Minimum energy path for a U6 RNA conformational change involving protonation, base pair rearrangement and base flipping (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 4.
Published in final edited form as: J Mol Biol. 2009 Jul 8;391(5):894–905. doi: 10.1016/j.jmb.2009.07.003
Abstract
The U6 RNA internal stem-loop (U6 ISL) is a highly conserved domain of the spliceosome that is important for pre-mRNA splicing. The U6 ISL contains an internal loop that is in equilibrium between two conformations controlled by the protonation state of an adenine (pKa = 6.5). Lower pH favors formation of a protonated C-A+ wobble pair and base flipping of the adjacent uracil. Higher pH favors stacking of the uracil, and allows an essential metal ion to bind at this position. Here, we define the minimal energy path for this conformational transition. To do this, we solved the U6 ISL structure at higher pH (8.0), in order to eliminate interference from the low pH conformer. This structure reveals disruption of the protonated C-A+ pair and formation of a new C-U pair, which explains the preference for a stacked uracil at higher pH. Next, we used Nudged Elastic Band (NEB) molecular dynamics simulations to calculate the minimum energy path between the two conformations. Our results indicate that the C-U pair is dynamic, which allows formation of the more stable C-A+ pair upon adenine protonation. After formation of the C-A+ pair, the unpaired uracil follows a minor groove base flipping pathway. Molecular dynamics simulations suggest that the extrahelical uracil is stabilized by contacts with the adjacent helix.
Keywords: U6 RNA, RNA Structure, NMR, Molecular Dynamics, Nudged Elastic Band, Base flipping
INTRODUCTION
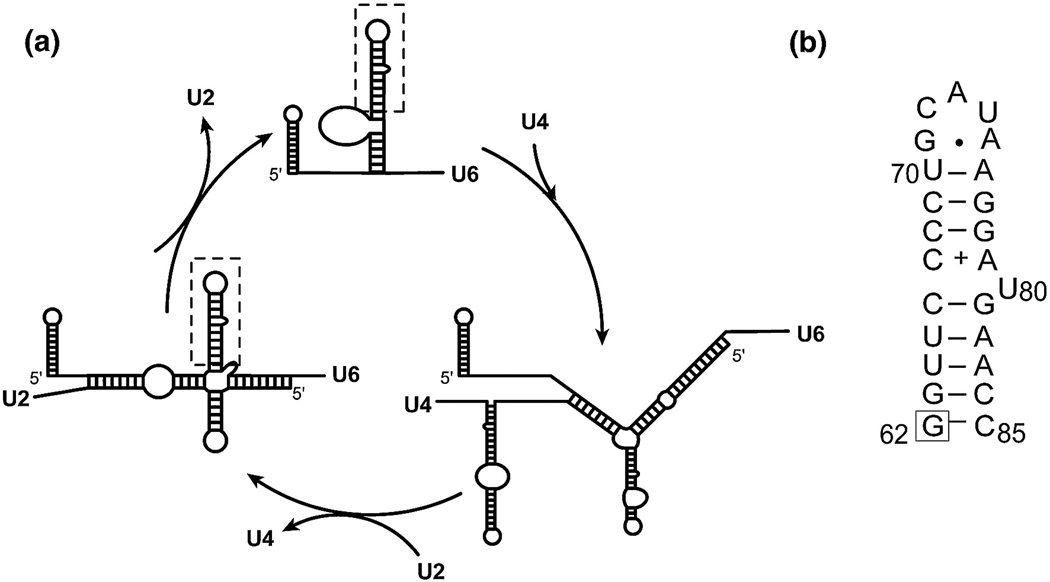
U6 RNA is one of five snRNAs (U1, U2, U4, U5 and U6) that, along with more than 70 proteins, assemble on the pre-mRNA to form a large and dynamic complex called the spliceosome (1, 2). The spliceosome catalyzes precise excision of introns and ligation of the flanking exons through a two-step phosphotransesterification reaction resulting in production of mature mRNA. Once fully assembled, the spliceosome is activated through a number of structural rearrangements in the snRNAs, facilitated by protein components (3). These rearrangements result in disruption of the U4/U6 complex and formation of the U2/U6 complex (Figure 1), which assists in splicing catalysis through substrate positioning and possibly chemistry (4). The association of U2 and U6 leads to formation of a highly conserved U6 Internal Stem Loop (U6 ISL) (3).
Figure 1.
A. Schematic diagram of Saccharomyces cerevisiae U6 snRNA and its conformations in the free U6 snRNP, the U4/U6 di-snRNP, and the U2/U6 snRNA complex in the activated spliceosome. The U6 ISL is indicated with a dashed box. B. Sequence and secondary structure of the U6 ISL used in this study. The previously described A62G substitution that does not affect the overall structure of the U6 ISL is indicated with a box. The secondary structure is shown in the low pH form, with a protonated A79 (indicated with a +) and a bulged U80.
The secondary structure of the yeast U6 RNA was investigated by in vivo and in vitro DMS probing experiments, and these data are consistent with the presence of an asymmetric 1×2 internal loop in the center of the U6 ISL (5) (Figure 1). This highly conserved internal loop seems to be crucial for spliceosome function, since it coordinates an essential Mg2+ ion at the U80 position (6, 7), mutations inside the bulge sequence generate growth defects that are not fully suppressed by compensatory mutations in U4 RNA (8), and hydroxyl radical probing experiments place the human ISL internal loop in the vicinity of the 5’ splice site in activated spliceosomes (9). In addition, the internal loop sequence found in the U6 ISL is also found in other RNAs. A database survey of 955 different RNA secondary structures found 12 occurrences of this 1×2 internal loop in 16S and 23S rRNAs and in RNase P (10).
The structure and dynamics of the U6 ISL have been investigated by NMR spectroscopy (7, 11–16). Consistent with the DMS probing results, the NMR solution structure determined at pH 7.0 shows that the U6 ISL contains a partially protonated C67-A79 wobble base pair and an unpaired U80 that is stacked in the helix (7, 11). In addition, comparison of the ISL structures at pH 7.0 and pH 5.7 indicates that the U80 nucleotide is in equilibrium between a stacked and a flipped-out conformation, with the latter predominating at low pH (12). The pH-dependent conformational switch appears to be induced by protonation of the A79-N1 atom (pKa = 6.5), which results in stabilization of the C67-A79 wobble pair and base-flipping of the neighboring U80 nucleotide (12). Analyses of NMR relaxation rates indicate that the lifetime of the protonated C67-A79 N1 base pair is 20 µs (12), while base-flipping of U80 occurs on the 80 µs timescale (13). Magnesium binding studies indicate that low pH inhibits metal ion binding, and the amount of metal ion binding is proportional to the fractional population of the stacked U80 (high pH) conformation (7, 13). Thus, the U6 ISL conformational equilibrium has the potential to regulate spliceosome activity (12, 13). However, the mechanism and associated energetics of this conformational transition have not been described, and in particular it is not clear why stacking of U80 is only favored upon disruption of the protonated C-A wobble pair. Although RNA is widely considered to be a dynamic molecule, the driving forces for structural rearrangements in RNA are poorly understood in general.
Since the population of protonated A79 is still significant at pH 7.0 (~24%), we hypothesized that the pH 7.0 U6 ISL NMR structure is affected by conformational averaging due to interference from the low pH conformation. If this hypothesis is true, then a high pH structure should reveal a different conformation of the U6 ISL internal loop. Here we determine the NMR structure of U6 ISL at pH 8.0, and indeed this NMR structure reveals a different loop conformation with a C67-U80 pair and an unpaired A79. The previously determined low pH (pH =5.7) structure and the pH 8.0 NMR structures were used as end-points for computation of the Minimum Energy Path (MEP) for the conformational change using Nudged Elastic Band (NEB) sampling (17). The results explain the physical basis for this conformational change, which involves a complex interplay of nucleobase protonation, alternative base pair formation, and base flipping.
RESULTS
NMR analysis
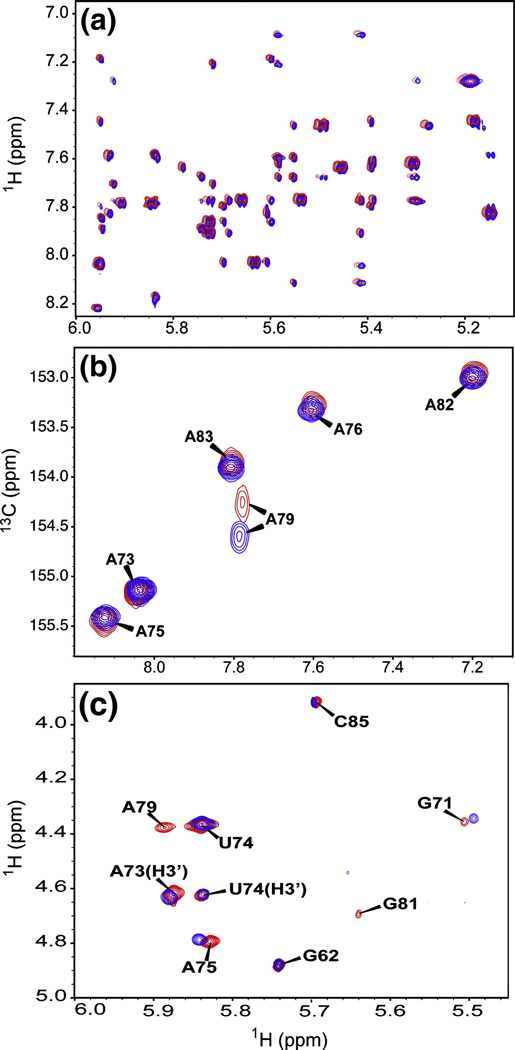
The U6 ISL RNA in this and previous studies incorporates an A62G substitution (Figure 1), which has no effect on the growth rate of yeast at 30°C (5) and no effect on the overall structure of the ISL (7), but allows for optimal in vitro transcription using T7 RNA polymerase. Comparison of 2D 1H-1H NOESY spectra at pH 8.0 and pH 7.0 reveals no detectable difference in the NOE pattern (Figure 2A). The high degree of similarity in the NMR spectra indicate that the overall fold is highly similar at both pH conditions, with two helices separated by an internal-loop (C67, A79 and U80) and a structured pentaloop (G71-A75). However, the chemical shift of the A79 C2 resonance and the intensities of H1’-H2’ correlations in 2D TOCSY spectra of U6 ISL are extremely sensitive to pH variations (12). It is apparent from HSQC spectra that at pH 8.0, there is an even higher population of deprotonated A79, as indicated by a downfield shift in the C2 resonance (Figure 2B). Additionally, there is a further shift towards a pure C3’ endo sugar pucker for A79 at pH 8.0, as evidenced by the fact that an H1’-H2’ cross-peak for A79 is visible at pH 7.0 but not at pH 8.0 (Figure 2C). These trends follow those previously observed when comparing NMR data at low pH (5.7) to pH 7 (12).
Figure 2.
Overlay of 750 MHz NMR spectra of the U6 ISL at pH 7.0 (red) and pH 8.0 (blue). A. Close up of NOE cross-peaks from 2D NOESY spectra. The pH 7.0 spectrum has been shifted upfield by 0.01 ppm to facilitate comparison of the two spectra. B. Adenine H2-C2 correlations in 2D 1H-13C HSQC spectra. C. 2D TOCSY spectra. All cross-peaks are H1’-H2’ except for H1’-H3’ where indicated.
The U6 ISL structures at pH 7.0 and 8.0 are different
Structures of the U6 ISL were refined using distance, dihedral, hydrogen-bond and residual dipolar coupling (RDC) restraints derived from NMR data (Table 1). RDCs report the angle of a bond vector relative to the external magnetic field, and are extremely sensitive to even very small structural perturbations (18, 19). Structure calculations were performed using the amber99 force field (20) and the generalized Born model for approximating the solvent effect. Inclusion of an implicit representation of solvent molecules in structure calculations has been shown to improve the NMR structures of macromolecules, providing better definition of dihedral angles and hydrogen-bonding in dynamic domains when experimental restraints are sparse (21). In order to get a realistic comparison between the ISL structures at pH 8.0 and pH 7.0, the previously determined pH 7.0 structures (PDB code 1SY4) (11) were refined using the same protocol. Owing to the extreme similarity of pH 8.0 and pH 7.0 NOESY spectra (Figure 2), the same distance restraints were used in the calculations, which therefore differed only in the RDC restraints measured at the two different pH values (Table 1). Dihedral and hydrogen-bond restraints were only included for the Watson-Crick paired regions.
Table 1.
Statistics for the U6 ISL RNA structures at pH 8.0 and pH 7.0.
| pH 8.0 | pH 7.0 | |
|---|---|---|
| Structures: | ||
| Refined | 12 | 12 |
| Accepted | 10 | 10 |
| NMR restraints: | ||
| NOE (per residue) | 343 (14.3) | 343 (14.3) |
| Dihedral | 162 | 162 |
| hydrogen-bond | 23 | 23 |
| RDC | 47 | 31 |
| RMSD (Å): | ||
| all heavy atoms (res. 62–73,75–84) | 0.8 ± 0.3 | 0.6 ± 0.3 |
| res. 62–70,76–85 (heavy atoms) | 0.3 ± 0.1 | 0.4 ± 0.2 |
| Internal loop res. 67,79,80 (heavy atoms) | 0.2 ± 0.1 | 0.2 ± 0.1 |
| Restraint violations: | ||
| NOE (>0.5 Å) | 0 | 0 |
| avg dihedral (>8.0°) | 0 | 0 |
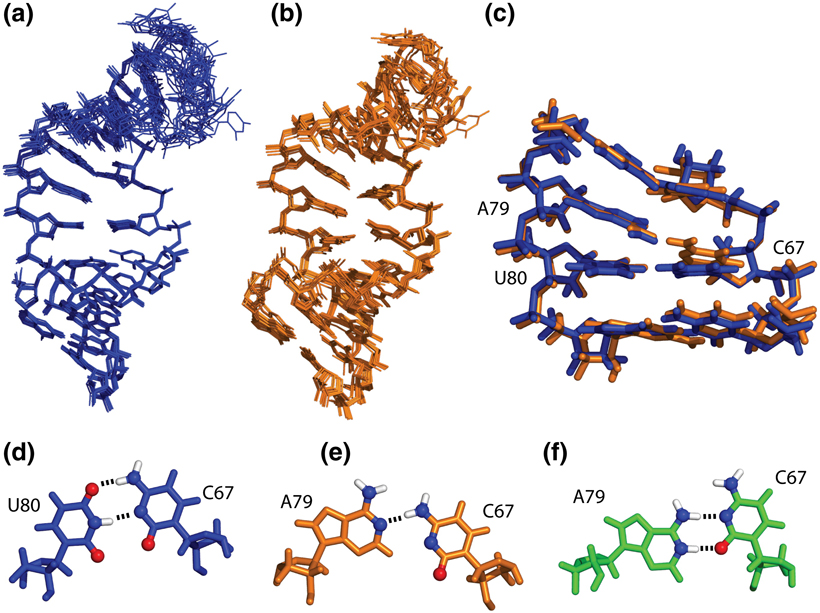
Both NMR structures consist of two A-form helices, separated by an internal-loop and capped by a flexible pentaloop that adopts a GNRA-tetraloop-like conformation (Figure 3A,B). The pH 8.0 and pH 7.0 structures superimpose with a pairwise r.m.s.d. of 0.9 ± 0.4 Å, which is close to the r.m.s.d. values for the separated ensembles (Table 1) and reflects the similarities in the NMR spectra (Figure 2). The r.m.s.d. of the pH 7 structures calculated previously in vacuum (11) to the same structures refined in implicit solvent is higher (1.6 ± 0.8 Å), owing to the differences in force fields and structure calculation protocols. Despite the similarity of the pH 8.0 and pH 7.0 structures, RDC analysis indicates a small yet systematic change in the orientation of the 1H-13C bond vectors relative to the alignment tensor throughout the molecule (Supporting Material, Figure S1). A close comparison of the structures shows that this change originates from slight variations in the orientation of bases rather than from global reorientation of the helical domains. In particular, the internal loop C67 nucleotide adopts different conformations at pH 8.0 and pH 7.0 (Figures 3CE). At pH 8.0, C67 forms a C67–U80 pair (C67 amino-U80 O4, C67 N3-U80 imino), leaving A79 unpaired. At neutral pH, the base of C67 appears to form one hydrogen bond with A79 (C67 amino-A79 N1), leaving U80 unpaired and stacked in the helix. In contrast, the previously determined structure at pH 5.7 (12) displayed a protonated C67-A79 wobble pair (C67 N3-A79 amino, C67 O2-A79 imino)(Figure 3F) with U80 flipped out of the helix (data not shown).
Figure 3.
NMR structure ensembles of the U6 ISL RNA. A. Structures determined at pH 8.0. B. Implicit solvent refinement of pH 7.0 structures. C. Superimposition of the internal loops and flanking Watson-Crick base pairs. The pH 8.0 structure is blue; the pH 7.0 structure is orange. D. C67-U80 base pair observed in the pH 8.0 structures. E. C67-A79 base pair observed in the pH 7.0 structures. F. C67-A79 base pair previously observed in the pH 5.7 structures (PDB code 1syz). Functional groups involved in base-pair hydrogen bonding are shown as spheres; hydrogen bonds are shown as dashed lines.
MD analysis of the high- and low-pH conformers of U6 ISL
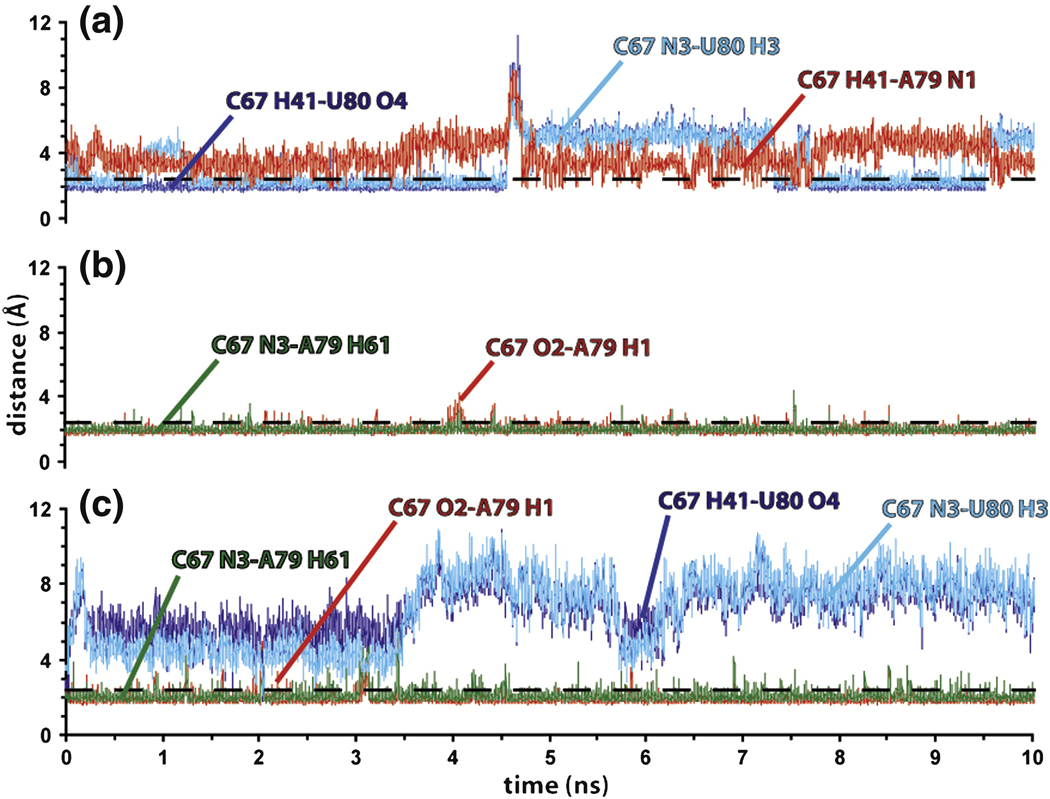
Stabilities of the high and low pH internal loop conformations were investigated by running 10 ns MD simulations performed in explicit solvent and counterions starting from the base-stacked (pH 8.0) and base-flipped (pH 5.7, PDB code 1SYZ) (12) NMR structures of the U6 ISL RNA. Analysis of the MD trajectory for the base stacked (pH 8.0) conformer reveals high stability for the internal-loop backbone dihedrals, which deviate only slightly from their original values in the NMR structure (Supporting Material, Figure S2). In addition, sugar puckers for C67, A79 and U80 are found in the N-type range during the overall simulation (Supporting Material, Figure S3), consistent with the absence of the H1’-H2’ correlations for these nucleotides in 2D DQF COSY spectra (data not shown) and the pH 8.0 TOCSY spectrum (Figure 2C). The MD simulation also supports formation of the C67-U80 pair. However, analysis of hydrogen-bond formation over time (Figure 4A) shows that the C67-U80 interaction breaks and reforms several times during the 10 ns trajectory. Such breaking events are associated with a close approach of the A79-N1 atom to the C67 amino protons (Figure 4A), allowing for transient formation of the single hydrogen-bonded C67-A79 pair observed in the pH 7.0 structure of the U6 ISL (Figure 3E).
Figure 4.
Distances between atoms during the MD simulations. A. U80 stacked (pH 8) structure. B. U80 flipped-out (pH 5.7) structure. C. The stacked pH 8.0 conformer incorporating the protonated form of A79. A dashed line at 2.4 Å is shown to indicate the distance required for hydrogen bond formation.
Compared to the MD data obtained for the base stacked conformer, the MD simulation performed on the base-flipped (pH 5.7) structure shows extensive variations in the internal-loop dynamics. In particular, the looped-out U80 nucleotide adopts multiple conformations during the MD sampling, inducing high backbone flexibility in the 3’-side of the internal-loop (Supporting Material, Figure S2). This is consistent with the disorder observed in the NMR structure (12). The increased disorder of U80 also results in conformational averaging of the A79 and U80 sugar puckers, that undergo several transitions between the S- and N-type range (Supporting Material, Figure S3). This finding is fully consistent with the NMR data that indicate conformational averaging of the A79 and U80 sugar puckers at low pH (Figure 2) and (12). The analysis of the hydrogen bond evolution with time (Figure 4B) confirms the existence of the double hydrogen bonded C67-A79+ wobble pair previously observed at low pH (12). Indeed, during the MD sampling the C67-A79+ pair undergoes only one, short (~100 ps) breathing event after ~4 ns, which is consistent with the high stability suggested for this base pair by thermodynamic and accessibility data (12, 16). In addition, the U80 base is observed to contact the upper helix of the U6 ISL for about one half of the MD trajectory (Supporting Material, Figure S4). Such interactions may stabilize the base-flipped conformation of U80 and are in agreement with the large decrease in surface accessibility observed for the adjacent A79 base at low pH (16).
In order to evaluate the effect of protonation at the A79-N1 atom on the structure and dynamics of the U80 stacked (high pH) conformer, an additional 10 ns MD simulation was performed starting from the pH 8.0 solution structure and incorporating the protonated form of A79. By analyzing the evolution of the base pairing pattern in the internal-loop nucleotides, a switch from the double hydrogen bond C67-U80 pair to a double hydrogen bond C67-A79+ wobble pair is observed at the beginning of the simulation (Figure 4C). The C67-A79+ wobble is stable for the overall simulation, leaving U80 unpaired and flexible in the helix. Thus, only part of the overall conformational transition is observed on the nanosecond timescale, which is consistent with previous NMR data (12, 13).
Minimum energy path for the base flipping transition
MD simulations provide important insights into conformational changes occurring on the ps-ns timescale. However, base-flipping transitions typically occur on the µs-ms timescale (13). Thus, timescale-independent methods must be used to examine transition states of such conformational changes. Here, we use a nudged elastic band (NEB) (17) approach to calculate the minimum energy path (MEP) for the U6 ISL base-flipping transition. In NEB, the endpoint structures are fixed in space and the path for a conformational change is quantified with a series of images of the molecule spaced along the path. Each image is connected to the next by virtual “elastic bands” that pull the system along the transition path. It is important to note that the obtained path is not pre-specified as a series of defined coordinates; rather, it represents the trajectory that a molecule follows through the conformational change and can be derived in a timescale-independent manner. The starting system is composed of I/2 images in the reactant configuration, followed by I/2 images in the product configuration (I = total number of images), and simulated annealing is implemented in order to find the low-energy path connecting the endpoints. This approach has been implemented in the Amber9 simulation package and employed for investigating the (syn)G-(anti)G to (anti)G-(syn)G transition of GG pairs in a model RNA (22, 23).
Here, the U80 stacked (pH 8.0) structure is referred to as the reactant configuration, while the U80 flipped out (pH 5.7) structure is the product configuration (Figure 5). The different conformations adopted by U80 during the transition path have been characterized by using a pseudo-dihedral angle θ, which is defined by the centers of mass of the following four units: 1) the A79 base, 2) the A79 sugar, 3) the U80 sugar, and 4) the U80 base. The θ angle describes the angular orientation of the U80 base with respect to A79, and is similar to other dihedral angles previously employed for investigating base-flipping transitions in nucleic acids (24–26). The θ angle adopts more positive values for motions toward the major groove, and more negative values for motions toward the minor groove. In order to eliminate the effect of minor transitions in the flexible pentaloop, the calculations have been run on a reduced construct of the ISL formed by fragments U64–U70 and A76–A83. To get a statistical sampling of the path, six NEB calculations were run on systems of 30 images by varying the seed number and the maximum temperature sampled by the simulated annealing protocol. In addition, for each calculation, two different starting systems have been prepared by including the A79 nucleotide in its familiar or protonated form.
Figure 5.
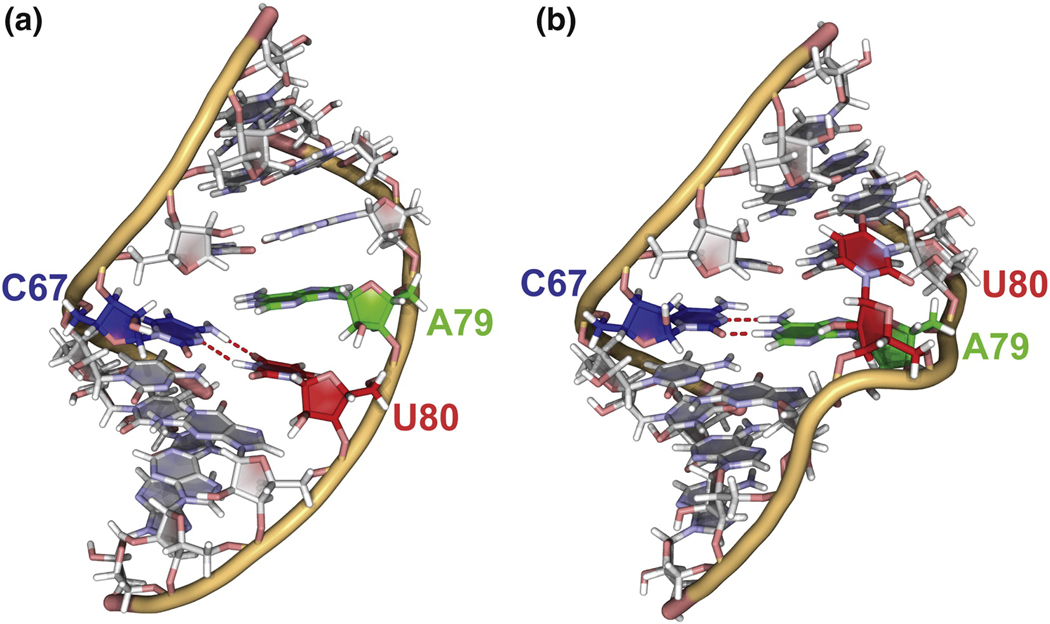
End point structures used for determining the minimum energy path (MEP). A. Stacked conformation (pH 8.0 structure). B. Energy minimized U80 flipped-out conformation. Both structures are shown with A79 protonated.
Since U80 is disordered when flipped-out of the helix, the product configuration is more ambiguous than the reactant configuration in this case. Therefore, the product configuration was set to the lowest energy structure sampled by the flipped-out conformation of U80. The lowest energy conformation was found from a 10 ns MD simulation on the pH 5.7 NMR structure (PDB code 1syz), during which 10 structures were extracted at regular intervals and energy minimized. This procedure identified the lowest energy conformation, in which the flipped U80 base makes van der Waals contacts in the minor groove with the 5’ nucleotides A79 and G78, and forms a hydrogen bond with the G78 amino group (G78 H22-U80 O4) (Figure 5B). This base-flipped conformation also allows for formation of an additional hydrogen bond between the G81 OH2’ and the A82 phosphate group. This conformation was dominant during the unrestrained MD (Figure S4), corresponds very closely to one of the low energy conformers observed in the NMR ensemble (12), and is consistent with surface accessibility studies (16).
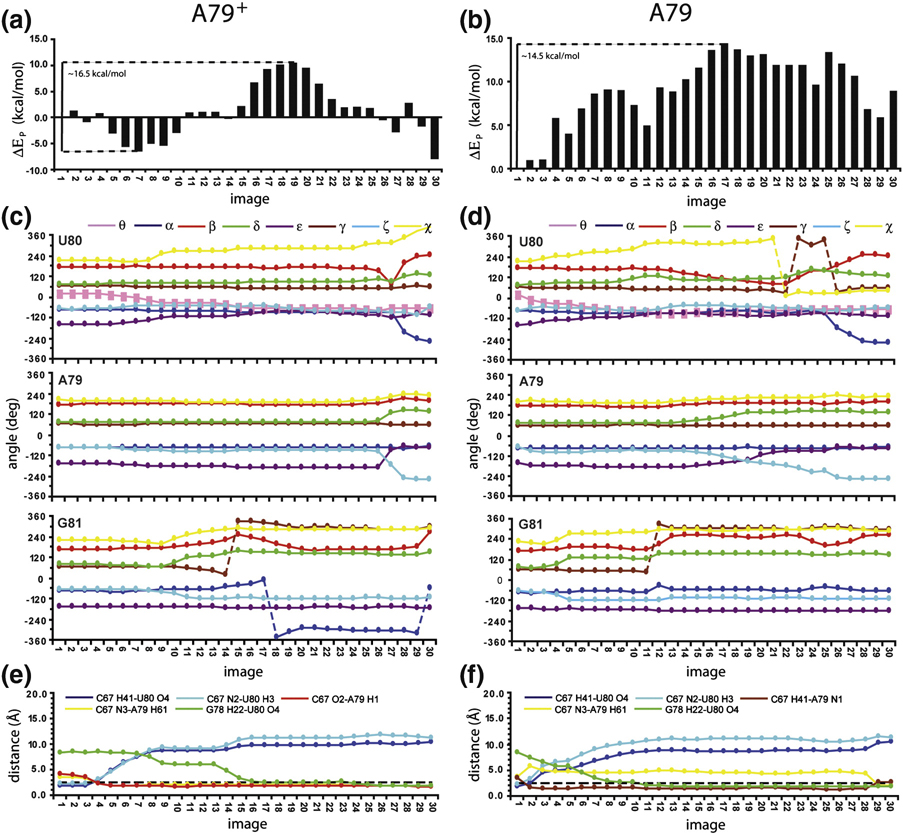
NEB simulations were performed in order to find the MEP connecting the U80 stacked structure (Figure 5A) to the U80 flipped-out structure (Figure 5B). The MEP obtained from the protonated A79 configuration (Figure 6A) begins with the disruption of the C67-U80 pair and the simultaneous formation of the double hydrogen bond C67-A79+ wobble (Figure 6A images 1–7). The new minimum for the stacked conformation (image 7) is reached at θ~−7.2° and is ~6.3 kcal/mol more stable than the starting structure. Interestingly, only minor variations in potential energy (EP) are found by flipping the U80 base to θ~−35.5° (image 9). Further flipping toward the looped-out conformation passes through two transition states (images 12 and 19; EA ~16.5 kcal/mol) describing the unstacking of the U80 base and transitions in the U80 and G81 backbone (Figure 6C). A new relative minimum is reached at image 27 in which the U80 base is completely flipped-out of the helix. Finally, other rearrangements in the dihedral angles of U80 and neighbor nucleotides lead to the structure for the looped-out conformer (image 30) which is ~1.6 kcal/mol more stable than image 7 and ~0.5 kcal/mol more favorable than the best energy minimum found for the stacked conformation among all the NEB calculations (image 5 in supplemental material Figure S5A). The MEP for base flipping is through the minor groove, and can be visualized in the accompanying movie (Supplemental Material).
Figure 6.
Potential energy (Ep) profiles for the base flipping conformational transition when the product configuration corresponds to the energy minimized U80 base-flipped conformation as depicted in Figure 5. Ep values are given as ΔEp values with respect to the starting configuration (image 1) for A79 in the protonated (A) and unprotonated (B) states. C and D. Torsion angles for the internal loop nucleotides as a function of the image for the A79 protonated and unprotonated states, respectively. Dashed lines indicate where corrections (±360°) are applied. E and F. Hydrogen bonds in the internal loop as a function of the image for the A79 protonated and unprotonated states, respectively. The dashed line is at 2.4 Å.
When A79 is not protonated (Figure 6B) the base flipping proceeds through the breaking of the C67-U80 pair and formation of a single hydrogen bond C67-A79 pair (C67 H41-A79 N1). This transition results in image 3 that is only 1.0 kcal/mol less stable than the starting structure. This result agrees well with the evolution of the base pairing pattern observed for internal-loop nucleotides during the 10 ns MD simulation of the U80 stacked (pH 8.0) conformer (Figure 4A). The base flipping transition occurs with EA ~14.5 kcal/mol, which is ~2.0 kcal/mol lower than the one obtained in the presence of protonated A79. Visualization of the transition paths reveals that the U80 base is able to hydrogen bond to the G78 amino group at image 11 when A79 is involved in the single hydrogen bond pair with C67 (Figure 6F). In contrast, formation of the stable C79-A79+ wobble pair locks the A79 base in a more exposed position hampering the flipping of the U80 nucleotide toward G78. In this case the G78-U80 hydrogen bond is formed at image 25 (Figure 6E) and does not contribute to stabilization of the absolute maximum in the transition path (image 19, Figure 6A). However, when A79 is not protonated, the best relative minimum for the looped-out conformation is reached at image 29 (Figure 6B) which is ~5.8 kcal/mol less stable than the starting structure. This finding is fully consistent with the preference for the stacked conformation reported for the U6 ISL at pH ≥ 7.0 (7, 12, 13, 16).
For comparison, the NEB simulation was also performed using a representative conformer from the NMR structure (1syz) as the product configuration, in which the flipped U80 is completely solvent exposed (Supporting Material, Figure S5). The MEPs derived from these NEB calculations are similar to the results in Figure 6, with the major difference being that the paths do not arrive at low energy conformations for the product configuration. The NEB calculations were repeated 6 times using both the energy minimized product configuration and the higher energy conformer from the NMR ensemble, both with A79 protonated and unprotonated, for a total of 24 NEB calculations (Supporting Material Figure S6). Interestingly, all the calculated paths indicate a minor groove direction for base flipping of the U80 nucleotide, irrespective of the protonation state of the A79 N1 position (Supporting Material Figure S6).
DISCUSSION
Structures of the U6 RNA ISL
Analyses of NMR line shapes and relaxation rates indicated that internal-loop residues (C67, A79 and U80) of the U6 ISL display motion on both the ps–ns and µs–ms timescales (12, 13). The U80 nucleotide adopts a stacked or looped-out conformation, depending on the protonation state of A79 (7, 11, 12). It was previously hypothesized that an improved stacking free energy between A79+ and G81 was a driving force for the base flipping transition (12).
We have solved the NMR structure of the U6 ISL at pH 8.0, which reveals the existence of a C67-U80 pair in the stacked conformer of the RNA (Figures 3A and D). The pH-dependent base pairing switch from C67-A79+ to C67-U80 in the internal loop provides a satisfying structural explanation for the preferred stacking of U80 at high pH. Interestingly, the high pH conformation is quite similar to the structure of the U6 ISL containing a lethal U80G mutation, which induces formation of a stable C67-G80 pair across the internal loop (14). These data suggest a requirement for either a uracil or conformational flexibility at position 80, in agreement with previous mutational results (8). Additionally, we note that stacking of U80 in the helix does not cause a bend, as previously hypothesized (11). Despite the fully stacked asymmetrical 1×2 internal loop in the high pH conformation, the helix is straight and has parameters that very closely match those of an ideal A-form helix. However, upon base flipping, the helix becomes more compact by approximately 1.5 Å. This helical motion could potentially alter tertiary contacts within the spliceosome, and can be visualized in the accompanying movie of the structure transition (Supporting Material).
MD simulations performed on the pH 7.0 (16) and pH 8.0 structures show that the C67-U80 pair is more stable than the single hydrogen bond C67-A79 pair (Figure 4A). In contrast, calculations incorporating the protonated A79 indicate that formation of a double hydrogen bond C67-A79+ wobble pair is preferred to the C67-U80 pair (Figure 4C). These simulations agree very well with the observed differences in the NMR structures at different pH (Figure 3). Clearly, the U6 ISL is in equilibrium between two conformations, and the equilibrium is dependent upon the protonation state of the A79 nucleotide. Since the protonated and unprotonated forms of A79 are in fast-exchange (~20 µs) (12), contributions from the two conformations are population-weighted in the NMR data. Thus, at pH 7.0 the significant population of protonated A79 (~24 %) (12) affects the experimental restraints used for the structure calculation, resulting in formation of a single hydrogen bond C67-A79 pair (Figure 3D) that should not be considered a valid ground state structure. However, this single hydrogen bond C67-A79 pair is observed as an intermediate in the MEP that defines the base flipping pathway when A79 is not protonated (Figure 6F).
Consistent with the low stability previously reported for CU pairs (27–29), our MD simulation reveals breathing of the C67-U80 pair on the ns timescale (Figure 4A). Such breathing events leave the C67, A79 and U80 bases flexible and available for intermolecular interactions. These internal-loop bases were observed to be highly accessible at high pH (16), making the internal-loop of the U6 ISL an attractive site for tertiary or intermolecular interactions. In contrast, the MD simulation of the U80 flipped-out (pH 5.7) structure shows high stability for the C67-A79+ wobble pair (Figure 4B,C) that decreases the number of available functional groups in the internal-loop. In addition, the flipped-out U80 base was found to adopt a preferential fold toward the G78 base (Figure 5B and Supporting Material, Figure S4), leading to a conformation that agrees well with previously reported accessibility data (16).
Pathway for an RNA conformational transition
Several NEB calculations were run in order to find the MEP for the U6 ISL conformational transition and to investigate the effect of A79 protonation on the energetics of the conformational change. When A79 is protonated, the absolute energy minimum for the U80 flipped-out conformation (image 30, Figure 6A) is found to be only slightly more favorable in energy than the absolute minimum for the stacked conformation (image 5, Figure 6A). This finding is inconsistent with the highly populated U80 looped-out conformation at low pH (12, 13, 16). In this respect, it should be noted that NEB results provide potential energy profiles for a conformational transition and only enthalpic contributions are accounted for in the obtained paths. The high disorder of the flipped-out U80 base may further stabilize the free energy of this conformer.
Our NEB calculations indicate that the minimum energy path for flipping out the U80 base is through the minor groove (Supporting Material, Figure S6). The minor groove base flipping path agrees well with a previous umbrella sampling molecular dynamics study, which reported a minor groove path for base flipping of a single uridine bulge (26). However, our results reveal a ~10 kcal/mol higher energy barrier for the conformational switch (figures 6A and Supporting Material Figure S5). The advantage of NEB is that the reaction coordinate is energy minimized over a multidimensional folding landscape. In umbrella sampling, the reaction coordinate is fixed and therefore may not properly describe all the transition states along the path. On the other hand, the disadvantage of NEB is that the entropic contribution to the minimum energy path is not considered. Therefore, NEB may overestimate the energy barrier for base flipping, if favorable entropic contributions from base flipping reduce the energy barrier for the transition.
Interestingly, when A79 is unprotonated, NEB data suggest that the U80 flipping through the minor groove may still be accessible at room temperature (Figures 6B and 7B). This finding agrees well with previously reported NMR data showing only a small decrease in accessibility of the U80 base at higher pH (16). However, the absolute energy minimum is observed corresponding to the stacked (pH 8.0) structure in all NEB calculations incorporating the unprotonated form of A79 (image 1, Figures 6B and Supporting Material Figures S6B and D), which is consistent with NMR data suggesting higher stability for the U80 stacked conformation than the looped out conformation at high pH (12).
A versatile internal-loop
The U6 ISL internal-loop has been shown to function in multiple processes in the splicing cycle (8) (Figure 1). The internal-loop nucleotides of this RNA are in close proximity to the spliceosome active site (8, 9) and may contact the splicing factors Prp8 (8) and Prp24 (30) during spliceosome assembly and activation. In addition, this 1×2 internal loop is also found in the secondary structures of other functional RNAs (ribosomal RNAs and RNase P) (10). We have shown that the U6 ISL is in equilibrium between two conformations, both of which are present at neutral pH (Figure 5). Thus, at physiological pH this equilibrium could be dramatically shifted by the approach of protein, RNA and/or metal ion cofactors. The dynamic conformation of the internal loop may be required for the multiple functions attributed to the U6 ISL internal-loop during spliceosome assembly and activation, and may modulate the binding of an essential metal ion cofactor at this site (6, 7). Finally, this work shows that RNA structures can change significantly in response to small changes in pH, and illustrates that nucleobase pKa values must be considered with great care when interpreting RNA structure and dynamics.
METHODS
RNA sample preparation
The U6 ISL RNA was prepared by in vitro transcription as previously described (7, 11). Sample conditions for NMR experiments were 1 mM RNA and 50 mM NaCl at pH 7.0 and 8.0. 2D TOCSY spectra were obtained in 50 mM NaCl and also in 15 mM KCl, and no detectable differences were observed between the two ionic conditions. Partial alignment of 13C-labeled U6 ISL for residual dipolar coupling measurements was achieved by addition of 17 mg/mL Pf1 filamentous bacteriophage (ASLA Biotech, Ltd.) in deuterium oxide.
NMR spectroscopy
All spectra were acquired at the National Magnetic Resonance Facility at Madison (NMRFAM) on Bruker DMX 600 and 750 MHz spectrometers. Exchangeable proton resonances were assigned by reference to 2D NOESY spectra, and collected with a 150 ms mixing time at 283 K in 90% H2O/10% D2O using a 1–1 spin-echo water suppression scheme. Non-exchangeable proton resonances were assigned as previously described (7, 11) and by reference to 2D NOESY (mixing times: 50, 150 and 300 ms) and 2D (1H–1H) TOCSY (mixing time 40 ms) in 99.99% D2O at 303 K. RDCs for 13C-labeled RNA were measured using J-modulated (13C–1H) CT-HSQC spectra (20 planes) (31). Data were processed by using XWINNMR or NMRPipe software (32). Resonance assignments were completed by using the program Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Structure calculations
NOE distance restraints were qualitatively categorized as strong (1.8–3.0 Ǻ), medium (2.0– 4.5 Ǻ), or weak (3.0–5.5 Ǻ) based on NOESY spectra with mixing times of 50, 150, and 300 ms. Torsion angle restraints were set to standard A-form values in all Watson–Crick regions (±30°) (7, 11, 12). Nucleotides with strong H1’-H2’ couplings (C72, U74) were constrained as C2’-endo (S-type; 145° ± 30°), whereas all other nucleotides were either constrained as C3’-endo or unrestrained. All glycosidic torsion angles (χ) were constrained to fall into the anti range (χ =−160° ± 30°) (7, 11), except for A79 and U80 which were left unrestrained. Hydrogen bond restraints were used for all Watson–Crick pairs as well as for the sheared G71-A75 base pair in the pentaloop.
Previously determined U6 ISL structures (11) were refined by using a generalized Born implicit solvent model (33) in Amber 9 (34). In all calculations, the amber99 force field (20) was used. The salt concentration was fixed at 0.05 M, and a 25-ps simulated annealing protocol was used for the refinement. The U6 ISL was refined by using distance, hydrogen-bond, and torsion angle constraints as summarized in Table 1. Square-well penalty functions were used for all NMR restraints with the force constants 32 kcal mol−1 Å−2 and 32 kcal mol−1rad−2 for distance and angle restraints, respectively. A 0.5 fs time step was used for the integration of the Newton motion equations, and a 15 Å cutoff was used for nonbonded atoms. The relative weights of the valence-angle energy, van der Waals terms, torsion energy, and improper torsional terms were increased gradually during the simulated annealing to maintain the planarity of the aromatic rings and proper local geometries. The resulting structures were further refined with the addition of RDC restraints. The initial alignment tensor was obtained using a 4000 step restrained energy minimization against the RDC restraints keeping the RNA structure fixed. This initial alignment tensor was then used in conjunction with the RDCs and all other structural restraints for further refinement using the same simulated annealing protocol employed for the calculation without RDCs. The force constant for the RDC restraints was set to 0.1 kcal mol−1 Hz−2 and the force constants for distance and dihedral restraints were increased to 300 kcal mol−1 Å−2 and 300 kcal mol−1 rad−2, respectively. Structures were viewed and analyzed using MOLMOL (35). Figures were produced using Pymol (http://www.pymol.org).
MD simulations
MD simulations were performed in explicit solvent starting from the lowest energy solution structures of the U6 ISL at pH 8.0 and pH 5.7 (PDB code 1SYZ) (12) using the Amber 9 package (34) and the parmbsc0 force field (36). The RNA was centered in a truncated octahedron and the initial shortest distance between the RNA atoms and the box boundaries was set to 10 Ǻ. Then, an optimal amount of sodium ions was added in order to generate a neutral system. The remaining box volume was filled with TIP3P type water molecules (37). Water and ion positions were minimized with 500 steps of steepest descent plus 500 steps of conjugate gradient by keeping the RNA fixed. Then, the entire system was energy minimized with 1500 steps of steepest descent followed by 1000 steps of conjugate gradient minimization. Each system was first equilibrated in a 20 ps run where temperature was gradually raised from 0 to 300 K. Hence, the equilibrated systems were simulated by keeping the temperature (300 K) and pressure (1 atm) constant. A weak coupling to external pressure baths was applied (relaxation time 1.0 ps) (38), while Langevin thermostat was used to control the temperature (collision frequency 1.0 ps−1) (39). Bonds to hydrogen atoms were constrained by the SHAKE algorithm (40). Non-bonded interactions were accounted for by using the PME method (41). An integration time step of 2 fs was used and trajectories were saved every 1 ps. Angle and bond parameters as well as atomic charges for the protonated adenine were calculated using the software Gaussian version 03 (http://www.gaussian.com) at the Hartree–Fock level with the 6–31G* basis set (20). Restrained electrostatic potential (RESP) atomic charges were derived by applying the RESP module of the AMBER 7.0 package to the Gaussian electrostatic potential (ESP) charges. The backbone root mean square deviation (rmsd) of investigated systems over the MD runs (Supporting Material, Figure S7) level off after equilibration periods shorter than 500 ps in all the cases. Thus all the subsequent analyses of the trajectories were carried out from 500 ps onward. All MD simulations were performed twice with different seed numbers, and the results were very similar for each simulation. Therefore, the results of single 10 ns MD simulations are shown.
NEB sampling
Nudged Elastic Band (NEB) calculations were run by using the energy minimized structures of the U6 ISL in the stacked and looped-out conformation (see Results) as end-points for the base-flipping conformational transition. All calculations used the Amber 9 software (34) and the parmbsc0 force field (36). The generalized Born implicit solvation model (33) was used, and the salt concentration was set to 0.05 M. The maximum distance for the pairwise summation in calculating the effective Born radii was set to 15 Ǻ. The non-bonded cutoff was 15 Ǻ. SHAKE (40) was used to constrain the position of hydrogen atoms and the time step was set to 2 fs. Langevin dynamics was used to control temperature (collision frequency 1000 ps−1) (39).
To sample the transition path, a 540 ps simulated annealing protocol was used. The first step involved 20 ps of MD with spring constants of 10 kcal mol−1 Ǻ−2 during which the temperature of the system is increased from 0 K to 300 K. Then a 100 ps MD step was applied at constant temperature (300 K) during which the spring constants are gradually increased to 50 kcal mol−1 Ǻ−2. The system was next heated to either 600 K or 800 K over the next 150 ps, and then cooled to 0 K (270 ps). Finally, 200 ps of quenched MD at 0K was performed.
Supplementary Material
01
02
Acknowledgements
The authors thank Professors David A. Brow, David H. Mathews, Brent Znosko and Qiang Cui for helpful comments and suggestions. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301 (BRTP/ NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR- 9214394), and the USDA. This work was supported by NIH grant R01 GM65166 (SEB) from the National Institutes of Health.
Footnotes
Accession numbers
Structure coordinates have been deposited into the Worldwide Protein Data Bank (http://www.pdb.org), ID codes 2KF0 (pH 7.0) and 2KEZ (pH 8.0).
References
- 1.Nilsen TW. In: RNA structure and function. Somons R, Grunberg-Manago M, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1998. pp. 279–307. [Google Scholar]
- 2.Burge CB, Tuschi TA, Sharp PA. In: RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 3.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 4.Valadkhan S, Manley JL. RNA. Vol. 9. New York, N.Y: 2003. Characterization of the catalytic activity of U2 and U6 snRNAs; pp. 892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortner DM, Troy RG, Brow DA. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes. Dev. 1994;8:221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- 6.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 8.McManus CJ, Schwartz ML, Butcher SE, Brow DA. RNA. Vol. 13. New York, N.Y: 2007. A dynamic bulge in the U6 RNA internal stem-loop functions in spliceosome assembly and activation; pp. 2252–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhode BM, Hartmuth K, Westhof E, Luhrmann R. Proximity of conserved U6 and U2 snRNA elements to the 5' splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badhwar J, Karri S, Cass CK, Wunderlich EL, Znosko BM. Thermodynamic characterization of RNA duplexes containing naturally occurring 1 × 2 nucleotide internal loops. Biochemistry. 2007;46:14715–14724. doi: 10.1021/bi701024w. [DOI] [PubMed] [Google Scholar]
- 11.Reiter NJ, Nikstad LJ, Allmann AM, Johnson RJ, Butcher SE. RNA. Vol. 9. New York, N.Y: 2003. Structure of the U6 RNA intramolecular stem-loop harboring an S(P)-phosphorothioate modification; pp. 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiter NJ, Blad H, Abildgaard F, Butcher SE. Dynamics in the U6 RNA intramolecular stem-loop: a base flipping conformational change. Biochemistry. 2004;43:13739–13747. doi: 10.1021/bi048815y. [DOI] [PubMed] [Google Scholar]
- 13.Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE. Dynamics and metal ion binding in the U6 RNA intramolecular stem-loop as analyzed by NMR. J. Mol. Biol. 2005;353:540–555. doi: 10.1016/j.jmb.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Sashital DG, Allmann AM, Van Doren SR, Butcher SE. Structural basis for a lethal mutation in U6 RNA. Biochemistry. 2003;42:1470–1477. doi: 10.1021/bi027137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- 16.Venditti V, Niccolai N, Butcher SE. Measuring the dynamic surface accessibility of RNA with the small paramagnetic molecule TEMPOL. Nucleic Acids Res. 2008;36:e20. doi: 10.1093/nar/gkm1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonson H, Mills G, Jacobsen KW. Nudged elastic band method for finding minimum energy paths of transitions. In: Berne BJ, Ciccoti G, Coker DF, editors. Classical and quantum dynamics in condensed phase simulations. Singapore: World Scientific; 1998. pp. 385–404. [Google Scholar]
- 18.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 19.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. Journal of the American Chemical Society. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 20.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 21.Xia B, Tsui V, Case DA, Dyson JH, Wright PE. Comparison of protein solution structures refined by molecular dynamics simulation in vacuum, with a generalized Born model, and with explicit water. J. Biomol. NMR. 2002;22:317–331. doi: 10.1023/a:1014929925008. [DOI] [PubMed] [Google Scholar]
- 22.Burkard ME, Turner DH. NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- 23.Mathews DH, Case DA. Nudged eleastic band calculation of minimum energy paths for the conformational change of a GG non-canonical pair. J. Mol. Biol. 2006;357:1683–1693. doi: 10.1016/j.jmb.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 24.Varnai P, Lavery R. Base flipping in DNA: pathway and energetics studied with molecular dynamic simulations. J. Am. Chem. Soc. 2002;124:7272–7273. doi: 10.1021/ja025980x. [DOI] [PubMed] [Google Scholar]
- 25.Banavali NK, M. A. D Free energy and structural pathways of base flipping in a DNA GCGC containing sequence. J. Mol. Biol. 2002;319:141–160. doi: 10.1016/S0022-2836(02)00194-8. [DOI] [PubMed] [Google Scholar]
- 26.Barthel A, Zacarias M. Conformational transitions in RNA single uridine and adenosine bulge structures: a molecular dynamics free energy simulation study. Biophys. J. 2006;90:2450–2462. doi: 10.1529/biophysj.105.076158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Kojima C, Yamazaki T, Kodama TS, Yasuno K, Miyashita S, Ono AM, Ono AS, Kainosho M, Kyogoku Y. Solution structure of an RNA duplex including a C-U base pair. Biochemistry. 2000;39:7074–7080. doi: 10.1021/bi000018m. [DOI] [PubMed] [Google Scholar]
- 28.Wu M, McDowell JA, Turner DH. A Periodic Table of Tandem Mismatches in RNA. Biochemistry. 1995;34:3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J, Wartell RM. The relative stabilities of base pair stacking interactions and single mismatches in long RNA measured by temperature gradient gel electrophoresis. Biochemistry. 1997;36:15326–15335. doi: 10.1021/bi9716783. [DOI] [PubMed] [Google Scholar]
- 30.Kwan SS, Brow DA. RNA. Vol. 11. New York, N.Y: 2005. The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 RNA binding; pp. 808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ottiger M, Delaglio F, Marquardt JL, Tjandra N, Bax A. Measurement of dipolar couplings for methylene and methyl sites in weakly oriented macromolecules and their use in structure determination. J. Magn. Reson. 1998;134:365–369. doi: 10.1006/jmre.1998.1546. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2001;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 34.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 36.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohoney MW, Jorgensen WL. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112:8910–8922. [Google Scholar]
- 38.Berendsen HJC, Postma JPM, van Gunsteren WF, Di, Nola A. Molecular dynamics with coupling to an external bath. J. Phys. Chem. 1984;91:6269–6271. [Google Scholar]
- 39.Pastor RW, Brooks BR, Pastor RW. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988;65:1409–1419. [Google Scholar]
- 40.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Chem. 1977;23:327–341. [Google Scholar]
- 41.Darden T, Perera L, Li L, Pedersen L. New tricks for modellers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Struct. Fold Des. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02