | 10158871 G>C | 340 G>C | 1 | G114R | MS Spl? | Y |
| 284 | RCC Conv | G2 T1a,N0,M0 | + | + | + | 10163229 delA | 372 delA | 2 | H125fs (T124fs) | FS | N* |
| 286 | RCC Conv | G3 T3b,N1,M1 | + | − | + | 10163287 G>T | 430 G>T | 2 | G144X | N | N |
| 290 | RCC Conv + sc | G4 T3a,N1,M0 | − | + | − | − | − | − | − | − | − |
| 291 | RCC Conv | G3 T3a,Nx,M1 | + | + | + | 10166488 C>T | 481 C>T | 3 | R161X | N | Y |
| 292 | RCC Conv + sc | G4 T3b,Nx,M0 | + | − | + | 10158833 T>C | 302 T>C | 1 | L101P | MS | Y |
| 294 | RCC Conv | G3 T1a,N0,M0 | + | − | + | 10163303 C>A | 446 C>A | 2 | A149D | MS | Y |
| 295 | RCC Conv | G2 T1a,N0,M0 | + | − | + | 10163270 C>G | 413 C>G | 2 | P138R | MS | Y |
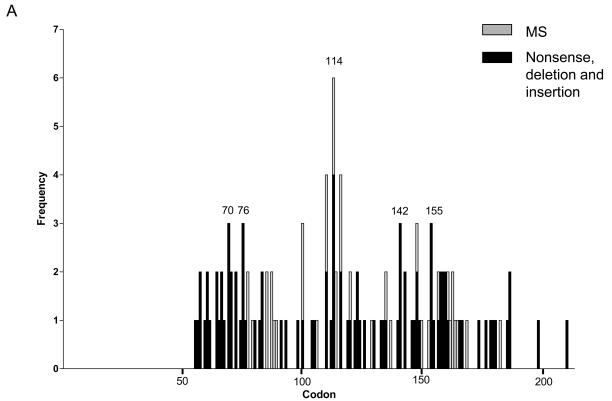
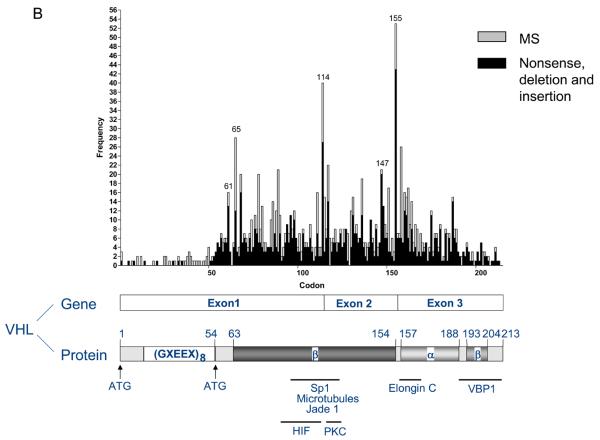
Of the 126 tumours with VHL mutations, all except one from the initial study (sample 98 (9)) were single mutations. Truncations (frameshift mutations (65), nonsense mutations (14) and mutations close to intron/exon boundaries that were predicted to remove splice sites (7) or potentially introduce an alternative splice site (1)) accounted for 87/127 (68.5%), missense mutations for 36/127 (28.3%) and in-frame deletions for 4/127 (3.1%). Six mutations were intronic with 54 (44.6%), 36 (29.8%) and 31 (25.6%) of exonic mutations occurring in exons 1 to 3 respectively. The distribution of mutations in our study and a composite of mutations from studies of sporadic conventional RCC is shown in Figure 2A and B.
Figure 2.
Spectrum of VHL mutations found in sporadic conventional RCC. A, mutations (n=126) reported from analysis of 177 samples in this study; B, cumulative data of 1244 mutations reported in the literature (5, 6, 8-45, 47). All studies have been checked to ensure that data published in multiple studies was not duplicated with some publications omitted for this reason or due to sequences not being available (4, 7, 46). Only conventional (clear cell) RCC cases were included except for one study where the specific pathological subtype of RCC was not indicated (44). Results from patients following trichloroethylene exposure or with end stage renal disease were excluded as previously (9).
Association of VHL status with clinical and pathological variables
Associations between VHL status and clinicopathological variables including sex, tumour grade and stage (pTNM as individual categories and overall stage), presence of rhabdoid/sarcomatoid features, maximum tumour diameter, MVI, coagulative tumour necrosis and disease-free, cancer-specific and overall survival were examined. There was a possible association between mutation and sex (84.6% of females had a mutation compared with 68.2% of males; p=0.0189) and methylation and grade (43.7% of grade 3 samples had methylation compared with 18.8% of grade 1 and 2 and 25.6% of grade 4; _p_=0.0158). Associations were also suggested between LOH and either grade (76.6% of grade 4 tumours had LOH, compared with 93.5% of grade 3 and 94.2% of grade 1/2 tumours; _p_=0.0050), rhabdoid/sarcomatoid features (9.6% of tumours with LOH had rhabdoid/sarcomatoid features compared with 26.3% of tumours with no LOH; _p_=0.046), nodal status (91.2% of patients with no nodal involvement had LOH compared with 75.0% of patients with nodal involvement; _p_=0.0227) and necrosis (81.0% of tumours with necrosis had LOH compared to 93.1% with no necrosis; _p_=0.0211).
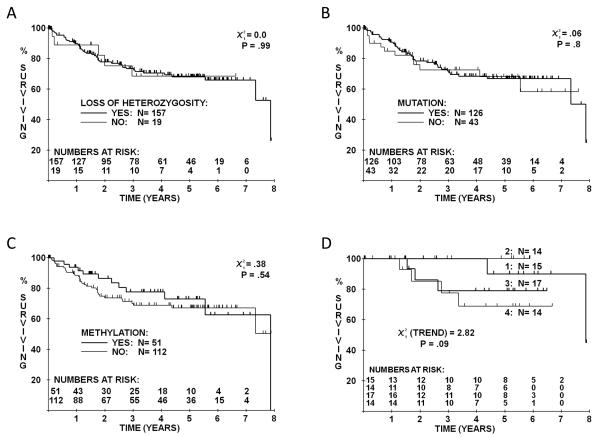
As expected, stage and grade showed a strongly significant association with cancer-specific survival, disease-free survival and overall survival (Supplementary Figure 1). There was no evidence of a significant difference in survival based on presence and absence of mutation, LOH or methylation (Figure 3A-C) or biallelic inactivation (mutation and LOH or methylation alone). There was a suggestion that LOH and mutation correlate with improved prognosis compared to patients with no VHL involvement and a similar putative relationship was seen with methylation; however sample numbers in the no VHL involvement group (n=6) were so small that this must be confirmed in an independent dataset. These findings were confirmed by Cox's Multivariate Proportional Hazards analysis, with none of the VHL comparisons being significant after accounting for grade, stage and MVI.
Figure 3.
Cancer-specific survival curves for conventional RCC patients based on presence or absence of A, LOH, B, mutation and C, methylation, for all patients and D, mutation truncation group for the subset of patients with stage I to III disease.
There were no significant associations between any of the clinicopathological variables and the different types of mutations (truncation, missense or none) or the location of the mutations in terms of truncation groups (group 1, codons 60-83; group 2, codons 84-122; group 3, codons 123-156; group 4, codons 157-213). However there was some evidence of a trend by truncation group category with truncation groups 3 and 4 possibly having a worse prognosis than groups 1 and 2, particularly in stage I to III patients (Figure 3D).
Discussion
One of the most significant findings of this large and comprehensive genetic and epigenetic analysis of the VHL gene in sporadic conventional RCC was the very small numbers of tumours (<5%) with no VHL involvement. Furthermore these six cases were difficult to classify accurately in most cases due to dedifferentiation. This is consistent with a recent study analysing 205 cases of conventional RCC where 91% of cases had mutation or methylation (34), and a smaller study where biallelic inactivation of VHL was found in 49/57 (86%) cases (22) with the corresponding figures for our study being 88% and 86% respectively. Much lower figures have been reported previously in many of the published studies. The higher levels currently detected may be due to the improved sensitivity of mutation detection, greater stringency in tissue selection for nucleic acid extraction and elimination of non-conventional RCC subtypes following pathological review. These findings confirm that that VHL involvement may provide a molecular basis for classification as conventional RCC (49).
The rate of LOH (89.2%) observed in our series was comparable with other published data (15, 16, 21, 24). In 14 samples LOH alone was observed which could potentially be due to mutations outside the region screened in this study such as deep intronic mutations that have an effect on splicing or mutations in the promoter that alter transcription. Alternatively these RCCs may represent a subgroup involving other tumour suppressor genes on chromosome 3p. The mutation rate (74.6%) found in this study was also high, being second only to a recently published paper which employed endonuclease scanning and sequencing for high throughput and sensitive mutation detection and reported mutations in 82.4% of 205 cases (34). The distribution of our mutations showed a high frequency of mutations around codons 65-76, 86-90 and 158-168 as well as at intron-exon boundaries (114 and 155), consistent with mutations reported in the literature in sporadic conventional RCC. No mutations were found in the first 54 codons of the VHL gene in this study and in the literature only 54 mutations affecting codons 1-54 have been reported, the majority of which result from three studies (31, 39, 43). The rate of methylation (31.3%) reported here was notably higher than that found in other studies with most reporting ~5-15% and the previous highest being 20.4% in our initial study (9, 11, 28, 29, 34, 38, 46). Different methods have been adopted for methylation analysis and different CpG islands analysed and this together with possible differences in sensitivity in the PCR make comparability between studies difficult.
The lack of consistency between studies in terms of associations between VHL changes and clinical variables probably arises from differences in study stringency (methodology, accurate histological subclassification and tissue selection) and the relatively small size of many studies which with multiple testing can lead to false positives which fail to replicate. Few studies have reported on the significance of VHL changes and patient outcome. The presence of VHL mutations has been associated with improved cancer-free and cancer-specific survival in patients with stage I to III disease treated with radical nephrectomy (_p_=0.0024 and _p_=0.023); this held for subsets of patients with higher grade (G2 to 4 or G3 and 4) and stage (II and III or III) tumours (46). The absence of mutation was associated with more aggressive tumours in a study of 100 patients that also examined CAIX expression. High CAIX was associated with VHL mutation and the data allowed stratification of patients into 3 groups according to the presence of VHL mutation status and/or strong CAIX expression (36). Although we did not find these relationships between VHL mutation itself and outcome, our study does suggest that no VHL involvement confers a worse prognosis than having either methylation or LOH and mutation, although these associations were not statistically significant, possibly given the very small numbers without VHL involvement. Similarly tumours with no LOH or with no VHL involvement were associated with necrosis, higher grade and sarcomatoid features and there was a trend towards the group with mutations potentially encoding the least truncated VHL protein having a worse prognosis, although again this was not significant.
Loss of function mutations have been observed to associate with poor prognosis compared to missense or no mutation (28, 39). A further study showed no overall association between the presence of mutation and patient characteristics, but did find relationships with the prevalence of particular subtypes of mutation such as between nonsense mutations and grade and nodal status and metastasis (34). Our study failed to confirm these findings, possibly due to the small number of events in some subcategories, even in a relatively large study. Preliminary studies have also suggested that VHL mutation status correlates with response to VEGF-directed therapies (38, 50) and corroboration of these observations in independent cohorts is now of great importance.
Examination of the VHL protein itself and downstream proteins/pathways may provide more insight into the functional consequences of particular genetic changes and their clinical relevance. In the case of truncations resulting in early termination, nonsense mediated decay may occur resulting in abrogation of protein expression similar to the potential effect of promoter methylation. Other mutations such as missense mutations or truncations not resulting in nonsense-mediated decay may encode proteins which have different effects on particular VHL functions. A recent study subclassifying conventional RCCs on the basis of the activity of downstream pathways showed that VHL-involved tumours expressing HIF-2α showed increased c-Myc activity whereas RCCs with no apparent VHL involvement (HIF-α negative) or VHL-involved tumours expressing HIF-1α/HIF-2α showed increased Akt/mTOR and ERK/MAPK signalling (22). The potential significance of this molecular stratification remains to be determined, but clearly indicates the need to analyse the functional consequences of the VHL changes at the protein level and is an exciting area of research that warrants future investigation.
Statement of translational relevance.
The central role played by the VHL tumour suppressor in the development of sporadic conventional RCC has been clearly demonstrated, but the clinical significance of particular VHL defects remains to be determined, with a lack of agreement between initial studies. Larger more stringent studies using robust methods for LOH, mutation and methylation detection applied to good quality tumour samples with more extensive clinical data including clinical outcome are required. This study extends our previous report to 177 patients with conventional RCC. The results indicate VHL gene involvement in almost all conventional RCCs and suggest that subclassification into groups that correlate with prognosis or response to therapy will be made on the basis of the biological effects of specific VHL alterations rather than their presence per se.
Supplementary Material
Supplementary Figure 1
Cancer-specific survival curves for conventional RCC patients based on A, stage and B, grade.
Supplementary Data
Acknowledgements
The funding support of Cancer Research UK, the assistance of staff in Urology and Oncology and the agreement of participating patients is gratefully acknowledged.
Abbreviations
RCC
Renal cell carcinoma
VHL
von-Hippel Lindau
VEGF
vascular endothelial growth factor
HIF
Hypoxia inducible factor
LOH
loss of heterozygosity
MLPA
multiplex ligation-dependent probe amplification
Footnotes
References
- 1.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–90. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 2.Drucker BJ. Renal cell carcinoma: current status and future prospects. Cancer Treat Rev. 2005;31:536–45. doi: 10.1016/j.ctrv.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Rini BI. Vascular endothelial growth factor-targeted therapy in metastatic renal cell carcinoma. Cancer. 2009;115:2306–12. doi: 10.1002/cncr.24227. [DOI] [PubMed] [Google Scholar]
- 4.Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–20. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 5.Ashida S, Furihata M, Tanimura M, et al. Molecular detection of von Hippel-Lindau gene mutations in urine and lymph node samples in patients with renal cell carcinoma: potential biomarkers for early diagnosis and postoperative metastatic status. J Urol. 2003;169:2089–93. doi: 10.1097/01.ju.0000063589.52935.84. [DOI] [PubMed] [Google Scholar]
- 6.Ashida S, Okuda H, Chikazawa M, et al. Detection of circulating cancer cells with von hippel-lindau gene mutation in peripheral blood of patients with renal cell carcinoma. Clin Cancer Res. 2000;6:3817–22. [PubMed] [Google Scholar]
- 7.Atkins DJ, Gingert C, Justenhoven C, et al. Concomitant deregulation of HIF1alpha and cell cycle proteins in VHL-mutated renal cell carcinomas. Virchows Arch. 2005;447:634–42. doi: 10.1007/s00428-005-1262-y. [DOI] [PubMed] [Google Scholar]
- 8.Bailly M, Bain C, Favrot MC, Ozturk M. Somatic mutations of von Hippel-Lindau (VHL) tumor-suppressor gene in European kidney cancers. Int J Cancer. 1995;63:660–4. doi: 10.1002/ijc.2910630510. [DOI] [PubMed] [Google Scholar]
- 9.Banks RE, Tirukonda P, Taylor C, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66:2000–11. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 10.Barnabas N, Amin MB, Pindolia K, Nanavati R, Worsham MJ. Mutations in the von Hippel-Lindau (VHL) gene refine differential diagnostic criteria in renal cell carcinoma. J Surg Oncol. 2002;80:52–60. doi: 10.1002/jso.10086. [DOI] [PubMed] [Google Scholar]
- 11.Brauch H, Weirich G, Brieger J, et al. VHL alterations in human clear cell renal cell carcinoma: association with advanced tumor stage and a novel hot spot mutation. Cancer Res. 2000;60:1942–8. [PubMed] [Google Scholar]
- 12.Brauch H, Weirich G, Klein B, Rabstein S, Bolt HM, Bruning T. VHL mutations in renal cell cancer: does occupational exposure to trichloroethylene make a difference? Toxicol Lett. 2004;151:301–10. doi: 10.1016/j.toxlet.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 13.Brieger J, Weidt EJ, Gansen K, Decker HJ. Detection of a novel germline mutation in the von Hippel-Lindau tumour-suppressor gene by fluorescence-labelled base excision sequence scanning (F-BESS) Clin Genet. 1999;56:210–5. doi: 10.1034/j.1399-0004.1999.560305.x. [DOI] [PubMed] [Google Scholar]
- 14.Charbotel B, Gad S, Caiola D, et al. Trichloroethylene exposure and somatic mutations of the VHL gene in patients with Renal Cell Carcinoma. J Occup Med Toxicol. 2007;2:13. doi: 10.1186/1745-6673-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster K, Prowse A, van den Berg A, et al. Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum Mol Genet. 1994;3:2169–73. doi: 10.1093/hmg/3.12.2169. [DOI] [PubMed] [Google Scholar]
- 16.Gallou C, Joly D, Mejean A, et al. Mutations of the VHL gene in sporadic renal cell carcinoma: definition of a risk factor for VHL patients to develop an RCC. Hum Mutat. 1999;13:464–75. doi: 10.1002/(SICI)1098-1004(1999)13:6<464::AID-HUMU6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 17.Gallou C, Longuemaux S, Delomenie C, et al. Association of GSTT1 non-null and NAT1 slow/rapid genotypes with von Hippel-Lindau tumour suppressor gene transversions in sporadic renal cell carcinoma. Pharmacogenetics. 2001;11:521–35. doi: 10.1097/00008571-200108000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Gemmill RM, Zhou M, Costa L, Korch C, Bukowski RM, Drabkin HA. Synergistic growth inhibition by Iressa and Rapamycin is modulated by VHL mutations in renal cell carcinoma. Br J Cancer. 2005;92:2266–77. doi: 10.1038/sj.bjc.6602646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gervais ML, Henry PC, Saravanan A, et al. Nuclear E-cadherin and VHL immunoreactivity are prognostic indicators of clear-cell renal cell carcinoma. Lab Invest. 2007;87:1252–64. doi: 10.1038/labinvest.3700684. [DOI] [PubMed] [Google Scholar]
- 20.Gimenez-Bachs JM, Salinas-Sanchez AS, Sanchez-Sanchez F, et al. Determination of vhl gene mutations in sporadic renal cell carcinoma. Eur Urol. 2006;49:1051–7. doi: 10.1016/j.eururo.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 22.Gordan JD, Lal P, Dondeti VR, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–46. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamano K, Esumi M, Igarashi H, et al. Biallelic inactivation of the von Hippel-Lindau tumor suppressor gene in sporadic renal cell carcinoma. J Urol. 2002;167:713–7. doi: 10.1016/S0022-5347(01)69132-8. [DOI] [PubMed] [Google Scholar]
- 24.He Z, Liu S, Guo M, Mao J, Hughson MD. Expression of fibronectin and HIF-1alpha in renal cell carcinomas: relationship to von Hippel-Lindau gene inactivation. Cancer Genet Cytogenet. 2004;152:89–94. doi: 10.1016/j.cancergencyto.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Hughson MD, He Z, Liu S, Coleman J, Shingleton WB. Expression of HIF-1 and ubiquitin in conventional renal cell carcinoma: relationship to mutations of the von Hippel-Lindau tumor suppressor gene. Cancer Genet Cytogenet. 2003;143:145–53. doi: 10.1016/s0165-4608(02)00856-7. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi H, Esumi M, Ishida H, Okada K. Vascular endothelial growth factor overexpression is correlated with von Hippel-Lindau tumor suppressor gene inactivation in patients with sporadic renal cell carcinoma. Cancer. 2002;95:47–53. doi: 10.1002/cncr.10635. [DOI] [PubMed] [Google Scholar]
- 27.Kenck C, Wilhelm M, Bugert P, Staehler G, Kovacs G. Mutation of the VHL gene is associated exclusively with the development of non-papillary renal cell carcinomas. J Pathol. 1996;179:157–61. doi: 10.1002/(SICI)1096-9896(199606)179:2<157::AID-PATH557>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 28.Kim JH, Jung CW, Cho YH, et al. Somatic VHL alteration and its impact on prognosis in patients with clear cell renal cell carcinoma. Oncol Rep. 2005;13:859–64. [PubMed] [Google Scholar]
- 29.Kondo K, Yao M, Yoshida M, et al. Comprehensive mutational analysis of the VHL gene in sporadic renal cell carcinoma: relationship to clinicopathological parameters. Genes Chromosomes Cancer. 2002;34:58–68. doi: 10.1002/gcc.10054. [DOI] [PubMed] [Google Scholar]
- 30.Lemm I, Lingott A, Pogge v, Strandmann E, et al. Loss of HNF1alpha function in human renal cell carcinoma: frequent mutations in the VHL gene but not the HNF1alpha gene. Mol Carcinog. 1999;24:305–14. [PubMed] [Google Scholar]
- 31.Ma X, Yang K, Lindblad P, Egevad L, Hemminki K. VHL gene alterations in renal cell carcinoma patients: novel hotspot or founder mutations and linkage disequilibrium. Oncogene. 2001;20:5393–400. doi: 10.1038/sj.onc.1204692. [DOI] [PubMed] [Google Scholar]
- 32.Meyer AJ, Hernandez A, Florl AR, et al. Novel mutations of the von hippel-lindau tumor-suppressor gene and rare DNA hypermethylation in renal-cell carcinoma cell lines of the clear-cell type. Int J Cancer. 2000;87:650–3. [PubMed] [Google Scholar]
- 33.Na X, Wu G, Ryan CK, Schoen SR, di'Santagnese PA, Messing EM. Overproduction of vascular endothelial growth factor related to von Hippel-Lindau tumor suppressor gene mutations and hypoxia-inducible factor-1 alpha expression in renal cell carcinomas. J Urol. 2003;170:588–92. doi: 10.1097/01.ju.0000074870.54671.98. [DOI] [PubMed] [Google Scholar]
- 34.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh RR, Park JY, Lee JH, et al. Expression of HGF/SF and Met protein is associated with genetic alterations of VHL gene in primary renal cell carcinomas. APMIS. 2002;110:229–38. doi: 10.1034/j.1600-0463.2002.100305.x. [DOI] [PubMed] [Google Scholar]
- 36.Patard JJ, Fergelot P, Karakiewicz PI, et al. Low CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. Int J Cancer. 2008;123:395–400. doi: 10.1002/ijc.23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rad FH, Ulusakarya A, Gad S, et al. Novel somatic mutations of the VHL gene in an erythropoietin-producing renal carcinoma associated with secondary polycythemia and elevated circulating endothelial progenitor cells. Am J Hematol. 2008;83:155–8. doi: 10.1002/ajh.21019. [DOI] [PubMed] [Google Scholar]
- 38.Rini BI, Jaeger E, Weinberg V, et al. Clinical response to therapy targeted at vascular endothelial growth factor in metastatic renal cell carcinoma: impact of patient characteristics and Von Hippel-Lindau gene status. BJU Int. 2006;98:756–62. doi: 10.1111/j.1464-410X.2006.06376.x. [DOI] [PubMed] [Google Scholar]
- 39.Schraml P, Struckmann K, Hatz F, et al. VHL mutations and their correlation with tumour cell proliferation, microvessel density, and patient prognosis in clear cell renal cell carcinoma. J Pathol. 2002;196:186–93. doi: 10.1002/path.1034. [DOI] [PubMed] [Google Scholar]
- 40.Shuin T, Kondo K, Torigoe S, et al. Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res. 1994;54:2852–5. [PubMed] [Google Scholar]
- 41.Suzuki H, Ueda T, Komiya A, et al. Mutational state of von Hippel-Lindau and adenomatous polyposis coli genes in renal tumors. Oncology. 1997;54:252–7. doi: 10.1159/000227697. [DOI] [PubMed] [Google Scholar]
- 42.Tsutsumi H, Miyamoto C, Furuichi Y, Yoshiike M, Nozawa S, Iwamoto T. VHL tumor suppressor gene: its mutation and protein level in renal cell carcinoma. Oncol Rep. 2003;10:1357–61. [PubMed] [Google Scholar]
- 43.van Houwelingen KP, van Dijk BA, Hulsbergen-van de Kaa CA, et al. Prevalence of von Hippel-Lindau gene mutations in sporadic renal cell carcinoma: results from The Netherlands cohort study. BMC Cancer. 2005;5:57. doi: 10.1186/1471-2407-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whaley JM, Naglich J, Gelbert L, et al. Germ-line mutations in the von Hippel-Lindau tumor-suppressor gene are similar to somatic von Hippel-Lindau aberrations in sporadic renal cell carcinoma. Am J Hum Genet. 1994;55:1092–102. [PMC free article] [PubMed] [Google Scholar]
- 45.Wiesener MS, Seyfarth M, Warnecke C, et al. Paraneoplastic erythrocytosis associated with an inactivating point mutation of the von Hippel-Lindau gene in a renal cell carcinoma. Blood. 2002;99:3562–5. doi: 10.1182/blood.v99.10.3562. [DOI] [PubMed] [Google Scholar]
- 46.Yao M, Yoshida M, Kishida T, et al. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. J Natl Cancer Inst. 2002;94:1569–75. doi: 10.1093/jnci/94.20.1569. [DOI] [PubMed] [Google Scholar]
- 47.Pavlovich CP, Padilla-Nash H, Wangsa D, et al. Patterns of aneuploidy in stage IV clear cell renal cell carcinoma revealed by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer. 2003;37:252–60. doi: 10.1002/gcc.10209. [DOI] [PubMed] [Google Scholar]
- 48.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 49.Kovacs G, Akhtar M, Beckwith BJ, et al. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183:131–3. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 50.Choueiri TK, Vaziri SA, Jaeger E, et al. von Hippel-Lindau gene status and response to vascular endothelial growth factor targeted therapy for metastatic clear cell renal cell carcinoma. J Urol. 2008;180:860–5. doi: 10.1016/j.juro.2008.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Cancer-specific survival curves for conventional RCC patients based on A, stage and B, grade.
Supplementary Data