Genetic Loci Influencing C-reactive Protein Levels and Risk of Coronary Heart Disease (original) (raw)
. Author manuscript; available in PMC: 2010 Jan 7.
Published in final edited form as: JAMA. 2009 Jul 1;302(1):37–48. doi: 10.1001/jama.2009.954
Abstract
Context:
Plasma levels of C-reactive protein (CRP) are independently associated with risk of coronary heart disease, but whether CRP is causally associated with coronary heart disease or merely a marker of underlying atherosclerosis is uncertain.
Objective:
To investigate association of genetic loci with CRP levels and risk of coronary heart disease.
Design, setting and participants:
We first carried out a genome-wide association (n=17,967) and replication study (n=14,747) to identify genetic loci associated with plasma CRP concentrations. Data collection took place between 1989 and 2008 and genotyping between 2003 and 2008. We carried out a Mendelian randomisation study of the most closely associated SNP in the CRP locus and published data on other CRP variants involving a total of 28,112 cases and 100,823 controls, to investigate the association of CRP variants with coronary heart disease. We compared our finding with that predicted from meta-analysis of observational studies of CRP levels and risk of coronary heart disease. For the other loci associated with CRP levels, we selected the most closely associated SNP for testing against coronary heart disease among 14,365 cases and 32,069 controls.
Main outcome measure:
Risk of coronary heart disease.
Results:
Polymorphisms in five genetic loci were strongly associated with CRP levels (% difference per minor allele): SNP rs6700896 in LEPR (−14.7% [95% Confidence Interval {CI}], −17.5 – −11.9, _P_=1.6×10−21), rs4537545 in IL6R (−10.8% [95% CI, −13.8 – −7.7], _P_=5.1×10−11), rs7553007 in CRP locus (−20.7% [95% CI, −23.5 – −17.9], _P_=3.3×10−38), rs1183910 in HNF1A (−13.6% [95% CI, −16.4 – −10.6], _P_=1.2×10−17) and rs4420638 in APOE-CI-CII (−21.8% [95% CI, −25.4 – −18.1], _P_=2.1×10−25). Association of SNP rs7553007 in the CRP locus with coronary heart disease gave odds ratio (OR) 0.98 (95% CI, 0.94 – 1.01) per 20% lower CRP. Our Mendelian randomisation study of variants in the CRP locus showed no association with coronary heart disease: OR 1.00 (95% CI, 0.97 – 1.02) per 20% lower CRP, compared with OR 0.94 (95% CI, 0.94 – 0.95) predicted from meta-analysis of the observational studies of CRP levels and coronary heart disease (Z-score −3.45, P<.001). SNPs rs6700896 in LEPR (OR 1.06 [95% CI, 1.02 – 1.09] per minor allele), rs4537545 in IL6R (OR 0.94 [95% CI, 0.91 – 0.97]) and rs4420638 in the APOE-CI-CII cluster (OR 1.16 [95% CI, 1.12 – 1.21]) were all associated with risk of coronary heart disease.
Conclusions:
The lack of concordance between the effect on coronary heart disease risk of CRP genotypes and CRP levels argues against a causal association of CRP with coronary heart disease.
INTRODUCTION
Coronary heart disease (CHD) is the leading cause of death worldwide.1 Inflammation plays a key role in the pathogenesis of CHD at every stage from initiation, to progression and rupture of the atherosclerotic plaque.2 C-reactive protein (CRP), an acute phase protein synthesised primarily by the liver, is currently the most widely used biomarker of inflammation.3 Observational studies have consistently demonstrated that higher plasma levels of CRP are associated with higher risk of CHD4,5, and measurement of CRP has been advocated as a means of improving risk prediction.6 There is considerable interest in establishing whether CRP has a causal role in CHD, or whether CRP is merely a marker of underlying atherosclerosis.7,8 While previous studies have addressed this question, it is unclear if they included a sufficient number of cases to have adequate statistical power to confirm or refute associations of CHD with genetically determined differences in CRP levels.8 Resolution of this question will improve understanding of inflammatory mechanisms in atherosclerosis.
The “Mendelian randomisation” concept has been used to investigate possible causal relationships of an intermediate trait (such as CRP levels) with disease, taking advantage of the random allocation of alleles at conception.9 If the intermediate trait is causally linked to disease, then genetic variants influencing the trait should also influence disease risk.10 Mendelian randomisation studies should be unaffected by confounding from environmental factors, e.g., smoking, and reverse causation bias, i.e., where the disease itself (atherosclerosis) influences the trait (CRP levels).9
The aims of the present study were to conduct a genome-wide association study to identify common genetic loci that influence CRP levels, and use the concept of Mendelian randomisation to improve understanding of the possible causal relationship of CRP levels with CHD.
DESIGN AND METHODS
Study design and rationale
The study involved four inter-related components:
- a genome-wide association and replication study to identify genetic loci associated with CRP levels, and at each locus, selection of the most closely associated SNP;
- a Mendelian randomisation study of CHD risk for the most associated SNP in the CRP locus in our data, and published data on CRP variants, to assess the potential causal association of CRP with CHD;
- comparison of the finding from the Mendelian randomisation study with that predicted from meta-analysis of the relationship of CRP levels with CHD from observational studies;
- a genetic association study of CHD for the most associated SNPs in genetic loci outside the CRP locus, using the concept of CRP as an intermediate phenotype10 to identify putative pathways linking inflammation with CHD.
Population cohorts
Genome-wide and replication study for CRP
Genome-wide association to identify variants related to CRP levels, measured using high sensitivity assays, was carried out in 17,967 participants from five studies: the London Life Sciences Population study (LOLIPOP: N=5,502), a population based cohort of European white and Indian Asian men and women, aged 35-75 yrs, living in West London, UK11 (data collection 2001 – 2007, genome-wide genotyping 2003 – 2008); the 1966 Northern Finnish Birth Cohort (NFBC: N=4,761), a prospective birth cohort of persons born in 1966 in the two northernmost provinces of Finland12,13 (data collection 1997 – 1998, genotyping 2007 – 2008); the Lausanne Cohort (CoLaus: N=5,226), a cross-sectional study of a random sample of European men and women, aged 35–75 yrs, living in Lausanne, Switzerland14 (data collection 2003 – 2006, genotyping 2006 – 2007); the Genetic Epidemiology of Metabolic Syndrome study (GEMS: N=1,781), a case-control study of dyslipidaemic cases (age 20–65 years) matched with normolipidaemic controls by sex and recruitment site15 (data collection 2003 – 2006, genotyping 2006 – 2007); and the Data from an Epidemiological Study on the Insulin Resistance syndrome study (DESIR: N=697), a longitudinal French general population cohort of persons aged 30-64 years recruited through the French social security system16 (data collection 1994 – 2004, genotyping 2006 – 2007). Replication of SNPs associated with CRP, identified in the genome-wide association study, was performed in a further 14,747 LOLIPOP participants who were not included in the genome-wide association study and were free of known CHD.
Mendelian randomisation and genetic association studies with CHD
Variants related to CRP were tested for association with CHD among 14,365 CHD cases and 32,069 controls. The participating studies comprised the Precocious Coronary Artery Disease study (PROCARDIS: N=8,328), a case-control study of premature CHD before age 66 years17 (data collection 1999 – 2006, genotyping 2008); the International Studies of Infarct Survival (ISIS: N=3,624), comprising men aged 30–54 yrs and women aged 30–64 yrs, with non-fatal MI, and their spouse controls18,19 (data collection 1989 – 1992, genotyping 2008); the British Heart Foundation Family Heart Study comprising individuals with myocardial infarction or coronary revascularisation before the age of 66 years and at least one first-degree relative with premature CHD, who were also studied as part of the Wellcome Trust Case Control Consortium (WTCCC: N=3,249 to 4,863)20 (data collection 1998 – 2006, genotyping 2006 – 2008); the German MI Family Studies (GerMIFS I): N=2,519; GerMIFS II: N=2,520, comprising persons with myocardial infarction before the age of 60 years and at least one first-degree relative with premature CHD, and matched controls20,21 (data collection 1996 – 2008, genotyping 2006 – 2008); the INTERHEART study, a multinational case-control study of persons presenting with first myocardial infarction (N=4,043)22, (data collection 1999 – 2003, genotyping 2008); and the LOLIPOP study (N=20,475) (data collection 2001 – 2007, genotyping 2008).
Genotyping
Genome-wide association scans were performed using the Affymetrix 500K mapping array, the Illumina 317K array and Perlegen Sciences customised arrays. To combine data across genotyping platforms, imputation was done using a Hidden Markov Model algorithm implemented in MACH v1.0 software (Center for Statistical Genetics, University of Michigan, MI, USA) (LOLIPOP) or IMPUTE v0.5.0 (Genetics Software Suite, University of Oxford, UK) v0.5.0 (other studies), and phased haplotypes from HapMap build35, dbSNP build 125. For the European datasets, the HapMap CEU sample was used for reference haplotypes; Indian Asian datasets were imputed based on a combination (mixed) of HapMap populations. Imputed SNPs with minor allele frequency (MAF)<.01, or low quality score (r2<0.30 in MACH, or information score <0.5 in IMPUTE) were removed. This left ~1.4 million directly genotyped or imputed autosomal SNPs per participant with data available in all samples. Genotyping for replication testing and for evaluation against CHD was performed using KASPar (LOLIPOP), Sequenom (INTERHEART, PROCARDIS), TaqMan (ISIS, WTCCC), or Affymetrix mapping arrays (GerMIFS1&2, WTCCC).
Statistical analyses
Genome-wide association and replication study for CRP
Genome-wide SNP associations for CRP were tested in multiple linear regression analyses, using an additive genetic model. CRP levels were log transformed to achieve approximate normal distribution, and analysed as a quantitative trait with adjustment for age and sex (analysis of residuals showed good adherence to normality assumptions). To account for heterogeneity in population structure, principal components derived from EIGENSTRAT v2.0 (Reich Lab, Harvard University, MA, USA) were included as covariates in age, sex adjusted analyses for CoLaus, GEMs, NFBC, and LOLIPOP Indian Asian Illumina analyses (the number of principal components included varied from 4 to 10, depending on the population structure of the specific cohort). For other LOLIPOP data sets genomic control factors were used to correct for any inflation. No principal components were included for DESIR as the population was recruited from a geographically restricted area. Statistical software used for genome-wide associations comprised SNPTEST (Genetics Software Suite, University of Oxford, UK) v1.1.3 (DESIR), v1.1.4 (CoLaus, GEMS, NFBC); and MACH2QTL (Center for Statistical Genetics, University of Michigan, MI, USA) v.1.0 (LOLIPOP). Results of the separate genome-wide association studies were combined using weighted Z-scores, and a fixed rather than random effects model to maximise discovery, since random effects estimates are associated with larger variance. QQ plots showed good adherence to null expectations (Lambda for combined data = 1.0625). We used P<5×10−8 to designate genome-wide significance, taking account of the ~ 1 million independent tests for common variants across the genome.23 For five genetic loci associated with CRP at genome-wide significance, we selected the single most closely associated SNP (i.e., smallest _P_-value) for replication against CRP.
By use of QUANTO24 v1.2 for quantitative traits, we estimated that the genome-wide association study had 80% power to detect SNPs associated with 0.2% of population variation in CRP levels, or an 11% difference in CRP levels per allele copy, at MAF 30% and genome-wide level of significance (P<5×10−8).
Mendelian randomisation study of genetic variants in the CRP locus and CHD
We analysed relationship of dbSNP rs7553007 in the CRP locus, the SNP most strongly associated with CRP levels in our data, for association with CHD risk using logistic regression under an additive genetic model, as part of a Mendelian randomisation study. To identify published data on the relationship of CRP variants with CHD, two electronic databases (Medline and EMBASE) were searched up to and including November 2008 for all prospective studies (including cohort, nested case-control and case-cohort studies), and case-control studies, with no threshold sample size. For the search, the MeSH terms ‘C-reactive protein’ and ‘polymorphism, single nucleotide’ or ‘polymorphism, genetic’ or ‘haplotype’ in combination with ‘coronary disease’ or ‘heart disease’, were used and the search was limited by the terms ‘human’ and ‘English language’. We also scanned reference lists of previous reports. Eight studies of 18 cohorts were identified examining. Two studies25,26 examining nine cohorts reported results for a single SNP (dbSNP rs1130864); for these studies, odds ratios (ORs) were reported under a recessive model comparing homozygotes (TT) for the minor allele with CT/CC genotype, and mean effect on CRP was obtained from published ratio of geometric means (1.21).25,26 For the remaining studies8,27,28,29,30,31, we selected dbSNP rs1205 on the basis of MAF>0.1 and largest per allele effect size on CRP levels (−0.35 mg/L) reported in the study of CRP variants and CRP levels by Verzilli et al.32 We used per allele OR of rs1205 with CHD where available27,29 or where this could be estimated directly from the data8,31; otherwise ORs were estimated from averaging published effect sizes for minor allele homozygotes (TT) and heterozygotes (CT) compared to wild type (CC).28,30 We estimated standard errors of the effect sizes of CRP variants on CHD from the reported 95% confidence intervals (CIs), assuming normality. Estimated ORs for CRP variants on CHD were converted to a common scale by standardising to 20% lower CRP, i.e., the approximate effect per minor allele of rs7553007 on CRP levels. Results were combined across studies by SNP, and across the three SNPs weighted by the inverse of variance. We used 95% CIs and assessed heterogeneity with standard χ2 statistics, expressed as I2, the proportion of variability between studies due to heterogeneity.33 In the absence of heterogeneity, we used a fixed effects model.
We also investigated the three-way associations between CRP genetic variants, CRP levels, and CHD in prospective studies which reported all three sets of data in the same cohorts25,26,27,30; we did not include retrospective (case-control) studies since these could be biased by treatment effects.34
Comparison of the result of the Mendelian randomisation study with meta-analysis of the CRP-CHD relationship
We compared the result from our Mendelian randomisation study with that predicted from a meta-analysis of the observational studies of CRP levels and CHD.5,8,26,35,36,40,50-72 This was obtained from a systematic review of the CRP-CHD relationship published by Shah et al.5, updated with all studies published from August 2007 until November 2008. For the search, the MeSH terms ‘C-reactive protein’ and ‘CRP’ in combination with ‘coronary’, ‘coronary heart disease’, ‘CHD’ and ‘CVD’ were used, and the search was limited by the terms ‘human’ and ‘English language’. Studies in which total mortality was the only outcome reported were excluded; if more than one paper was published on the same cohort/ population the most recent one was used in the meta-analysis. Five new population studies were identified from four reports.8,26,35,36 Risk ratios for CHD associated with CRP levels (logarithmically transformed) were extracted from each study. Two studies were excluded: one37 which cited risk ratios per unit increase in CRP (i.e., not logarithmically transformed) and one38 which did not provide CIs for the association of CRP with CHD. The studies reported risk ratios based on different comparisons of CRP (tertiles, quintiles, or quartiles), or as differences in risk for a given increase in CRP; these were converted to common unit of 1 standard deviation (SD) change. The risk ratio per SD change was converted to per 20% lower CRP by multiplying coefficients (and 95% CIs) on the logarithmic scale by −0.223 assuming 20% reduction in CRP is equivalent to a 0.223SD reduction in log CRP. Multivariate adjusted risk ratios, controlled for conventional cardiovascular risk factors, were used when available; two studies reported risk ratios adjusted for age, and age and smoking only.39,40 Because of significant heterogeneity, random effects meta-analysis was used to combine the risk ratios from the individual studies. The overall OR for CHD was used to test the observed vs. predicted association of CRP variants with CHD, standardised to 20% lower CRP.
Genetic association of variants outside the CRP locus and CHD
The relationships with risk of CHD of the most associated SNP in the four genetic loci other than the CRP locus in our data, were analysed by logistic regression under an additive genetic model. Results were combined across studies by SNP, using a fixed effects model weighted by the inverse of variance, as there was no significant heterogeneity. By use of the Genetic Power Calculator41, we estimated that the genetic association study with CHD had 80% power to detect an OR of 1.04 per allele copy at P<0.05 and MAF 30%.
With the exception of the genome-wide association analyses described above, all statistical analyses were done with STATA v10 (Statacorp LP, TX, USA). A significance level of P<0.05 was used; all tests were two-sided.
Ethics approval was obtained locally for each of the participating cohorts for analyses of genetic markers of cardiovascular disease risk. No additional ethics approvals were required for this study.
RESULTS
Genome-wide association and replication study for CRP
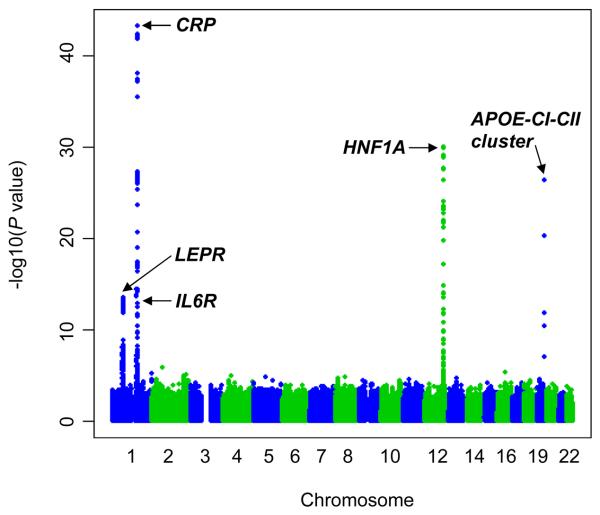
We found 160 SNPs to be associated with CRP levels at P<5×10−8, distributed in the following five loci: LEPR (GeneID 3953; GenBank NC_000001.9, region 65658906 to 65875410), IL6R (GeneID 3570, GenBank NC_000001.9, region 152644293 to 152706812), CRP (GeneID 1401, GenBank NC_000001.9, region 157951003 to 157948703), HNF1A (GeneID 6927, GenBank NC_ NC_000012.10, region 119900932 to 119924698) and APOE-CI-CII (GeneID 348, 341, 344, GenBank NC_000019.8, region 50100879 to 50104490). A Manhattan plot of results from the combined analysis of genome-wide association data is shown in Figure 1. Genomic context and _P_-values for the most associated SNP at each of the five loci are shown in Table 1. The association of these SNPs with CRP was confirmed in replication testing (all _P_≤10−10); for all five SNPs, the minor allele was associated with reduced levels of CRP (Table 1). For the most associated SNP in the CRP locus (rs7553007), CRP levels were lower by 21% (95 % CI, −23.5–−17.9) per minor allele. For the other four SNPs, per minor allele differences in CRP levels ranged from −10.8% (95% CI, −13.8–−7.7) for dbSNP rs4537545 in IL6R to −21.8% (95% CI, −25.4–−18.1) for dbSNP rs4420638 in the APOECI-CII cluster (Table 1).
Figure 1.
Manhattan plot of results from the combined analysis of genome-wide association data (9 cohorts)
Table 1.
Genomic context†, alleles, minor allele frequency and association test results for most associated SNP at each locus. Effect size is change in CRP (%, 95% Confidence Interval) per copy of minor allele, under an additive genetic model
| SNP | |||||
|---|---|---|---|---|---|
| rs6700896 | rs4537545 | rs7553007 | rs1183910 | rs4420638 | |
| Genomic context | |||||
| Chromosome | 1 | 1 | 1 | 12 | 19 |
| Position | 65862370 | 152685503 | 157965173 | 119905190 | 50114786 |
| Locus | LEPR | IL6R | CRP | HNF1A | APOE-CI-CII cluster |
| Alleles | |||||
| Reference | C | C | G | C | A |
| Minor | T | T | A | T | G |
| Minor allele frequency* | |||||
| Europeans | 0.38 | 0.43 | 0.33 | 0.32 | 0.19 |
| Indian Asians | 0.46 | 0.31 | 0.29 | 0.39 | 0.12 |
| Genome-wide association | |||||
| P value | 3.1×10−14 | 1.8×10−14 | 7.6×10−44 | 1.2×10−30 | 4.5×10−27 |
| Replication | |||||
| CRP effect,%‡ | −14.7 | −10.8 | −20.7 | −13.6 | −21.8 |
| 95% CI | −17.5 to −11.9 | −13.8 to −7.7 | −23.5 to −17.9 | −16.4 to −10.6 | −25.4 to −18.1 |
| P value | 1.6×10−21 | 5.1×10−11 | 3.3×10−38 | 1.2×10−17 | 2.1×10−25 |
In the replication study (LOLIPOP), the percent variance of CRP explained ranged from 0.2% (dbSNP rs11839910 in HNF1A) to 1.3% (rs7553007 in the CRP locus). SNP rs4420638 in the APOE-CI-CII cluster was strongly associated with total cholesterol (0.18 [95% CI, 0.13–0.22] mmol/L per minor allele), LDL cholesterol (0.16 [95% CI, 0.12–0.2] mmol/L), triglycerides (0.11 [95% CI, 0.07–0.16] mmol/L) and HDL cholesterol (−0.03 [95% CI, −0.04–−0.02] mmol/L) (Table 2). SNP rs1183910 in HNF1A was also associated with total cholesterol (0.03 [95% CI, 0.00–0.06] mmol/L per minor allele), LDL cholesterol (0.03 [95% CI, 0.01–0.05] mmol/L) and HDL cholesterol (0.01 [95% CI, 0.00–0.02] mmol/L). The remaining three SNPs were not significantly related to plasma lipids or with any of the other phenotypes tested (Table 2).
Table 2.
Relationships of most associated SNPs in the five genetic loci influencing CRP levels, with cardiovascular risk factors in replication sample (LOLIPOP), from regression analyses adjusted for age, sex and ethnic group, under an additive genetic model. Data presented as unit change (continuous traits) or odds ratio (diabetes) per copy of minor allele
| Cardiovascular risk factors | SNP (Locus) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs6700896 (LEPR) | P | rs4537545 (IL6R) | P | rs7553007 (CRP) | P | rs1183910 (HNF1A) | P | rs4420638 (APOE-CI-CII) | P | |
| Weight (kg) | −0.35 (−0.74 to 0.04) | 0.08 | 0.01 (−0.39 to 0.42) | 0.93 | −0.06 (−0.48 to 0.36) | 0.79 | 0.08 (−0.32 to 0.47) | 0.68 | 0.11 (−0.43 to 0.65) | 0.68 |
| Body mass index (kg/m2) | −0.06 (−0.19 to 0.08) | 0.45 | −0.08 (−0.21 to 0.06) | 0.30 | −0.03 (−0.18 to 0.11) | 0.68 | −0.02 (−0.16 to 0.11) | 0.77 | 0.02 (−0.16 to 0.21) | 0.77 |
| Systolic BP (mm Hg) | −0.01 (−0.5 to 0.47) | 0.98 | 0.28 (−0.22 to 0.79) | 0.26 | 0.24 (−0.28 to 0.76) | 0.36 | 0.19 (−0.3 to 0.68) | 0.44 | −0.05 (−0.72 to 0.62) | 0.89 |
| Diastolic BP (mm Hg) | 0.03 (−0.25 to 0.32) | 0.79 | 0.19 (−0.11 to 0.48) | 0.21 | −0.08 (−0.38 to 0.23) | 0.65 | 0.06 (−0.23 to 0.35) | 0.65 | 0.00 (−0.39 to 0.40) | 0.97 |
| Cholesterol (mmol/L) | 0.01 (−0.02 to 0.04) | 0.45 | 0.00 (−0.04 to 0.03) | 0.99 | −0.03 (−0.07 to 0.00) | 0.08 | 0.03 (0.00 to 0.06) | 0.02 | 0.18 (0.13 to 0.22) | 7.5×10−17 |
| Triglycerides (mmol/L) | −0.01 (−0.05 to 0.02) | 0.58 | −0.04 (−0.07 to 0.00) | 0.05 | −0.01 (−0.05 to 0.02) | 0.71 | −0.03 (−0.06 to 0.01) | 0.23 | 0.11 (0.07 to 0.16) | 3.0×10−7 |
| HDL cholesterol (mmo/L) | 0.00 (−0.01 to 0.01) | 0.75 | 0.00 (−0.01 to 0.01) | 0.34 | 0.00 (−0.01 to 0.01) | 0.72 | 0.01 (0.00 to 0.02) | 0.03 | −0.03 (−0.04 to −0.02) | 1.7×10−5 |
| LDL cholesterol (mmol/L) | 0.01 (−0.01 to 0.04) | 0.21 | 0.00 (−0.03 to 0.03) | 0.69 | −0.03 (−0.06 to 0.00) | 0.08 | 0.03 (0.01 to 0.05) | 0.008 | 0.16 (0.12 to 0.20) | 4.7×10−20 |
| Diabetes (OR) | 1.01 (0.92 to 1.10) | 0.90 | 1.01 (0.92 to 1.10) | 0.89 | 1.03 (0.94 to 1.14) | 0.48 | 1.04 (0.95 to 1.13) | 0.39 | 0.92 (0.81 to 1.04) | 0.19 |
Mendelian randomisation study of genetic variants in the CRP locus and CHD
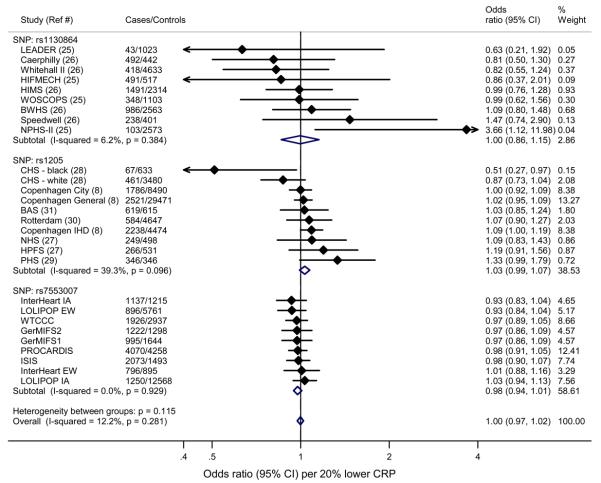
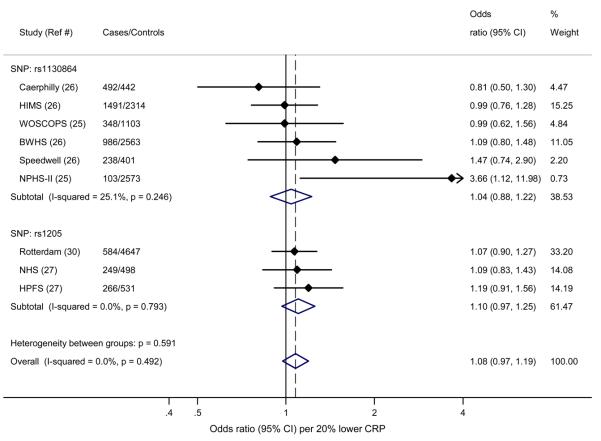
Figure 2 shows the results of the Mendelian randomisation experiment of CHD with variants in the CRP locus: SNP rs7553007 (our data) and published data for SNPs rs1130864 and rs1205 for 18 cohorts. SNP rs7553007 was not significantly associated with CHD; estimated OR was 0.98 (95% CI, 0.94-1.01) per 20% lower CRP. For rs1130864, OR was 1.00 (95% CI, 0.86-1.15) and for rs1205, OR was 1.03 (95% CI, 0.99-1.07). There was no association of CHD with CRP variants (per 20% lower CRP) when results for all three SNPs were combined, OR 1.00 (95% CI, 0.97-1.02) (Figure 2).
Figure 2.
Results of Mendelian randomisation experiment of rs7553007 in the CRP locus (present study), with rs1130864 and rs1205 from published studies, with CHD, among 28,112 cases and 100,823 controls. Effects are given as Odds Ratios (95% Confidence Intervals) per 20% lower CRP
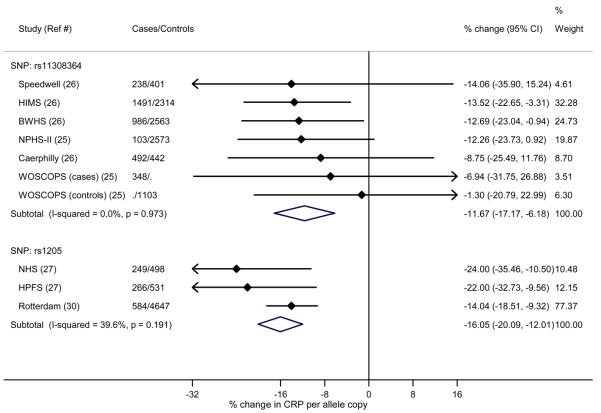
In the three-way comparison of CRP genetic variants, CRP levels, and CHD risk reported in prospective studies, there was significant association of CRP variants with CRP levels, CRP levels with CHD, but not CRP variants with CHD (Figure 3).
Figure 3.
Meta-analysis of associations of SNPs rs1130864 and rs1205 with CRP levels, CRP levels with CHD, and SNPs with CHD, in prospective studies where all three analyses have been reported. (a) Associations of SNPs rs1130864 and rs1205 with CRP levels. Effects are given as % change (95% Confidence Intervals) per minor allele (rs1205) and per major allele (rs1130864). (b) Associations of CRP levels with CHD. Effects are given as Odds Ratios (95% Confidence Intervals) per 20% lower CRP. (c) Associations of SNPs rs1130864 and rs1205 with CHD. Effects are given as Odds Ratios (95% Confidence Intervals) per 20% lower CRP
Comparison of the result of the Mendelian randomisation study with meta-analysis of the CRP-CHD relationship
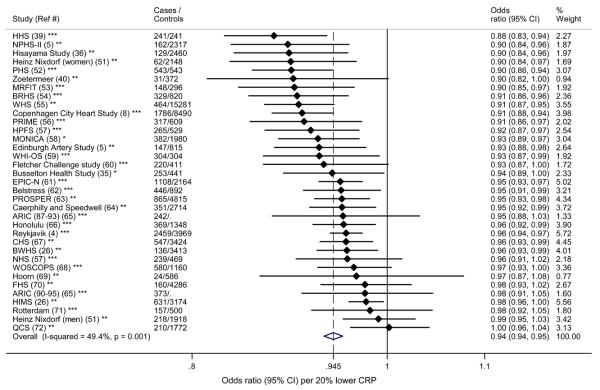
The meta-analysis of observational studies of CRP levels and CHD gave a predicted OR 0.94 (95% CI, 0.94–0.95) per 20% lower CRP (Figure 4). This is significantly different to the estimated effect on CHD (OR 1.00) of genetically determined differences in CRP levels obtained from our Mendelian randomisation study (Z=−3.45, P<0.001).
Figure 4.
Meta-analysis of the relationship of CRP levels with CHD from prospective observational studies. Effects are given as Odds Ratios (95% Confidence Intervals) per 20% lower CRP
Footnote to Figure 4.
Random effects estimate presented in Figure, fixed effects estimate: OR 0.951 (95% CI, 0.945-0.957). For cohort studies, number of ‘controls’ represents the number of event-free individuals. Number of controls was not available for ARIC (Ref 65).
*Case-cohort **Cohort ***Nested case-control
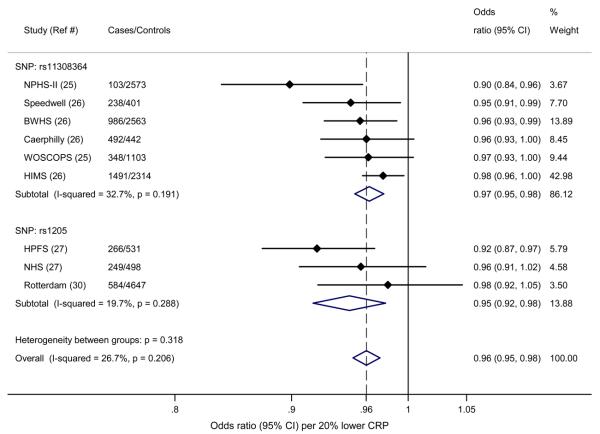
Genetic association of variants outside the CRP locus and CHD
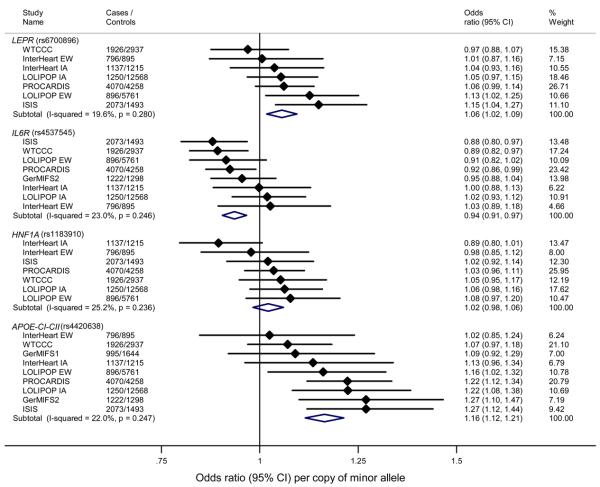
Minor alleles of dbSNP rs6700896 in LEPR and rs4420638 in APOE-CI-CII cluster were associated with significantly increased risk of CHD (OR 1.06 [95% CI, 1.02–1.09] and 1.16 [95% CI, 1.12–1.21] respectively), SNP rs4537545 in IL6R with a decreased risk of CHD (OR 0.94 [95% CI, 0.91, 0.97]), while SNP rs1183910 in HNF1A was not significantly associated with CHD (Figure 5). The effects of SNPs rs6700896 in LEPR and rs4420638 in the APOE-CI-CII cluster on CHD were in the opposite direction to that predicted from the relationship of CRP levels with CHD (Figure 4); for rs4420638 the finding was consistent with the effects on blood lipid concentrations (Table 2).
Figure 5.
Associations of SNPs in LEPR (rs6700896), IL6R (rs4537545), HNF1A (rs1183910) loci and APOE-CI-CII (rs4420638) cluster in the genetic association study with CHD. Effects are given as Odds Ratios (95% Confidence Intervals) per copy of minor allele
COMMENT
The present genome-wide association study confirms the associations of common genetic variants in the LEPR, IL6R, CRP, HNF1A loci and APOE-CI-CII cluster with CRP levels.42,43 However, minor allele of SNP rs7553007 and other variants in the CRP locus included in our Mendelian randomisation study were not associated with CHD risk.
The variants included in our Mendelian randomisation study are associated with ~20% lower CRP levels32, corresponding to 6% reduction in CHD risk predicted by the meta-analysis of observational studies of CHD risk in relation to differences in CRP levels. The lack of association with CHD of genetic variants in the CRP locus suggests that the observational data linking CRP levels and CHD may be confounded by association with other CHD risk factors, or reflect a secondary inflammatory response associated with atherosclerosis (‘reverse causation’), rather than indicating a causal relationship.
Our analysis of SNP rs7553007 with CHD risk includes more cases than all previously published studies of CRP variants and CHD combined8,25,26,27,28,29,30,31, yielding a total of over 28,000 cases and 100,000 controls for the Mendelian randomisation study. The largest previous study, of three cohorts in Copenhagen, included 6,545 cases.8 In addition to rs1205 included in our Mendelian randomisation study, it provides data on the tri-allelic SNP rs3091244 and SNP rs3093077, both having variants with larger effects on CRP levels (20-30%) than rs1205; however, both variants are rare, and neither was associated with CHD in the Copenhagen study.8
The JUPITER trial recently reported a benefit of treatment with rosuvastatin on CHD risk among men and women with elevated CRP levels (2.0 mg/L or higher) and with low-density lipoprotein (LDL) cholesterol levels below 130 mg/dL (3.4 mmol/L).34 In the treatment group, there was 54% reduction in rates of myocardial infarction compared with placebo; LDL levels were reduced by 50% and CRP levels by 37%. Although the JUPITER trial demonstrates the benefits of statin therapy in people with LDL levels below current treatment threshold, the results may simply reflect the benefits of lipid-lowering therapy in people who would not currently be considered for pharmacotherapy, rather than the benefits of CRP-lowering per se.44
Investigation of genetic variants underlying an intermediate phenotype (such as CRP) has been advocated as a means of discovering new disease susceptibility loci.10 In our study, minor alleles of SNPs rs6700896 in LEPR and rs4420638 in the APOE-CI-CII cluster showed significantly increased risk of CHD. However, both variants were associated with reduced levels of CRP, suggesting that the links with CHD are not mediated by CRP. LEPR has not previously been reported to increase risk of CHD, though associations of variants in the APOE-CI-CII cluster with CHD have been observed.45,46 While the association of genetic variation in APOE-CI-CII with CHD can be explained in large part by its effects on blood lipids, this is not the case for SNP rs6700896 in LEPR, which was not associated with major CHD risk factors. SNP rs6700896 is located in intron 19 of LEPR, the gene encoding the leptin receptor, a member of the class I cytokine receptor family.47 LEPR is expressed in the hypothalamus and vascular endothelial cells48, and LEPR signalling has a role in appetite control, weight regulation, glucose homeostasis, blood pressure regulation and angiogenesis.47,48,49 The minor allele of SNP rs4537545 in IL6R was also associated with reduced CRP levels, and reduced risk of CHD. SNP rs4537545 in IL6R is in high LD (r2= 0.96 in the HapMap CEU reference population) with a non-synonymous SNP (rs8192284, Asp358Ala) associated with increased IL6R expression, and alterations of IL6R membrane binding50, providing a potential mechanism linking rs4537545 to biological function. Further studies will be needed to confirm LEPR and IL6R as new susceptibility loci for CHD.
Our study has limitations. Because of the relatively small effect of common genetic variants in CRP locus on CRP levels, large sample size is needed to detect associations with CHD. In order to combine data across studies in our Mendelian randomisation study, we assumed a common effect on risk of CHD from different variants in the CRP locus, by standardising to a common difference on the log CRP scale. This approach is valid to the extent that the hypothesised effect of CRP variants on CHD risk reflects circulating levels of CRP. Though we found no association of CRP variants with CHD risk, it is not possible to exclude a small effect, despite the large sample size. However, we can effectively exclude the size of association predicted from observational data on the relationship of CRP levels to CHD. The Mendelian randomisation approach makes a number of assumptions concerning possible causality. These include the potential for pleiotropic effects of the genetic variants under study (or variants in high LD with them) giving an alternative pathway to CHD, or confounding through associations with disease risk factors.9 Neither rs7553007 or the other two variants included from the CRP locus correlate with CHD risk factors8,26, satisfying an important condition for a valid Mendelian randomisation experiment.9
In summary, our Mendelian randomisation study of over 28,000 cases and 100,000 controls found no association of variants in the CRP locus and CHD, arguing against a causal role for CRP in atherosclerosis. Moreover, this study suggests that development of therapeutic strategies targeting specific reductions in plasma levels of CRP are unlikely to be fruitful.
ACKNOWLEDGEMENTS
We thank Maria de Iorio, PhD and Pimphen Charoen, MPhil (Imperial College London) for help with the statistical analyses. Individuals acknowledged here by name were not compensated for their contributions to this paper.
Funding and support. With the exception of GlaxoSmithKline, whose scientists were involved in the planning, data collection and analysis for the GEMS and CoLaus studies, funders had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review or approval of the manuscript.
DESIR study. DESIR has been supported by INSERM, CNAMTS, Lilly, Novartis Pharma and Sanofi-Aventis, the Association Diabète Risque Vasculaire, the Fédération Française de Cardiologie, La Fondation de France, ALFEDIAM, ONIVINS, Ardix Medical, Bayer Diagnostics, Becton Dickinson, Cardionics, Merck Santé, Novo Nordisk, Pierre Fabre, Roche, Topcon. We acknowledge funding to PF by the European Union (Integrated Project EURODIA LSHM-CT-2006-518153 in the Framework Programme 6 [FP06] of the European-Community).
GEMS Study. Detailed information on the GEMS study design, sampling frame, and recruitment procedures has been published previously.15 We acknowledge the work of the GEMS investigators: Camperdown, Sydney, NSW, Australia (Prof. P. Barter, Ph.D.); Department of Internal Medicine and Biocenter Oulu, University of Oulu, Oulu, Finland (Prof. Y. A. Kesäniemi, Ph.D.); Gladstone Institute of Neurological Disease and Gladstone Institute of Cardiovascular Disease, San Francisco, CA, U.S.A. (Prof. R. W. Mahley, Ph.D.); Division of Cardiology, University of Ottawa Heart Institute, Ottawa, ON, Canada (Prof. R. McPherson, F.R.C.P.); Center for Human Nutrition, Department of Clinical Nutrition, University of Texas Southwestern Medical Center, Dallas, TX, U.S.A. (Prof. S. M. Grundy, Ph.D.); G.W. was also a GEMS investigator. The study was sponsored in part by GlaxoSmithKline, and all participants were duly informed about this sponsorship, and consented for the use of biological samples and data by GlaxoSmithKline and its subsidiaries; the study was approved by the local ethics committees.
German MI Family Study. The study was supported by grants from the Deutsche Forschungsgemeinschaft and the Deutsche Herzstiftung, the National Genome Research Network 2 of the German Federal Ministry of Education and Research, and the Cardiogenics project of the European Union.
INTERHEART Study. SSA holds the Michael G. DeGroot and Heart and Stroke Foundation of Ontario Chair in Population Health and the May Cohen Eli Lilly Endowed Chair in Women's Health Research, McMaster University. JCE is a research scholar of the Fonds de la Recherche en Santé du Québec. We thank Ron Do, MSc for help with statistical analyses. We acknowledge the contribution of Prof Salim Yusuf who initiated and together with the Steering Committee members, supervised the conduct of the INTERHEART study. We acknowledge all of the clinical centres in INTERHEART which are named in the INTERHEART 2004 report. We thank the members of the project office of the Population Health Research Institute, Sumathy Rangarajan and Laura Joldersma (study coordination), and Changchun Xie PhD (statistical support). The INTERHEART study was funded by the Canadian Institutes of Health Research, the Heart and Stroke Foundation of Ontario, the International Clinical Epidemiology Network (INCLEN), and through unrestricted grants from several pharmaceutical companies (with major contributions from AstraZeneca, Novartis, Sanofi Aventis, Knoll Pharmaceuticals [now Abbott], Bristol Myers Squibb, and King Pharma), and by various national bodies in different countries Chile: Universidad de la Frontera, Sociedad Chilena de Cardiologia Filial Sur; Colombia: Colciencias, Ministerio de Salud; Croatia: Croatian Ministry of Science & Technology; Guatemala: Liga Guatemalteca del Corazon; Hungary: Astra Hassle, National Health Science Council, George Gabor Foundation; Iran: Iran Ministry of Health; Italy: Boehringer-Ingelheim, Japan: Sankyo Pharmaceutical Co., Banyu Pharmaceutical Co., Astra Japan; Kuwait: Endowment Fund for Health Development in Kuwait; Pakistan: ATCO Laboratories; Philippines: Philippine Council for Health Research & Dev., Pfizer Philippines Foundation, Inc., Astra Pharmacetuicals, Inc. & the Astra Fund for Clinical Research & Continuing Medical Education, Pharmacia & Upjohn Inc.; Poland: Foundation PROCLINICA; Singapore: Singapore National Heart Association; South Africa: MRC South Africa, Warner-Parke-Davis Pharmaceuticals, Aventis; Sweden: Grant from the Swedish State under LUA Agreement, Swedish Heart and Lung Foundation; Thailand: The Heart Association of Thailand, Thailand Research Fund. This project was supported by the ECOGENE-21 project from the Center for Applied Health Research (CAHR) program (grant CAR43283).
ISIS Study. We gratefully acknowledge the patients and their relatives who collaborated, their general practitioners, and the medical and nursing staff from more than 100 hospitals in the U.K. A full list of the participating centers and collaborators is given in the ISIS-3 report.19 The ISIS trials and epidemiological studies were supported by the manufacturers of the study drugs, and by the British Heart Foundation, Medical Research Council, Cancer Research UK, Tobacco Products Research Trust of the UK Department of Health Independent Scientific Committee on Smoking and Health, and Oxford NHS Genetic Knowledge Park.
Lausanne Cohort (CoLaus) study. Investigators: Jacques S Beckmann, Sven Bergmann, Murielle Bochud, Toby Johnson, NL, VM, Kijoung Song, PV, GW, DMW, Xin Yuan. Principal Investigators: VM, PV. Study design: JSB, VM, PV, GW, DMW. Assembly of the cohort: GW Data analysis: NL. Project management: JSB, SB, MB, VM, PV, DMW. TJ was supported by the Giorgi-Cavaglieri Foundation. JSB was supported by UNIL. SB is supported by the Giorgi-Cavaglieri Foundation and the Swiss National Science Foundation (Grant # 3100AO-116323/1). MB was supported by the Swiss National Science Foundation. PV and GW received financial support from GlaxoSmithKline to build the CoLaus study. This work has been supported by GlaxoSmithKline and the Faculty of Biology and Medicine of Lausanne, Switzerland.
London Life Sciences Population (LOLIPOP) study. Investigators: JCC, PE, JS, JSK. Principal investigator: JSK. Data analysis: JCC, WZ, Delilah Zabeneh, Robin Walters, Maria de Iorio, David Balding. LOLIPOP is supported by the British Heart Foundation Grant SP/04/002.
North Finland Birth Cohort of 1966. Investigators: PE, NBF, Anna-Liisa Hartikainen, M-RJ, MIM, LP, Anneli Pouta. Data analysis: LC, Pimphen Charoen. Biochemical Analysis: AR. We acknowledge the support of NHLBI grant 5R01HL087679-02 through the STAMPEED program, the MRC of the UK, EURO-BLCS, QLG1-CT-2000-01643, Biocenter of University of Oulu, and Academy of Finland, and NIMH grant 1RL1MH083268-01.
PROCARDIS. Investigators: RC, MF, Anuj Goel, AH, Simon C Heath, G Mark Lathrop, JFP, Udo Seedorf, Ann-Christine Syvänen, Giovanni Tognoni, H.W. Principal investigator for project: HW. Principal investigators for collection centre: RC, AH, US, GT. Genotyping: SCH, GML, A-CS. Quality control: SCH. Data analysis: MF, AG, JFP. Project management: JFP, HW. See www.procardis.org for full membership of PROCARDIS consortium. We are grateful to the participants and to the medical and nursing staff who assisted in this project. This work was funded by the British Heart Foundation, EC Sixth Framework Programme (LSHM33 CT- 2007- 037273), AstraZeneca AB and the Knut and Alice Wallenberg Foundation.
WTCCC-CAD. Recruitment for the WTCCC study was supported by grants from the British Heart Foundation and the UK Medical Research Council. We also acknowledge support from the Wellcome Trust Functional Genomics Initiative in Cardiovascular Genetics. Dr. Samani holds a chair funded by the British Heart Foundation.
Footnotes
Financial disclosures
None declared.
REFERENCES
- 1.Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet. 1997;349(9061):1269–76. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- 2.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–95. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 3.Verma S, Szmitko PE, Ridker PM. C-reactive protein comes of age. Nat Clin Pract Cardiovasc Med. 2005;2(1):29–36. doi: 10.1038/ncpcardio0074. [DOI] [PubMed] [Google Scholar]
- 4.Danesh J, Wheeler JG, Hirschfield GM, et al. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350(14):1387–97. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 5.Shah T, Casas JP, Cooper JA, et al. Critical appraisal of CRP measurement for the prediction of coronary heart disease events: new data and systematic review of 31 prospective cohorts. Int J Epidemiol. 2009;38(1):217–31. doi: 10.1093/ije/dyn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook NR, Buring JE, Ridker PM. The effect of including C-reactive protein in cardiovascular risk prediction models for women. Ann Intern Med. 2006;145(1):21–9. doi: 10.7326/0003-4819-145-1-200607040-00128. [DOI] [PubMed] [Google Scholar]
- 7.Pepys MB. C-reactive protein is neither a marker nor a mediator of atherosclerosis. Nat Clin Pract Nephrol. 2008;4(5):234–5. doi: 10.1038/ncpneph0778. [DOI] [PubMed] [Google Scholar]
- 8.Zacho J, Tybjaerg-Hansen A, Jensen JS, et al. Genetically elevated C-reactive protein and ischaemic vascular disease. New Engl J Med. 2008;359(18):1897–908. doi: 10.1056/NEJMoa0707402. [DOI] [PubMed] [Google Scholar]
- 9.Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5(8):e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter DJ, Altshuler D, Rader DJ. From Darwin's finches to canaries in the coal mine--mining the genome for new biology. N Engl J Med. 2008;358(26):2760–3. doi: 10.1056/NEJMp0804318. [DOI] [PubMed] [Google Scholar]
- 11.Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(6):716–8. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 12.Tzoulaki I, Jarvelin MR, Hartikainen AL, et al. Size at birth, weight gain over the life course, and low-grade inflammation in young adulthood: northern Finland 1966 Birth Cohort study. Eur Heart J. 2008;29(8):1049–56. doi: 10.1093/eurheartj/ehn105. [DOI] [PubMed] [Google Scholar]
- 13.Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan X, Waterworth D, Perry J, et al. Population-Based Genome-Wide Association Study Reveals Four Novel Trans- and Two Cis-Acting Loci Influencing Plasma Levels of Liver Function Tests. Am J Hum Genetics. 2008;83(4):520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyszynski DF, Waterworth DM, Barter PJ, et al. Relation between atherogenic dyslipidemia and the Adult Treatment Program-III definition of metabolic syndrome (Genetic Epidemiology of Metabolic Syndrome Project) Am J Cardiol. 2005;95(2):194–8. doi: 10.1016/j.amjcard.2004.08.091. [DOI] [PubMed] [Google Scholar]
- 16.Bouatia-Naji N, Rocheleau G, Van Lommel L, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320(5879):1085–8. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 17.Farrall M, Green FR, Peden JF, et al. Genome-wide mapping of susceptibility to coronary artery disease identifies a novel replicated locus on chromosome 17. PLoS Genet. 2006;2(5):e72. doi: 10.1371/journal.pgen.0020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke R, Xu P, Bennett D, et al. Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet. 2006;2(7):e107. doi: 10.1371/journal.pgen.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ISIS-3 (Third International Study of Infarct Survival) Collaborative Group ISIS-3: a randomised comparison of streptokinase vs tissue plasminogen activator vs anistreplase and of aspirin plus heparin vs aspirin alone among 41 299 cases of suspected acute myocardial infarction. Lancet. 1992;339(8796):573–770. [PubMed] [Google Scholar]
- 20.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schunkert H, Götz A, Braund P, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117(13):1675–84. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pare G, Serre D, Brisson D, et al. Genetic analysis of 103 candidate genes for coronary artery disease and associated phenotypes in a founder population reveals a new association between endothelin-1 and high-density lipoprotein cholesterol. Am J Hum Genet. 2007;80(4):673–82. doi: 10.1086/513286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 24.Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006. http://hydra.usc.edu/gxe, accessed 1 December 2008.
- 25.Casas JP, Shah T, Cooper J, et al. Insight into the nature of the CRP-coronary event association using Mendelian randomization. Int J Epidemiol. 2006;35(4):922–31. doi: 10.1093/ije/dyl041. [DOI] [PubMed] [Google Scholar]
- 26.Lawlor DA, Harbord RM, Timpson NJ, et al. The association of C-reactive protein and CRP genotype with coronary heart disease: findings from five studies with 4,610 cases amongst 18,637 participants. PLoS ONE. 2008;3(8):e3011. doi: 10.1371/journal.pone.0003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pai JK, Mukamal KJ, Rexrode KM, Rimm EB. C-reactive protein (CRP) gene polymorphisms, CRP levels, and risk of incident coronary heart disease in two nested case-control studies. PLoS ONE. 2008;3(1):e1395. doi: 10.1371/journal.pone.0001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange LA, Carlson CS, Hindorff LA, et al. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. JAMA. 2006;296(22):2703–11. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- 29.Miller DT, Zee RY, Suk Danik J, et al. Association of common CRP gene variants with CRP levels and cardiovascular events. Ann Hum Genet. 2005;69(6):623–38. doi: 10.1111/j.1529-8817.2005.00210.x. [DOI] [PubMed] [Google Scholar]
- 30.Kardys I, de Maat MPM, Uitterlinden AG, et al. C-reactive protein gene haplotypes and risk of coronary heart disease: the Rotterdam Study. Eur Heart J. 2006;27(11):1331–7. doi: 10.1093/eurheartj/ehl018. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Zhao J, Huang J, Su S, Qiang B, Gu D. −717A>G polymorphism of C-reactive protein gene associated with coronary heart disease in ethnic Han Chinese: the Beijing atherosclerosis study. J Mol Med. 2005;83(1):72–8. doi: 10.1007/s00109-004-0585-5. [DOI] [PubMed] [Google Scholar]
- 32.Verzilli C, Shah T, Casas JP, et al. Bayesian meta-analysis of genetic association studies with different sets of markers. Am J Hum Genet. 2008;82(4):859–72. doi: 10.1016/j.ajhg.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 34.Ridker PM, Danielson E, Fonseca FAH, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein levels. N Engl J Med. 2008;359(21):2195–207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 35.Hung J, Knuiman MW, Divitini ML, Langton PE, Chapman CL, Beilby JP. C-reactive protein and interleukin-18 levels in relation to coronary heart disease: prospective cohort study from Busselton Western Australia. Heart Lung Circ. 2008;17(2):90–5. doi: 10.1016/j.hlc.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Arima H, Kubo M, Yonemoto K, et al. High-sensitivity C-reactive protein and coronary heart disease in a general population of Japanese: the Hisayama study. Arterioscler Thromb Vasc Biol. 2008;28(7):1385–91. doi: 10.1161/ATVBAHA.107.157164. [DOI] [PubMed] [Google Scholar]
- 37.Agewall S, Wikstrand J, Fagerberg B. Prothrombin fragment 1+2 is a risk factor for myocardial infarction in treated hypertensive men. J Hypertens. 1998;16(4):537–41. doi: 10.1097/00004872-199816040-00016. [DOI] [PubMed] [Google Scholar]
- 38.Folsom AR, Chambless LE, Ballantyne CM, et al. An Assessment of Incremental Coronary Risk Prediction Using C-Reactive Protein and Other Novel Risk Markers: The Atherosclerosis Risk in Communities Study. Arch Intern Med. 2006;166(13):1368–73. doi: 10.1001/archinte.166.13.1368. [DOI] [PubMed] [Google Scholar]
- 39.Roivainen M, Viik-Kajander M, Palosuo T, et al. Infections, inflammation, and the risk of coronary heart disease. Circulation. 2000;101(3):252–7. doi: 10.1161/01.cir.101.3.252. [DOI] [PubMed] [Google Scholar]
- 40.Störk S, Feelders RA, van den Beld AW, et al. Prediction of mortality risk in the elderly. Am J Med. 2006;119(6):519–25. doi: 10.1016/j.amjmed.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 41.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 42.Ridker PM, Pare G, Parker A, et al. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am J Hum Genet. 2008;82(5):1185–92. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reiner AP, Barber MJ, Guan Y, et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet. 2008;82(5):1193–201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hlatky MA. Expanding the orbit of primary prevention – Moving beyond JUPITER. N Engl J Med. 2008;359(21):2280–2. doi: 10.1056/NEJMe0808320. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Zhou X, Ye S, et al. Combined effects of apoE-CI-CII cluster and LDL-R gene polymorphisms on chromosome 19 and coronary artery disease risk. Int J Hyg Environ Health. 2006;209(3):265–73. doi: 10.1016/j.ijheh.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–6. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers MG, Cowley MA, Münzberg H. Mechanisms of leptin action and leptin resistance. Annu Rev Physiol. 2008;70:537–56. doi: 10.1146/annurev.physiol.70.113006.100707. [DOI] [PubMed] [Google Scholar]
- 48.Sierra-Honigmann MR, Nath AK, Murakami C, et al. Biological action of leptin as an angiogenic factor. Science. 1998;281(5383):1683–6. doi: 10.1126/science.281.5383.1683. [DOI] [PubMed] [Google Scholar]
- 49.Rosmond R, Chagnon YC, Holm G, et al. Hypertension in obesity and the leptin receptor gene locus. J Clin Endocrinol Metab. 2000;85(9):3126–31. doi: 10.1210/jcem.85.9.6781. [DOI] [PubMed] [Google Scholar]
- 50.Rafiq S, Frayling TM, Murray A, et al. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun. 2007;8(7):552–9. doi: 10.1038/sj.gene.6364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erbel R, Möhlenkamp S, Lehmann N, et al. Sex related cardiovascular risk stratification based on quantification of atherosclerosis and inflammation. Atherosclerosis. 2008;197(2):662–72. doi: 10.1016/j.atherosclerosis.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 52.Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH. Inflammation, asparin, and the risk of cardiovascular disease in apparently healthy men. New Engl J Med. 1997;336(14):973–9. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- 53.Kuller LH, Tracy RP, Shaten J, Meilahn EN. Relation of c-reactive protein and coronary heart disease in the MRFIT nested case-control study. Am J Epidemiol. 1996;144(6):537–47. doi: 10.1093/oxfordjournals.aje.a008963. [DOI] [PubMed] [Google Scholar]
- 54.Danesh J, Whincup P, Walker M, et al. Low grade inflammation and coronary heart disease: prospective study and updated meta-analyses. Brit Med J. 2000;321(7255):199–204. doi: 10.1136/bmj.321.7255.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridker PM, Rifai N, Cook NR, Bradwin G, Buring JE. Non-HDL cholesterol, apolipoproteins A-I and B100, standard lipid measures, lipid ratios, and CRP as risk factors for cardiovascular disease in women. JAMA. 2005;294(3):326–33. doi: 10.1001/jama.294.3.326. [DOI] [PubMed] [Google Scholar]
- 56.Luc G, Bard J-M, Juhan-Vague I, et al. C-reactive protein, interleukin-6, and fibrinogen as predictors of coronary heart disease: the PRIME study. Arterioscler Thromb Vasc Biol. 2003;23(7):1255–61. doi: 10.1161/01.ATV.0000079512.66448.1D. [DOI] [PubMed] [Google Scholar]
- 57.Pai JK, Pischon T, Ma J, et al. Inflammatory markers and the risk of coronary heart disease in men and women. New Engl J Med. 351(25):2599–610. doi: 10.1056/NEJMoa040967. [DOI] [PubMed] [Google Scholar]
- 58.Koenig W, Khuseyinova N, Baumert J, et al. Increased concentrations of c-reactive protein and IL-6 but not IL-18 are independently associated with incident coronary events in middle-aged men and women. Results from the MONICA/KORA Augsburg case-cohort study, 1984-2002. Arterioscler Thromb Vasc Biol. 2006;26(12):2745–51. doi: 10.1161/01.ATV.0000248096.62495.73. [DOI] [PubMed] [Google Scholar]
- 59.Pradhan AD, Manson JE, Rossouw JE, et al. Inflammatory biomarkers, hormone replacement therapy, and incident coronary heart disease. Prospective analysis from the Women's Health Initiative Observational Study. JAMA. 2002;288(8):980–7. doi: 10.1001/jama.288.8.980. [DOI] [PubMed] [Google Scholar]
- 60.Woodward M, Rumley A, Welsh P, MacMahon S, Lowe G. A comparison of the associations between seven hemostatic or inflammatory variables and coronary heart disease. J Thromb Haemost. 2007;5(9):1795–800. doi: 10.1111/j.1538-7836.2007.02677.x. [DOI] [PubMed] [Google Scholar]
- 61.Boekholdt SM, Hack CE, Sandhu MS, et al. C-reactive protein levels and coronary artery disease incidence and mortality in apparently healthy men and women: the EPIC-Norfolk prospective population study 1993–2002. Atherosclerosis. 2006;187(2):415–22. doi: 10.1016/j.atherosclerosis.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 62.De Backer J, Mak R, De Bacquer D, et al. Parameters of inflammation and infection in a community based case-control study of coronary heart disease. Atherosclerosis. 2002;160(2):457–63. doi: 10.1016/s0021-9150(01)00602-5. [DOI] [PubMed] [Google Scholar]
- 63.Sattar N, Murray HM, McConnachie A, et al. C-reactive protein and prediction of coronary heart disease and global vascular events in the Prospective Study of Provastatin in the Elderly at Risk (PROSPER) Circulation. 2007;115(8):981–9. doi: 10.1161/CIRCULATIONAHA.106.643114. [DOI] [PubMed] [Google Scholar]
- 64.Lowe GDO, Sweetnam PM, Yarnell JWG, et al. C-reactive protein, fibrin d-dimer, and risk of ischemic heart disease: the Caerphilly and Speedwell studies. Arterioscler Thromb Vasc Biol. 2004;24(10):1957–62. doi: 10.1161/01.ATV.0000141842.27810.a9. [DOI] [PubMed] [Google Scholar]
- 65.Folsom AR, Aleksic N, Catellier D, Juneja HS, Wu KK. C-reactive protein and incident coronary heart disease in the Atherosclerosis Risk In Communities (ARIC) study. Am Heart J. 2002;144(2):233–8. doi: 10.1067/mhj.2002.124054. [DOI] [PubMed] [Google Scholar]
- 66.Sakkinen P, Abbott RD, Curb JD, Rodriguez BL, Yano K, Tracy RP. C-reactive protein and myocardial infarction. J Clin Epidemiol. 2002;55(5):445–51. doi: 10.1016/s0895-4356(01)00502-9. [DOI] [PubMed] [Google Scholar]
- 67.Cushman M, Arnold AM, Psaty BM, et al. C-reactive protein and the 10-year incidence of coronary heart disease in older men and women: the Cardiovascular Health Study. Circulation. 2005;112(1):25–31. doi: 10.1161/CIRCULATIONAHA.104.504159. [DOI] [PubMed] [Google Scholar]
- 68.Packard CJ, O'Reilly DSJ, Caslake MJ, et al. Lipoprotein-associated phospholipase A2 as an independent predictor of coronary heart disease. New Engl J Med. 2000;343(16):1148–55. doi: 10.1056/NEJM200010193431603. [DOI] [PubMed] [Google Scholar]
- 69.Jager A, van Hinsbergh VWM, Kostense PJ, et al. von Willebrand factor, c-reactive protein, and 5-year mortality in diabetic and nondiabetic subjects: the Hoorn study. Arterioscler Thromb Vasc Biol. 1999;19(12):3071–8. doi: 10.1161/01.atv.19.12.3071. [DOI] [PubMed] [Google Scholar]
- 70.Wilson PWF, Nam B-H, Pencina M, D'Agostino RB, Benjamin EJ, O'Donnell CJ. C-reactive protein and risk of cardiovascular disease in men and women from the Framingham Heart Study. Arch Intern Med. 2005;165(21):2473–8. doi: 10.1001/archinte.165.21.2473. [DOI] [PubMed] [Google Scholar]
- 71.van der Meer IM, de Maat MPM, Kiliaan AJ, van der Kuip DAM, Hofman A, Witteman JCM. The value of c-reactive protein in cardiovascular risk prediction. The Rotterdam study. Arch Intern Med. 2003;163(11):1323–8. doi: 10.1001/archinte.163.11.1323. [DOI] [PubMed] [Google Scholar]
- 72.St-Pierre AC, Cantin B, Bergeron J, et al. Inflammatory markers and long-term risk of ischemic heart disease in men. A 13-year follow-up of the Quebec Cardiovascular Study. Atherosclerosis. 2005;182(2):315–21. doi: 10.1016/j.atherosclerosis.2005.02.009. [DOI] [PubMed] [Google Scholar]