Tethering σ70 to RNA polymerase reveals high in vivo activity of σ factors and σ70-dependent pausing at promoter-distal locations (original) (raw)
Abstract
Bacterial σ factors compete for binding to RNA polymerase (RNAP) to control promoter selection, and in some cases interact with RNAP to regulate at least the early stages of transcript elongation. However, the effective concentration of σs in vivo, and the extent to which σ can regulate transcript elongation generally, are unknown. We report that tethering σ70 to all RNAP molecules via genetic fusion of rpoD to rpoC (encoding σ70 and RNAP's β′ subunit, respectively) yields viable Escherichia coli strains in which alternative σ-factor function is not impaired. β′::σ70 RNAP transcribed DNA normally in vitro, but allowed σ70-dependent pausing at extended -10-like sequences anywhere in a transcriptional unit. Based on measurement of the effective concentration of tethered σ70, we conclude that the effective concentration of σ70 in E. coli (i.e., its thermodynamic activity) is close to its bulk concentration. At this level, σ70 would be a bona fide elongation factor able to direct transcriptional pausing even after its release from RNAP during promoter escape.
Keywords: RNA polymerase, σ factor, transcriptional regulation, E. coli, pausing
A fundamental paradigm in transcriptional regulation is the cyclical association of RNA polymerase (RNAP) with dissociable initiation, elongation, and termination factors (Burgess et al. 1969; Chamberlin 1976). In bacteria, σ factors bind tightly to free RNAP to program promoter recognition and initiation (Gill et al. 1991; Maeda et al. 2000), but are thought to release stochastically from elongation complexes (ECs) as, or shortly after, RNAP escapes promoters (Shimamoto et al. 1986). The avidity of σ-factor binding decreases dramatically in part because RNA chains >8 nt compete with σ for sites on RNAP (Krummel and Chamberlin 1989; Daube and von Hippel 1999; Murakami et al. 2002). The elongation factor NusA also may assist σ release by competitive binding to the EC (Gill et al. 1991). A similar cycle of initiation factor binding and release operates for eukaryotic RNAPs, although the details are less well understood (Kimura et al. 2002; Pokholok et al. 2002).
During development, differentiation, and changes in environment, binding, and release of initiation factors allow cells to alter patterns of gene expression by reprogramming RNAP. In bacteria, reprogramming is accomplished by switching among σ factors associated with RNAP (e.g., seven σs in Escherichia coli, 18 in Bacillus subtilis, and at least 65 in Streptomyces coelicolor; Ishihama 2000; Kunst et al. 1997; Bentley et al. 2002). Among these, one σ is the major or housekeeping σ and is typically present at the highest level (σ70 in E. coli).
Selection among σs for RNAP binding is thought to be mediated by concentration-dependent competition among available σ molecules for available RNAP molecules (Zhou and Gross 1992; Hicks and Grossman 1996; Farewell et al. 1998; Gross et al. 1998; Kolesky et al. 1999; Ishihama 2000; Maeda et al. 2000). The concentrations of available σs are in turn highly regulated by their expression levels, rates of degradation, and sequestration via binding to inhibitory proteins called anti-σs (Ishihama 2000). Although RNAP outnumbers total σ in E. coli (∼1.7:1; Materials and Methods), σs must compete for the smaller pool of available RNAP (∼0.6 per σ), which includes newly synthesized RNAP and RNAP not engaged in transcription or sequestered nonspecifically (Ishihama 2000; see Discussion).
The idea that σs are released from RNAP during transcript elongation has been challenged recently (Bar-Nahum and Nudler 2001; Mukhopadhyay et al. 2001). In one view, permanent association of σ70 with some RNAP is proposed to accelerate recycling of this RNAP for new rounds of transcription by circumventing the need to rebind σ70, which could be rate-limiting in the transcription cycle (Bar-Nahum and Nudler 2001).
σ70 plays at least one regulatory role during transcript elongation. σ70 can remain bound to RNAP long enough to stimulate pausing at promoter-proximal sites that resemble -10 promoter elements (Ring et al. 1996). In this case, σ70-dependent pausing was lost when the pause site was moved 20 bp downstream and tested in vitro (Ring et al. 1996). This could be explained by stochastic σ70 release after promoter escape (Shimamoto et al. 1986) or by resistance of mature ECs to σ70-stimulated pausing.
To assess the effective concentration of σ70 in vivo and to gain insight into σ70's effect on the EC, we created an rpoC::rpoD gene fusion that tethered σ70 to all RNAP in cells via a covalent polypeptide linkage. Tethering proteins by genetic fusion fixes the local concentration of interacting proteins (Raag and Whitlow 1995; Timpe and Peller 1995; Robinson and Sauer 1998). Depending on the length of the tether and the location of binding sites relative to the tether-attachment points, tethering can generate local protein concentrations of 10-5 to 10 M (Robinson and Sauer 1998). The bulk concentration of σ70 in vivo is ∼15 μM (Materials and Methods). However, a variety of factors, including macromolecular crowding, can dramatically alter the effective concentration of σ70 in cells (i.e., its thermodynamic activity). Tethering σ70 to RNAP makes it possible to examine the effects in vivo of known local concentrations of tethered σ70, and thus gain insight into the effective concentration of σ70 in cells.
Results
E. coli is viable with σ70 fused to RNAPat the β′ C terminus
We initially tested the in vivo function of σ70 tethered to the C terminus of β′ or β when corresponding gene fusions were conditionally expressed from plasmids (Table 1). (RNAP tolerates alterations at the ends of β′ or β; σ70 tolerates alterations at its N terminus; Severinov et al. 1997; Sharp et al. 1999.) β::σ70 was unable to complement for loss of σ70 or β function. However, β′::σ70 complemented for loss of σ70 and, to a limited extent, β′. We considered this result promising and concentrated further study on β′::σ70. As an initial control that β′::σ70 was incorporated into RNAP, we replaced region 4 of the tethered σ70 with the corresponding region of σ32 (Kumar et al. 1995). This eliminated the ability of β′ in the fusion to provide even weak β′ function (β′::σ70/32; Table 1), suggesting that the weak complementation of _rpoC_TS by β′;::σ70 reflected incorporation into a functional RNAP. Based on these results, we next asked if the rpoC;::rpoD fusion could replace rpoC in the chromosome (Materials and Methods).
Table 1.
Plating efficiency of tethered σ70 expressed from plasmids in wild-type and mutant strains
| _rpoD_- straina | _rpoB_- strainb | _rpoC_- strainb | |||
|---|---|---|---|---|---|
| Plasmid | Subunit | —IPTG | +IPTG | +IPTG | +IPTG |
| pTrc99c | None | <10-6 | <10-6 | <10-6d | <10-6d |
| pRL663 | β′ | — | — | — | 1 |
| pRL706 | β | — | — | 1 | — |
| pCL391 | σ70 | 1 | 1 | — | — |
| pRM314 | β′::σ70 | <10-6 | 0.5—1 | — | Weak, <10-6 scc |
| pRM328 | β::σ70 | <10-6 | <10-6 | <10-6d | — |
| pRM317 | β′::σ70/32 | <10-6 | <10-6 | — | <10-6d |
The resulting strain, in which σ70 was tethered to all RNAPs, proved viable. We transduced the rpoC;::rpoD allele into a strain in which rpoD expression can be shut off (Lonetto et al. 1998). This strain exhibited no defect in either growth or recovery from stationary phase when forced to rely on β′;::σ70 for both β′ and σ70 function (Fig. 1A,B). The simplest interpretation of these results is that β′;::σ70 RNAP is a fully functional enzyme and that the weak complementation of _rpoC_TS by the plasmid-encoded β′;::σ70 reflected partial overexpression toxicity (Table 1).
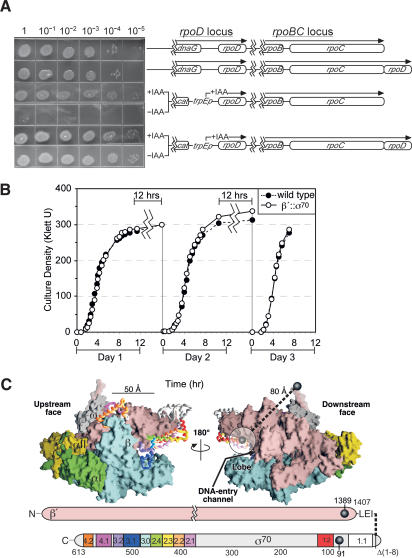
Figure 1.
Strains and growth phenotypes. (A) Early log-phase cultures of strains with the rpoD and rpoC loci illustrated were serially diluted and plated onto LB plates or LB plates containing indole acrylic acid (IAA). Strains (top to bottom) are RL301, RL1374, CAG20153, and RL1094 (Supplementary Table 1). The efficiencies of plating minus IAA versus plus IAA were ∼10-5 for the wild-type strain (CAG20153) and ∼1 for the β′;::σ70 strain (RL1094). (B) Wild-type or β′;::σ70 with TrpR-repressed rpoD strains (C600K- and RL1094) were monitored during growth in LB at 37°C using a Klett colorimeter (data are averages for two independent cultures). After residing in stationary phase for ∼12 h, the strains were diluted back to Klett <5. (C) Model of σ70 tethering to β′ based on the Thermus thermophilis holoenzyme structure (Vassylyev et al. 2002; β′ NCD removed). The upstream and downstream faces of RNAP are shown above schematics of the E. coli β′ and σ70 subunits. RNAP subunits are shown in spacefill; σ, is shown as a Cα-trace colored by regions shown in the schematic. The N-terminal and C-terminal residues of σ and β′ resolved in the structure are depicted as black spheres (corresponding to β′1389 and σ7091 in E. coli). σ region 1.1 (semitransparent white circle) is positioned based on the hydrated volume of an 82-amino acid globular protein attached to σ7091.
The structure of bacterial RNAP holoenzyme (Vassylyev et al. 2002) is consistent with this interpretation (Fig. 1C). Although σ region 1.1 is not resolved in the structure, if spherical, it would occupy a hydrated volume ∼30 Å in diameter on the downstream face of RNAP. At least 27 amino acids may flexibly connect region 1.1 to β′: three amino acids from the gene fusion, plus 19 C-terminal amino acids of E. coli β′ that correspond to the disordered C-terminal segment in the RNAP structure, plus the last five visible amino acids of β′, which form a random coil. These 27 amino acids (∼100 Å fully extended) are sufficient to span the 80 Å between the last resolved amino acids in β′ and the likely positions of region 1.1, proposed to be within the DNA-entry channel in the holoenzyme and at the outside edge of RNAP's lobe domain in initiation complexes (Mekler et al. 2002).
To verify that σ70 remained tethered to RNAP in the viable β′;::σ70 strain, we examined the subunits present in cell extracts by immunoblotting with anti-β′ and anti-σ70 antibodies (see Materials and Methods). Only β′;::σ70, not β′ or σ70, was present in a whole-cell extract from the β′;::σ70 strain when rpoD expression was shut off (Fig. 2A, cf. lanes 1 and 2). Therefore, β′;::σ70 was not cleaved to separate β′ and σ70 subunits in vivo, and this intact β′;::σ70 polypeptide could serve as the sole source of both β′ and σ70 in viable E. coli cells.
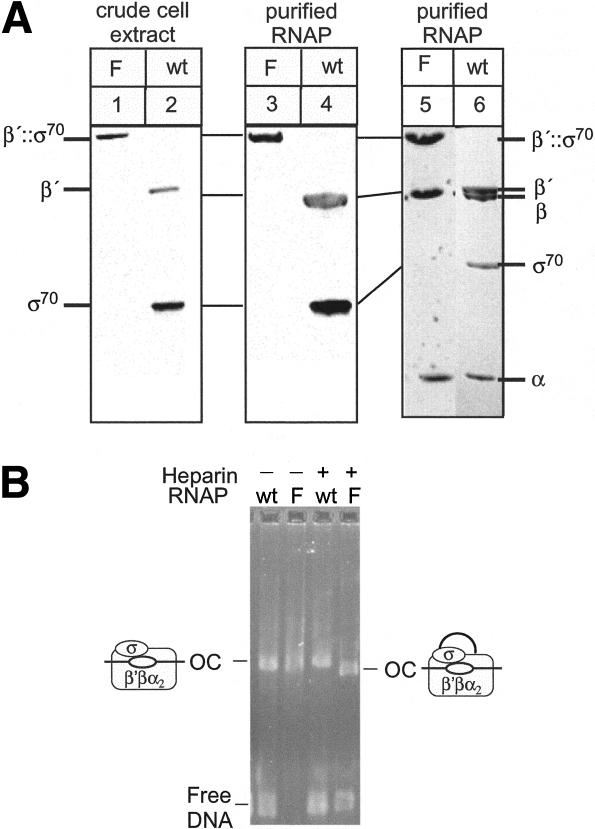
Figure 2.
Purified RNAP and immunoblot analysis. (A) (Left) Cellular extracts of wild-type (wt) or β′;::σ70 (F) strains (RL301 and RL1094) were separated by 3%-8% Tris-Acetate (Novex), transferred to nitrocellulose, and probed with a mixture of β′ and σ70 antibodies (Materials and Methods). Equal amounts of cellular protein were present in each lane. (Center) Western blot of wild-type (wt) and β′;::σ70 (F) RNAPs. Samples were separated by 3%-8% Tris Acetate (Novex) and probed as in the left panel. (Right) β′;::σ70 RNAP (F) purified from the strain containing the rpoC;::rpoD fusion and _Trp_-repressed rpoD (RL1094) separated by 4%-15% PAGE gel (Pharmacia) shown next to wild-type (wt) RNAP for size comparison. (B) β′;::σ70 (F) or wild-type (wt) RNAPs were mixed with λPR template, allowing the formation of open complexes at 37°C for 20 min (heparin was present where indicated for the last 10 min), separated by native gel electrophoresis (4% NuSieve agarose, 0.5× TBE), and stained with EtBr. The absence of heparin masked the slightly faster mobility of β′;::σ70 OCs (see legend to Fig. 4C) because β′;::σ70 OCs appeared to aggregate more than wild-type OCs.
A possible explanation for the viability of the β′;::σ70 strain would be that one β′;::σ70 polypeptide provides β′ and a second polypeptide provides σ70 in a single RNAP holoenzyme. If this were true, then purified, active β′;::σ70 RNAP should contain two β′;::σ70 polypeptides. However, highly purified, fully active β′;::σ70 RNAP gave a stoichiometry of one β′;::σ70 to one β and contained no β′ or σ70 (Fig. 2A, lanes 3,5; densitometry not shown). Another possibility would be that σ70 from one β′;::σ70 RNAP provided σ70 function to a second β′;::σ70 RNAP. If this were true, then open complexes (OCs) formed by β′;::σ70 RNAP would be of significantly greater mass (920 kD vs. 460 kD, not including DNA), and would exhibit significantly slower electrophoretic mobility. To test this, we formed OCs with wild-type and β′;::σ70 RNAPs on a λPR-containing DNA fragment and compared them by native gel electrophoresis (Fig. 2B). The β′;::σ70 OCs exhibited similar mobility to wild type (see legend to Fig. 4C, below). We conclude that β′;::σ70 RNAP and its OCs contain a single β′;::σ70 polypeptide.
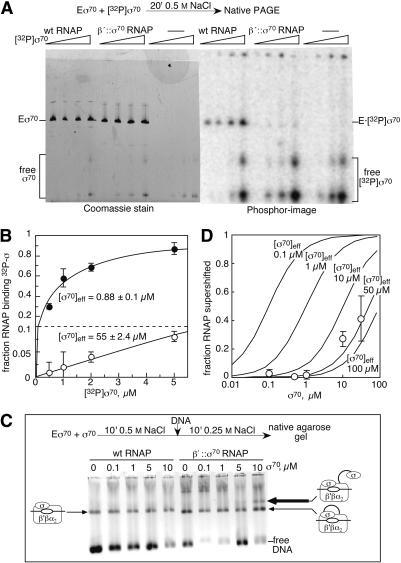
Figure 4.
Effective local concentration of tethered σ70 measured by competitive binding. (A) Equilibrium σ70-binding assay (see Materials and Methods). Samples are (left to right) 0.5, 1, 2, and 5 μM [32P]σ70 with 1 μM wild-type RNAP, β′;::σ70 RNAP, or no RNAP. The positions of holoenzyme (E σ70) and free σ70 are indicated; slower σ70 bands are σ70 dimers. (B) Quantitation of A. The fraction of RNAP binding 32P-labeled σ70 is plotted against the amount of 32P-labeled σ70 present in the reactions. (•) Wild-type RNAP; (○) β′::σ70 RNAP. Effective concentrations of σ70 were estimated by nonlinear regression (Materials and Methods) from the averages of three experiments. (C) OC mobility assay. β′::σ70 or wild-type RNAPs were incubated with the indicated amounts of σ70 and then with added promoter DNA (Materials and Methods). OCs and DNA were separated on a native agarose gel and stained with EtBr. The positions of the free DNA, the wild-type OC, the β′::σ70 OC, and the supershifted β′::σ70 OC formed with a second, untethered σ70 are indicated. Heparin was omitted in this experiment to minimize smearing of the supershifted OC band, although this caused variable nonspecific binding of free DNA (evident as aggregates near the top of the gel). Addition of heparin gave constant amounts of free DNA in gels and approximately the same amounts of supershifted OC, allowing use of up to 30 μM σ70 (Fig. 4D). The slightly increased mobility of β′::σ70 OCs (see also Fig. 2B) and of β′::σ70 RNAP (panel A) may reflect a slight structural change caused by tethering that is under study. (β′::σ70 RNAP does, however, contain ω.) (D) Quantitation of OC supershifting as a function of σ70 concentration. The fraction of RNAP binding a second σ70 (the supershifted species in C) is plotted against the concentration of added, untethered σ70 (0.1, 0.5, 1, 10, and 30 μM; average of three experiments that included heparin). Predicted binding curves (assuming all σ70 species to be equivalent for binding to RNAP) for different effective local concentrations of the tethered σ70 are shown for comparison (Materials and Methods).
The results described to this point confirm that RNAP tethered to σ70 supports cell growth and functions with the σ70 to which it is tethered. This suggests that β′::σ70 RNAP can use at least σ32 and σE, which are required for viability of E. coli at 37°C (Zhou et al. 1988; De Las Penas et al. 1997), and σS, which is necessary for wild-type recovery from the stationary phase (Ishihama 2000). We next examined use of alternative σs more carefully and tested for function of NusA, which also competes with σ70 for interaction with RNAP (Gill et al. 1991). In the presence or absence of chromosomally encoded, untethered σ70, β′::σ70 strains plated a λ phage that requires function of NusA (and other Nus proteins; Friedman et al. 1976) equivalently to wild type (Table 2). The same was true for survival at 45°C (an even more stringent requirement for σ32 and σE), for growth on medium requiring σN function, for σF-dependent swarming motility, and for σS-dependent formation of peroxidase in the stationary phase (Table 2). Thus, alternative σs and NusA appear to exhibit high in vivo activity, sufficient to allow normal function in β′::σ70 strains despite the presence of σ70 tethered to all RNAP molecules. This remains true even in β′::σ70 strains also containing chromosomally encoded, untethered σ70.
Table 2.
Properties of wild-type and β′::σ_70_ fusion strains in various conditions
| Phenotype | Required factor | Wild-type σ70 levela | β′::σ70, no add. σ70a | β′::σ70, plus σ70a |
|---|---|---|---|---|
| λ growthb | NusA, B, E, G | 1 | 0.8 ± 0.2 | 1.0 ± 0.4 |
| Growth at 45°Cc | σ32, σE | 1.0 ± 0.2 | 1.0 ± 0.1 | 1.0 ± 0.1 |
| Growth on Arg/Glnd | σN | 1.2 ± 0.3 | 1.3 ± 0.1 | 1.0 ± 0.4 |
| Motilitye | σF | 1 | 1.0 ± 0.1 | 1.0 ± 0.1 |
| Catalase productionf | σS | + | + | + |
β′::σ70 RNAPdisplays wild-type enzymatic properties and activities
We next sought to determine if β′::σ70 RNAP also behaved normally in vitro. For this purpose, we tested transcription by wild-type and β′::σ70 RNAPs of a linear DNA template encoding the well-characterized his pause site downstream of the T7 phage A1 promoter (Chan and Landick 1989). Upon initiation under conditions that allowed RNAP to transcribe only to A29, no difference was apparent between wild-type and β′::σ70 RNAPs in the rate of abortive initiation, the rate of productive initiation, or the ratio of abortive to productive products (as reflected by the AUC abortive RNA and A29 productive RNA; Fig. 3A). When transcription past A29 was allowed after 2 min, both RNAPs escaped the A29 position efficiently and exhibited indistinguishable kinetics of pausing and subsequent transcription to the end of the template. We also tested the ability of NusA protein to increase the duration of pausing at the his pause site and the efficiency of termination by β′::σ70 RNAP at several intrinsic terminators; no significant differences from wild-type RNAP were observed (data not shown).
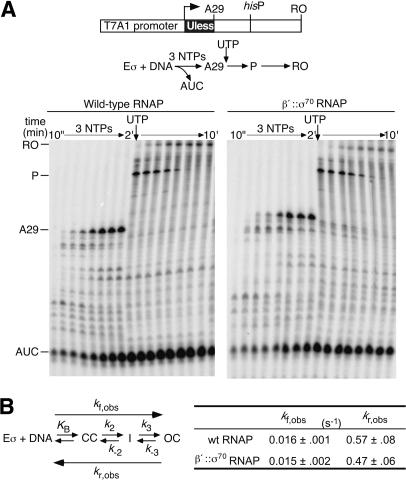
Figure 3.
(A) In vitro transcription assay. β′::σ70 or wild-type RNAPs (40 nM) were allowed to initiate transcription on a linear DNA template (25 nM) with ApU, ATP, [32P]CTP, and GTP. Samples were mixed with 2× STOP buffer at 10, 20, 30, 45, 60, 75, 90, and 120 sec. UTP was then added to allow transcription past A29, and additional samples were taken 10, 20, 30, 45, 60, 120, 240, and 480 sec later (final sample 10 min after reaction started). The samples were separated by 15% denaturing PAGE. (AUC) Trinucleotide abortive product; (A29) A29 EC; (P) his pause RNA; (RO) run-off RNA. (B) Kinetics of promoter association, following the mechanism defined by Saecker et al. (2002). _k_f,obs and _k_r,obs were obtained for β′::σ70 and wild-type RNAPs on the template shown in panel A (≥3 independent experiments; Materials and Methods).
To test whether σ70 tethering affects steps on the pathway of OC formation, we measured the overall rates of OC formation at the T7 A1 promoter (_k_f,obs) and of preformed OC dissociation (_k_r,obs; Fig. 3B; Materials and Methods). Both rates are composites of multiple steps (McClure 1980; Saecker et al. 2002) and would reveal any effects of the tether. Neither _k_f,obs nor _k_r,obs differed significantly between wild-type and β′::σ70 RNAPs (Fig. 3B), consistent with wild-type function of the tethered σ70.
Tethering σ70 to RNAPincreases σ70 local concentration to ∼55 μM
The lack of effect of tethering σ70 to RNAP caused us to consider whether tethering actually increased the local concentration of σ70 around RNAP. Such increased local concentrations have been measured for other cases of protein tethering (Timpe and Peller 1995; Robinson and Sauer 1998), and are well-grounded in polymer-chain theory (Flory 1969), but must be determined for each case.
To measure the local concentration of tethered σ70, we performed competition binding assays using a 32P-labeled derivative of σ70 (Materials and Methods). We compared the ability of [32P]σ70 to displace untethered σ70 from wild-type RNAP and to displace tethered σ70 from β′::σ70 RNAP using conditions in which σ70 binding to RNAP equilibrates (Sharp et al. 1999). To detect [32P]σ70 bound to RNAP, we separated the reactions by nondenaturing PAGE and visualized RNAP and σ70 by protein staining and [32P]σ70 using a PhosphorImager (Fig. 4A). [32P]σ70 displaced untethered σ70 from the wild-type holoenzyme as predicted for equivalent _K_ds of the prebound σ70 and [32P]σ70. However, only modest [32P]σ70 binding to β′::σ70 RNAP could be detected even at high concentrations of [32P]σ70 (Fig. 4A,B). Assuming that all 32P at or above the position of β′::σ70 RNAP in the gels arose from [32P]σ70 binding to β′::σ70 RNAP (because binding of a second σ70 would slow migration), we calculated a local concentration of tethered σ70 of ∼55 μM (Fig. 4B; Materials and Methods). This is >50-fold higher than the concentration of RNAP in the assay (1 μM), but significantly less than measured for optimal cases of tethering (Robinson and Sauer 1998). The extensive topography of σ70-RNAP contacts relative to the tether attachment points on σ70 and RNAP, interference of the tether with some contacts, or the tether length may limit tether enhancement of local concentration.
This experiment unambiguously demonstrated weaker binding of free σ70 to RNAP in the presence of tethered σ70. The local concentration estimate for tethered σ70 of 55 μM should be a lower limit because it included 32P that was nonspecifically retarded. However, our estimate conceivably could be inflated if the complex of [32P]σ70 and β′::σ70 RNAP dissociated during electrophoresis. This seemed unlikely because release of [32P]σ70 during electrophoresis would form a smear between the positions of holoenzyme and free σ70. No such smear was evident; rather, the amount of [32P]σ70 visible at the position of free σ70 was the same in the β′::σ70 lanes and the σ70 alone lanes, but was reduced in the wild-type RNAP lanes by the amount bound to RNAP (Fig. 4A, cf. the PhosphorImager densities at the bottom of the 1 μM and 2.5 μM lanes).
To confirm that tethered σ70 produced a local σ70 concentration of ∼55 μM, we sought to detect the binding of untethered σ70 to β′::σ70 RNAP in a manner that would prevent its release during electrophoresis. We reasoned that if the untethered σ70 were engaged in OC formation by β′::σ70 RNAP, two σ70s would be bound to RNAP because untethered σ70 would be trapped in the network of RNAP-σ70-DNA interactions in the OC (Murakami et al. 2002), and tethered σ70 would remain covalently attached to RNAP. To favor trapping untethered σ70 bound to β′::σ70 RNAP, we used a promoter on which OCs were stable for many hours (PUPFullcon; Materials and Methods). We equilibrated RNAPs with increasing concentrations of free σ70, incubated them with PUPFullcon
DNA for 10 min, and then separated the reactions on a nondenaturing agarose gel. After visualizing the locations of DNA by ethidium staining, OCs were readily apparent as retarded bands for both wild-type and β′::σ70 RNAPs (Fig. 4C). A second, more slowly migrating species appeared when 5-10 μM σ70 was added to β′::σ70 RNAP (Fig. 4C, thick arrow), but was not present with wild-type RNAP. Nonspecific binding and smearing prevented us from using concentrations of σ70 above 30 μM. Because the supershifted species was also not observed with DNA alone or DNA plus 5-10 μM σ70 (data not shown), we attribute it to β′::σ70 RNAP complexed with a second, untethered σ70 that is engaged in promoter contacts. To estimate the local concentration of the tethered σ70, we plotted the amounts of the supershifted species versus concentrations of added σ70 and compared the data with predicted curves (Materials and Methods; Fig. 4D). They matched reasonably to the prediction for ∼50 μM local concentration of tethered σ70 and were more than the prediction for 100 μM. Thus, both assays give an estimate of tethered σ70 local concentration consistent with ∼55 μM.
Tethering σ70 to RNAPmodestly increases σ70 local concentration in vivo
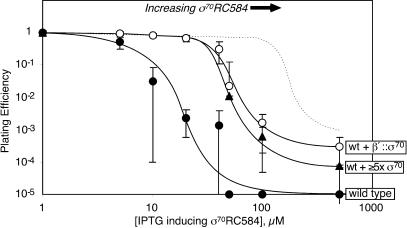
To compare the local concentration of tethered σ70 with the effective concentration of wild-type σ70 in vivo, we tested the ability of chromosomally encoded β′::σ70 or increased amounts of plasmid-encoded, untethered σ70 to protect wild-type cells (also containing chromosomally encoded σ70) against the toxic effects of a plasmid-encoded mutant σ70 (RC584). σ70RC584 binds RNAP and interferes with promoter recognition (Siegele et al. 1989). The presence of tethered σ70 significantly increased resistance to σ70RC584 (Fig. 5, cf. closed and open circles; wild-type σ70 is not toxic when expressed at comparable levels, dashed line). Expression of untethered σ70 from a low-copy-number, compatible plasmid to about fivefold above the normal level conferred a level of σ70RC584 resistance similar to that conferred by tethered σ70 (Fig. 5, triangles).
Figure 5.
β′::σ70 inhibits toxicity caused by σ70RC584. RL113 (wild type, •), RL113 + pBADσ70 (wt + ≥5× σ70; ▴), or RL1366 (wt + β′::σ70; ○) carrying pRM389 (σ70RC584 expressed under LacI control) were plated with or without IPTG as described in Materials and Methods (both strains were deleted for lacY). Data are mean values of three to six independent experiments. The dotted line shows the approximate effect of wild-type σ70 expressed from pCL391 for comparison.
These results confirm that β′::σ70 RNAP generates a local σ70 concentration higher than that of untethered σ70 in wild-type cells. A straightforward interpretation would place the effective concentration of wild-type σ70 at ∼11 μM because approximately fivefold overexpression gave σ70RC584 resistance similar to ∼55 μM local concentration of β′::σ70. However, overexpressed wild-type and RC584 σ70s could affect each other's levels in ways not measurable in plating experiments. Furthermore, we did not test whether a lower level of σ70 overexpression also could protect against RC584 σ70 as β′::σ70. Thus, we conclude wild-type cells contain σ70 at effective concentrations ≥11 μM and significantly less than 55 μM.
Tethering σ70 to RNAPonly modestly affects heat shock and σ32 function
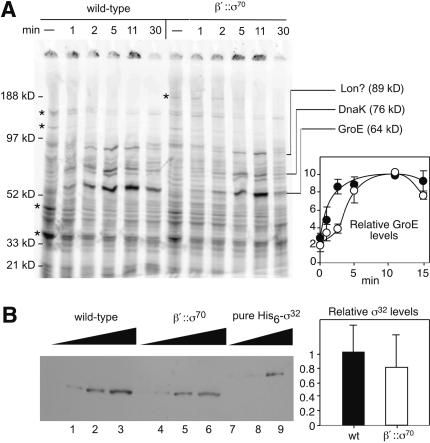
We next wanted to examine the effect of tethered σ70 on function of the alternate σ factor σ32, which is required for the well-defined heat-shock response in E. coli (Straus et al. 1987). When σ70 is overexpressed, σ32 function appears to be compromised by competition for binding to core RNAP (Zhou et al. 1992). However, σ32 is up-regulated in response, resulting in little if any inhibition of synthesis of heat-shock proteins or delay in the heat-shock response. We first confirmed that β′::σ70 RNAP could use σ32 in vitro (transcription of groE by β′::σ70 RNAP depended on added σ32; data not shown). We next tested whether β′::σ70 RNAP would affect expression of heat-shock proteins and σ32 levels similarly to overexpression of σ70.
The heat-shock response in β′::σ70 cells was slightly delayed relative to wild-type cells (Fig. 6A). The GroE synthesis rate was slower in β′::σ70 cells 1 or 2 min after shifting cells to 42°C, but reached the same level as wild-type by 11 min post-heat shock (Fig. 6A, plot). The characteristic suppression of σ70-dependent transcription was similar in β′::σ70 and wild-type cells (Fig. 6A, cf. * bands).
Figure 6.
Heat-shock response. (A) Cells were grown in minimal media at 30°C and then shifted to 42°C. At the times indicated, samples were incubated with [35S]methionine for 2 min. Samples were separated by 4%-12% PAGE. The positions of DnaK, GroE, and an 89-kD protein likely to be Lon are indicated. (*) Major proteins whose expression decreases on heat shock; the top band is β′::σ70, and the next-to-top band is β, β′. Relative GroE levels are averages of four experiments. (•) Wild-type (RL301); (○) β′::σ70 strain with chromosomally encoded σ70 (RL1374). (B) Immunoblot analysis using antibodies against σ32 (Materials and Methods). (Lanes _1_-3) Wild-type whole-cell lysate (MC1060). (Lanes _4_-6) β′::σ70 whole-cell lysate (RL1454). (Lanes _7_-9) His6-tagged σ32. Equal amounts of total cellular protein were electrophoresed.
To ask if heat shock was near normal because σ32 levels were up-regulated in the β′::σ70 strain, we measured σ32 levels by immunoblotting. The σ32 level in β′::σ70 cells was ∼80% of that in wild-type cells, a statistically insignificant difference (Fig. 6B; Materials and Methods). The slight delay in GroE expression in heat-shocked β′::σ70 cells is consistent with a local concentration of tethered σ70 higher than the wild-type σ70 effective concentration, but the nonelevated σ32 levels in β′::σ70 cells and the inhibition of σ70-directed transcription upon heat shock even when σ70 was tethered to RNAP were surprising (see Discussion).
Tethered σ70 causes σ70-dependent pausing independent of distance from the promoter
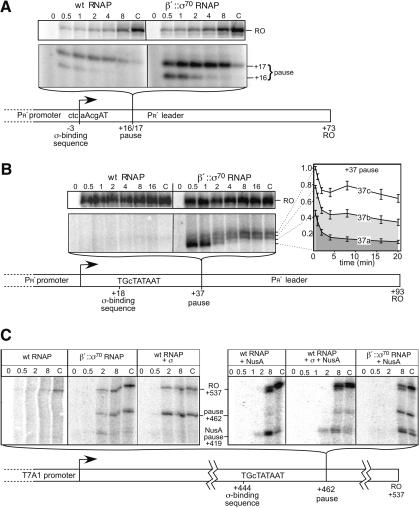
An additional consequence of tethering σ70 is that its high local concentration will be maintained during transcript elongation. Because untethered σ70 is able to stimulate pausing at -10-like sequences near a promoter (Ring et al. 1996), we wondered if tethering would allow σ70 to stimulate pausing at -10-like sequences generally. To test this possibility, we examined σ70-dependent pausing on templates originally studied by Roberts and colleagues (+16/17 and +37 pauses), and on an additional template encoding a consensus extended -10 sequence near +450 of an artificial transcription unit (+462 pause). β′::σ70 RNAP gave slightly increased pausing at the +16/17 site (ctcaAcgAT, -10-like σ70-binding sequence; Fig. 7A). This strong pausing also occurred at the +37 site (TGcTATAAT, consensus extended -10-like σ70-binding sequence; Fig. 7B), where wild-type RNAP pauses weakly if at all. This result confirms that wild-type RNAP fails to pause at +37 because its σ70 is released, rather than because the structure of the EC past +25 precludes pausing (Ring et al. 1996). Tethered σ70, in contrast, stimulates pausing because it remains attached to RNAP in a functional state after completion of the transition to an EC.
Figure 7.
The effective concentration of tethered σ70 is sufficient to cause promoter-distal, σ70-dependent pausing. The effective concentration of tethered σ70 is sufficient to cause promoter-distal, σ70-dependent pausing. Synchronous in vitro transcription with β′::σ70 or wild-type RNAPs (Materials and Methods; time of transcript elongation as indicated in min; C, incubation with additional 200 μM NTP for 8 min after the last indicated time point). (A) +16/17 pause template (Ring et al. 1996). The top panel shows the run-off (RO) product; the lower panel shows the pause. The schematic of the template indicates the σ70-binding sequence and the position of the pause. (B) +37 pause template (+20 in Ring et al. 1996). Gel panels and schematic are as in A. The three pause positions on this template have not been mapped precisely and therefore are designated 37a, 37b, and 37c; the third pause band (relative to the +16/17 site) is probably caused by the stronger consensus σ70-binding sequence. The time dependence of pause RNA levels (plot) is the average of three independent experiments. (C) +462 pause template. Gel panels and schematic are as in A. σ (1 μM) or NusA (10 μM) were added as indicated.
To ask if the tethered σ70 could act at even greater distances from a promoter, we tested the +462 pause template (TGcTATAAT σ70-binding sequence; Fig. 7C). As expected, wild-type RNAP was unable to recognize the +462 pause; however, β′::σ70 RNAP exhibited strong pausing. If pausing at +462 occurred simply because the tethered σ70 was present at high local concentration, then elevated concentrations of untethered σ70 should cause wild-type RNAP to pause. Consistent with this interpretation, wild-type RNAP recognized the +462 pause when additional σ70 (1 μM) was added to ECs (Fig. 7C). We concluded that this pause depended on the consensus extended -10-like sequence because a mutant TGcTgTAAg site nearly eliminated pausing (data not shown). This effect of nonconsensus substitutions may explain why Marr et al. (2001) found that additional σ70 (0.8 μM) did not direct pausing at the nonconsensus +16/17 pause. Pause escape by β′::σ70 RNAP was barely detectable at consensus extended -10-like sequences (e.g., plot in Fig. 7B). However, β′::σ70 RNAP was paused, not terminated, because addition of GreA or GreB protein reduced pausing dramatically (data not shown), as shown for the +16/17 pause by Marr and Roberts (2000).
Because NusA is thought to displace σ70 from the EC (Gill et al. 1991), we next asked how NusA affected recognition of the +462 pause. Addition of NusA to 10 μM slowed elongation overall, generated a new pause at 419 nt, and reduced, but did not eliminate, σ70-dependent pausing caused by untethered σ70 (Fig. 7C, wt RNAP + σ + NusA). However, NusA had little effect on +462 pausing by β′::σ70 RNAP (Fig. 7C) or by wild-type RNAP when untethered σ70 was added to 10 μM (1:1 stoichiometry with NusA; data not shown). These results establish that tethered σ70, and even untethered σ70 if added at higher concentration (but still well below that predicted to occur in vivo), can direct σ70-dependent pausing irrespective of pause site location in a transcriptional unit. The findings are consistent with the paradigm that NusA competes with σ70 for interaction with an EC (Gill et al. 1991), but suggest that NusA may not eliminate σ70-dependent pausing in vivo (see Discussion).
Discussion
Our study of tethering σ70 to RNAP leads to four main conclusions. First, a local σ70 concentration of ∼55 μM (the measured value for β′::σ70 RNAP) does not significantly perturb the physiology of E. coli, consistent with the effective concentration of untethered σ70 in wild-type cells being only slightly lower. Second, tethered σ70 at this modestly increased local concentration only slightly delays heat-shock gene expression and is still inactivated upon heat shock (like untethered σ70), even though σ32 levels are not elevated. Third, tethered σ70 or even 1 μM untethered σ70 can stimulate pausing in vitro anywhere in a transcriptional unit. Fourth, the effective concentration of free σ70 present in cells appears sufficient to allow it to interact transiently with an EC even after σ70 release, and to stimulate pausing at extended -10-like sequences in vivo.
Effective concentration of σ70 in vivo
An accurate understanding of σ-factor binding by RNAP requires knowing the effective concentrations of σs and RNAP in vivo. However, many factors can cause the effective concentrations of molecules in cells to differ from their bulk concentrations. First, the cytoplasm of E. coli is a highly concentrated mixture of macromolecules, small molecules, and water more akin to a gel than to a dilute solution. Macromolecular crowding will increase the effective concentrations of σ70 and RNAP significantly, quite likely by a factor of 10 or more (Record et al. 1998; Ellis 2001). The magnitude of crowding effects may depend on growth conditions, as the water content of cells changes significantly in response to osmolytes in the growth medium (Cayley et al. 1991).
Second, the effective concentrations of σ70 and RNAP will be reduced by interactions that sequester them from participation in the competitive binding equilibria among σ factors and RNAP. ECs sequester about two-thirds of RNAP (Ishihama 2000). Additional RNAP binds nonspecifically to DNA and possibly to RNA; these contributions depend on the binding constants, target sizes, and amounts of DNA or RNA available for nonspecific interaction. Most σs in E. coli also specifically bind anti-σs (FlgM for σF, RseA for σE, DnaK for σ32, and possibly Rsd for σ70; Ishihama 2000 and references therein). 6S RNA specifically sequesters holoenzyme (Wassarman and Storz 2000). Some free σ or RNAP-bound σ could be sequestered nonspecifically. Promoter complexes also will sequester some σs, but it is difficult to estimate how many because some σ may release slowly after initiation (Shimamoto et al. 1986; Bar-Nahum and Nudler 2001; Mukhopadhyay et al. 2001) and because some OCs appear unable to initiate transcription (Susa et al. 2002). The extent to which these various interactions reduce σ70 and RNAP availability depends on their avidity, but together they likely reduce substantially the effective concentrations of σ70 and RNAP molecules.
Finally, the association of σs with RNAP is unlikely to be in equilibrium in vivo. RNAP constantly enters the pool available for σ binding both by release from DNA at terminators and by new synthesis. σs bind RNAP tightly, which means that the off-rates may be slower than the time it takes RNAP to bind and initiate at a promoter.
Given this complexity, the prospects for calculating the effective concentration of σ70 in vivo are poor. However, the effects of tethering σ70 to RNAP at known local concentration provides some insight. Both enhanced competition against σ70RC584 (Fig. 5) and the slight delay in the heat-shock response (Fig. 6) of tethered σ70 suggest that the normal effective concentration of untethered, wild-type σ70 is modestly less than the local concentration of tethered σ70 (55 μM). Comparison to the effects of overproducing σ70 suggest it is ≥11 μM, close to σ70's bulk concentration (∼15 μM). This is reminiscent of a similar conclusion reached for a cellular protein ordinarily present at very different bulk concentration, lac repressor (Law et al. 1993). For lac repressor, the balance of crowding and sequestration effects also yields an effective concentration near its bulk concentration (∼1 nM in wild-type cells).
Competition of σ70 and σ32
Despite the modest increase in the effective concentration of σ70 caused by tethering, σ32 levels are not elevated in the β′::σ70 strain as expected for increased σ70 concentration (Zhou and Gross 1992), and the tethered σ70 is still inactivated upon heat shock. We cannot absolutely exclude the possibility that a small fraction of proteolytically cleaved β′::σ70 RNAP allows the near normal σ32 function. However, we favor σ32 function with intact β′::σ70 RNAP for three reasons. First, we did not detect cleavage of β′::σ70 RNAP after heat shock (data not shown). Second, even if some cleavage occurred, σ32 must compete in these strains against additional, chromosomally encoded σ70 that also could bind any available core RNAP. Third, σ32 functions with uncleaved β′::σ70 RNAP in vitro and this cannot be explained by proteolytic fragmentation of β′::σ70. Rather, we suggest the explanation for the lack of σ32 overexpression when σ70 is tethered to RNAP lies in the difference between increasing σ70 concentration locally versus globally. Overexpression of σ70 produces inclusion bodies and becomes toxic to E. coli at high levels of σ70 (data not shown; see Fig. 5). Aggregates of σ70 (the precursor to inclusion bodies) may be bound to the chaperone DnaK and thus could induce E. coli's stress response and elevate the σ32 level by releasing σ32 from DnaK. Increasing σ70 concentration locally for RNAP, as occurs in the β′::σ70 strain, would not provoke this stress response because it would not elevate the global σ70 level. In essence, competition of σ70 and σ32 may occur for both RNAP and DnaK upon global σ70 overexpression, but should be limited to RNAP for σ70 tethering.
The inactivation of tethered σ70 upon heat shock suggests something other than simple competition for σ32 shuts off σ70-dependent gene expression. Indeed, even in wild-type cells, something in addition to an increase in σ32 level seems necessary to explain the decrease in σ70-directed gene expression upon heat shock (Fig. 6A; Straus et al. 1987). Although σ32 levels are elevated fivefold 15 min after shift of E. coli to 42°C, σ70 levels themselves increase 2.5-fold (Taylor et al. 1984; Straus et al. 1987). This is equivalent to ∼60 σ32 and ∼900 σ70 per 1000 RNAP, suggesting that an unknown factor actively inhibits σ70 during heat shock. Whatever the mechanism of σ70 inhibition, it must also act on tethered σ70 and allow σ32 to dominate during heat shock despite its lower level. Such a requirement for additional factors has been a general conclusion in most studies of σ-factor switching (Fujita and Sadaie 1998; Kolesky et al. 1999; Lord et al. 1999; Maeda et al. 2000; Jishage et al. 2002; Rollenhagen et al. 2003). The fact that this mechanism operates on tethered σ70 rules out explanations involving physical segregation of σ70 away from sites of RNAP function because σ70 is fixed to RNAP by covalent linkage.
σ70 may function as an elongation factor in vivo
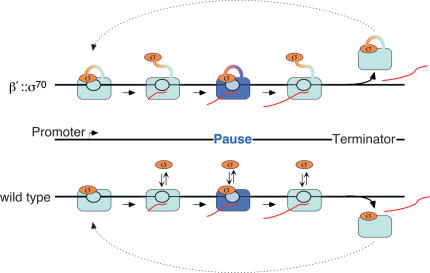
The textbook view of σ70 participation in the transcription cycle may need revision, although our results suggest that when or whether σ is released is not the relevant question. Even 1 μM σ70 can interact from solution with ECs in vitro and cause pausing anywhere in a transcriptional unit; this behavior is similar to that of σ70 permanently tethered to RNAP. Because σ70's effective concentration in cells is significantly higher than 1 μM, continuous transient interactions must occur with ECs regardless of when or whether it is released after promoter escape (Fig. 8).
Figure 8.
Model of σ70-RNAP interactions during transcription in E. coli.
Although NusA competes with σ70 for binding to the EC (Gill et al. 1991) and may temper the effects of σ70-stimulated pausing, it is unlikely to eliminate this pausing in vivo. In vitro, a 10-fold excess of NusA reduced, but did not eliminate, σ70-stimulated pausing (Fig. 7C). Although the in vivo concentration of NusA has not been reported, a 10-fold excess over σ70 would correspond to >110 μM or more than 2.6% of total cellular protein. Thus, it is unlikely there is enough NusA in cells to eliminate σ70-dependent pausing at extended -10-like sequences.
The view that emerges from this study is that the effective concentration of σ70 in vivo is sufficient to allow interaction with the EC and stimulation of pausing at extended -10-like sequences, making σ70 a bona fide elongation factor. Arndt and Chamberlin (1988) discovered that even 300 nM σ70 stimulates release of RNAP from terminators up to 20-fold during multiround transcription in vitro. They concluded that σ70 loads directly onto RNAP during the process of termination. Analysis of the time required for each round of RNA synthesis suggested that the rate-limiting step for recycling of RNAP for new rounds of transcription is an isomerization in this RNAP-σ70 complex (previously reported by Wu et al. 1976), rather than RNAP-σ70 reassociation. Given these observations and our results, we suggest that σ70 could be released from ECs upon promoter escape, but remain able to interact transiently with the EC during transcript elongation. If RNAP encounters an extended -10-like sequence (several of which are present within E. coli's transcriptional units), transiently interacting σ70 could rebind to stimulate pausing. When RNAP reaches a terminator, σ70 could rebind tightly, making the overall behavior of wild-type σ70 similar to the behavior of σ70 tethered to RNAP without requiring continuous physical retention of σ70 by the EC.
Materials and methods
Strains, strain construction, plasmids, protein purification, plating efficiencies, and strain phenotypes are described in the Supplemental Material and Supplementary Table 1.
Templates for in vitro transcription and mobility shift assays
Templates were generated by PCR amplification (Supplementary Table 1) and purified by phenol extraction and spermine precipitation (Hoopes and McClure 1981). The template for mobility-shift assays (Fig. 4C) was isolated from a 1.5% GTG low-melting agarose gel, melted at 65°C in 5 vol of TE, extracted twice with phenol, extracted with CHCl3, and EtOH-precipitated.
Cell extracts
Cellular extracts were prepared from early exponential-phase cells by centrifugation for 2 min at 10,000_g_, suspension in SDS buffer (4% SDS, 10% glycerol, 62.5 mM Tris at pH 6.8, 5% β-mercaptoethanol), and incubation at 100°C for 10 min.
Immunoblot analysis
Immunoblotting and detection were performed using ECL Plus reagents (Pharmacia), following the manufacturer's instructions and using primary antibodies against β′ (NT73; 1:10,000), σ70 (2G10; 1:5000), and σ32 (3RH1; 1:3000), which was kindly provided by R. Burgess (University of Wisconsin, Madison, WI, USA), and secondary antibody (horseradish peroxidase-coupled mouse IgG) from Pharmacia. Proteolytic cleavage of β′::σ70 to yield wild-type-length β′ or σ70 was undetectable in β′::σ70 strains, with detection limits of 0.5 ng of β′ and 1.2 ng of σ70 per microgram of cell lysate. Levels of σ32 in wild-type and β′::σ70 strains (Fig. 6B) were obtained from multiple independent cell lysates and blots (average ratio of σ32 in the β′::σ70 strain relative to wild type of 0.8 ± 0.4). The estimated error in protein concentration loaded on gels (determined by Bradford assay) was 20%.
Promoter kinetics
To determine _k_f,obs, 50 nM RNAP (β′::σ70 or wild-type) was incubated with 25 nM T7 A1 template at 22°C in transcription buffer (Artsimovitch and Landick 2002). Samples were removed from 10 sec to 8 min, mixed with 0.1 vol of 2.5 mM ApU; 25 μM each ATP, [32P]CTP, and GTP; and 1 mg/mL heparin; incubated at 37°C for 10 min; and then mixed with an equal volume of 2× STOP buffer (8 M urea, 98 mM Tris-borate at pH 8.3, 10 mM Na2 EDTA, 0.2% bromophenol blue, 0.2% xylene cyanol). The RNA products were separated by 15% denaturing PAGE and quantified using a PhosphorImager. The results were fit to a single-exponential of formation by nonlinear regression using Kaleidagraph. At the concentrations of RNAP and DNA used, this assay would detect a decrease in the equilibrium binding of RNAP and DNA to form CC, but likely not an increase. To determine _k_r,obs, OCs were first formed for 10 min as described for _k_f,obs determination. At time 0, heparin was added to 100 μg/mL, and sampled from 15 sec to 100 min. Samples were mixed with 0.1 vol of 2.5 mM ApU and 25 μM each ATP, [32P]CTP, and GTP; incubated at 37°C for 10 min; and then mixed with an equal volume of 2× STOP buffer. The RNA products were quantified as described for _k_f,obs determination, and the results were fit to a single-exponential decay of OC by nonlinear regression using Kaleidagraph.
Equilibrium binding assay
HMKσ70 (250 pmole) was 32P-labeled as described previously (Artsimovitch and Landick 2002). β′::σ70 or wild-type RNAPs (1 μM) were incubated with [32P]σ70 at 0.5, 1, 2.5, and 5 μM in equilibrating conditions using E buffer (0.5 M NaCl, 40 mM Tris-HCl at pH 8.0, 0.1 mM EDTA, 3 mM DTT, 0.2% Tween) plus 20% glycerol at 30°C for 20 min (Severinova et al. 1996; Sharp et al. 1999). Samples were then separated on a 4%-15% native Phast gel (Pharmacia) for 125 Vh at 15°C. Gels were Coomassie blue R-stained, analyzed using a PhosphorImager (Fig. 4A), and quantified (Fig. 4B). Effective σ70 concentration was obtained by nonlinear regression of fraction RNAP containing [32P]σ70 versus concentration of [32P] σ70 using the equation y = x/(x + E), where x is the concentration of [32P]σ70 and E is the effective concentration of tethered σ70.
Open complex mobility assay
β′::σ70 or wild-type RNAPs (100 nM) were mixed with increasing amounts of σ70 (amount added σ70 in micromolar indicated above gel panel) under equilibrium conditions (E buffer + 5% glycerol) at 37°C for 10 min. Linear DNA containing PUPFullcon (Supplementary Table 1) was added to 150 nM as the reaction was diluted to 0.25 M NaCl and incubated at 37°C for 10 min. OCs formed on this promoter are exceptionally stable (t1/2 > 18 h; T. Gaal, pers. comm.). Reactions were electrophoresed on 4% NuSieve (FMC), 0.5× TBE, and stained with EtBr. Effective σ70 concentration (Fig. 4D) was modeled using the equation y = x/(x + E), where x is the concentration of untethered σ70 and E is the effective concentration of tethered σ70 (set arbitrarily to 0.1, 1, 10, 50, and 100 μM).
Heat-shock response assay
Wild-type (RL301) or β′::σ70 strain (RL1374) cells were grown in supplemented glucose medium at 30°C and labeled with [35S]methionine (10 mCi/mL, Amersham; 0.2 mCi/mL final concentration) for 2 min at various times after shift to 42°C as described (Zhou et al. 1988). The cells were recovered by centrifugation, washed twice, suspended in SDS buffer, boiled for 10 min, and separated by 3%-8% Tris-Acetate (Novex) beside molecular weight markers. The gel was dried down and visualized using a PhosphorImager.
In vitro σ70-dependent pause assay
Transcription on the +16/17 and +37 pause templates was performed as described (Ring et al. 1996). For the +462 pause template, A29 ECs were formed at 37°C by mixing 7.5 nM RNAP, 250 nM template, 150 μM ApU, and 10 μM ATP, GTP, and 32P-CTP. The high excess of template was used to ensure only one RNAP binding per template. σ70 (1 μM), NusA (10 μM), or both were added to ECs as indicated. All 4 NTPs (150 μM) and 100 μg Rif/mL were then added to allow elongation to resume. Samples were removed at the times indicated, mixed with equal volume 2× STOP buffer, separated by denaturing PAGE (5%; 0.5× TBE), and analyzed using a PhosphorImager. The size of the σ70-dependent pause band (462 nt, predicted) was measured by comparison to size markers generated by transcription of restriction endonuclease-treated +462 pause templates and found to be 465 ± 0.5 nt (data not shown).
In vivo concentrations
The in vivo concentrations of σ70 and RNAP are central to understanding the effects of tethering σ70. Based on quantitative immunoblotting, Jishage and Ishihama (1995) report a constant in vivo concentration of σ70 of 50-80 fmole/μg cellular protein regardless of growth phase in rich medium at 37°C. At a growth rate of 40 min/generation in LB medium (Fig. 1), an E. coli cell contains ∼7 × 10-13 mL of water (composing 70% of the cell mass) and ∼2.3 × 10-13 g of protein (Cayley et al. 1991; Bremer and Dennis 1996). Assuming a partial specific volume for protein of 0.72 (Cantor and Schimmel 1980), this gives a total cell volume of ∼1 × 10-12 mL. Based on these numbers, we calculate a bulk protein concentration of 230 mg/mL [(2.3 × 10-13 g of protein)/(1 × 10-12 mL)] and a bulk concentration of σ70 of 12-19 μM (12-19 nmole/mL). The molar ratios of total σs to σ70 and of RNAP to σ70 are reported to be 1.7 and 2.8 in the same conditions, with 65% of RNAP stably bound to DNA (e.g., as ECs; Ishihama 2000). Thus, the bulk concentration of free RNAP (available to bind σ) is also 12-19 μM and of total σs, 19-31 μM. Based on this, we derive the average bulk concentrations of free RNAP, σ70, and total σs of ∼15 μM, ∼15 μM, and ∼25 μM, respectively.
Acknowledgments
We thank R. Saecker and T. Record for suggesting the experiment shown in Figure 4C and for many helpful discussions; and C. Gross, J. Roberts, W. Ross, R. Gourse, I. Artsimovitch, and members of the Gourse and Landick labs for sharing ideas and encouragement during the course of this work. We also thank C. Vrentas for assisting with some experiments not shown; R. Gourse, R. Saecker, W. Ross, H. Murray, M. Palangat, and S. Kyzer for insightful comments on the manuscript; and T. Gaal, C. Chan, M. Sharp, D. Friedman, C. Gross, T. Arthur, R. Burgess, M. Marr, and J. Roberts for gifts of plasmids, strains, phage, and antibodies. This work was supported by NIH grants GM38660 (to R.L.) and, for support of R.A.M, T32 GM008349.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1142203.
References
- Arndt K.M. and Chamberlin, M.J. 1988. Transcription termination in Escherichia coli. Measurement of the rate of enzyme release from Rho-independent terminators. J. Mol. Biol. 202**:** 271-285. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I. and Landick, R. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109**:** 193-203. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G. and Nudler, E. 2001. Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli. Cell 106**:** 443-451. [DOI] [PubMed] [Google Scholar]
- Bentley S.D., Chater, K.F., Cerdeno-Tarraga, A.M., Challis, G.L., Thomson, N.R., James, K.D., Harris, D.E., Quail, M.A., Kieser, H., Harper, D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417**:** 141-147. [DOI] [PubMed] [Google Scholar]
- Bremer H. and Dennis, P. 1996. Modulation of cell parameters by growth rate. In Escherichia coli and Salmonella: Cellular and molecular biology, 2nd ed. (eds. F. Neidhardt et al.), pp. 1553-1569. ASM press, Washington, DC.
- Burgess R.R., Travers, A.A., Dunn, J.J., and Bautz, E.K. 1969. Factor stimulating transcription by RNA polymerase. Nature 221**:** 43-46. [DOI] [PubMed] [Google Scholar]
- Cantor C.R. and Schimmel, P.R. 1980. Biophysical chemistry. Freeman, New York.
- Cayley S., Lewis, B.A., Guttman, H.J., and Record Jr., M.T. 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222**:** 281-300. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. 1976. RNA polymerase—An overview. In RNA polymerase (eds. R. Losick and M. Chamberlin), pp. 17-68. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Chan C. and Landick, R. 1989. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. J. Biol. Chem. 264**:** 20796-20804. [PubMed] [Google Scholar]
- Daube S.S. and von Hippel, P.H. 1999. Interactions of Escherichia coli σ70 within the transcription elongation complex. Proc. Natl. Acad. Sci. 96**:** 8390-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Penas A., Connolly, L., and Gross, C.A. 1997. σE is an essential σ factor in Escherichia coli. J. Bacteriol. 179**:** 6862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J. 2001. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 26**:** 597-604. [DOI] [PubMed] [Google Scholar]
- Farewell A., Kvint, K., and Nystrom, T. 1998. Negative regulation by RpoS: A case of σ factor competition. Mol. Microbiol. 29**:** 1039-1051. [DOI] [PubMed] [Google Scholar]
- Flory P. 1969. Statistical mechanics of chain molecules. Interscience, London.
- Friedman D.I., Baumann, M., and Baron, L.S. 1976. Cooperative effects of bacterial mutations affecting λ N gene expression. I. Isolation and characterization of a nusB mutant. Virology 73**:** 119-127. [DOI] [PubMed] [Google Scholar]
- Fujita M. and Sadaie, Y. 1998. Promoter selectivity of the Bacillus subtilis RNA polymerase σA and σH holoenzymes. J. Biochem. (Tokyo) 124**:** 89-97. [DOI] [PubMed] [Google Scholar]
- Gill S.C., Weitzel, S.E., and von Hippel, P.H. 1991. Escherichia coli σ 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J. Mol. Biol. 220**:** 307-324. [DOI] [PubMed] [Google Scholar]
- Gross C.A., Chan, C., Dombroski, A., Gruber, T., Sharp, M., Tupy, J., and Young, B. 1998. The functional and regulatory roles of σ factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 63**:** 141-155. [DOI] [PubMed] [Google Scholar]
- Hicks K.A. and Grossman, A.D. 1996. Altering the level and regulation of the major σ subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol. Microbiol. 20**:** 201-212. [DOI] [PubMed] [Google Scholar]
- Hoopes B.C. and McClure, W.R. 1981. Studies on the selectivity of DNA precipitation by spermine. Nucleic Acids Res. 9**:** 5493-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54**:** 499-518. [DOI] [PubMed] [Google Scholar]
- Jishage M. and Ishihama, A. 1995. Regulation of RNA polymerase σ subunit synthesis in Escherichia coli: Intracellular levels of σ 70 and σ 38. J. Bacteriol. 177**:** 6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M., Kvint, K., Shingler, V., and Nystrom, T. 2002. Regulation of σ factor competition by the alarmone ppGpp. Genes & Dev. 16**:** 1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Sugaya, K., and Cook, P.R. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159**:** 777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesky S., Ouhammouch, M., Brody, E.N., and Geiduschek, E.P. 1999. σ competition: The contest between bacteriophage T4 middle and late transcription. J. Mol. Biol. 291**:** 267-281. [DOI] [PubMed] [Google Scholar]
- Krummel B. and Chamberlin, M.J. 1989. RNA chain initiation by Escherichia coli RNA polymerase. Structural transition of the enzyme in early ternary complexes. Biochemistry 28**:** 7829-7842. [DOI] [PubMed] [Google Scholar]
- Kumar A., Grimes, B., Logan, M., Wedgwood, S., Williamson, H., and Hayward, R.S. 1995. A hybrid σ subunit directs RNA polymerase to a hybrid promoter in Escherichia coli. J. Mol. Biol. 246**:** 563-571. [DOI] [PubMed] [Google Scholar]
- Kunst F., Ogasawara, N., Moszer, I., Albertini, A.M., Alloni, G., Azevedo, V., Bertero, M.G., Bessieres, P., Bolotin, A., Borchert, S., et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390**:** 249-256. [DOI] [PubMed] [Google Scholar]
- Law S.M., Bellomy, G.R., Schlax, P.J., and Record Jr., M.T. 1993. In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J. Mol. Biol. 230**:** 161-173. [DOI] [PubMed] [Google Scholar]
- Lonetto M.A., Rhodius, V., Lamberg, K., Kiley, P., Busby, S., and Gross, C. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 284**:** 1353-1365. [DOI] [PubMed] [Google Scholar]
- Lord M., Barilla, D., and Yudkin, M.D. 1999. Replacement of vegetative σA by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J. Bacteriol. 181**:** 2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Fujita, N., and Ishihama, A. 2000. Competition among seven Escherichia coli σ subunits: Relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28**:** 3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr M.T. and Roberts, J.W. 2000. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell 6**:** 1275-1285. [DOI] [PubMed] [Google Scholar]
- Marr M.T., Datwyler, S.A., Meares, C.F., and Roberts, J.W. 2001. Restructuring of an RNA polymerase holoenzyme elongation complex by lambdoid phage Q proteins. Proc. Natl. Acad. Sci. 98**:** 8972-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W.R. 1980. Rate-limiting steps in RNA chain initiation. Proc. Natl. Acad. Sci. 77**:** 5634-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V., Kortkhonjia, E., Mukhopadhyay, J., Knight, J., Revyakin, A., Kapanidis, A.N., Niu, W., Ebright, Y.W., Levy, R., and Ebright, R.H. 2002. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108**:** 599-614. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay J., Kapanidis, A.N., Mekler, V., Kortkhonjia, E., Ebright, Y.W., and Ebright, R.H. 2001. Translocation of σ70 with RNA polymerase during transcription: Fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106**:** 453-463. [DOI] [PubMed] [Google Scholar]
- Murakami K.S., Masuda, S., Campbell, E.A., Muzzin, O., and Darst, S. 2002. Structural basis of transcription initiation: An RNA polymerase holoenzyme/DNA complex. Science 296**:** 1285-1290. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Hannett, N.M., and Young, R.A. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9**:** 799-809. [DOI] [PubMed] [Google Scholar]
- Raag R. and Whitlow, M. 1995. Single-chain Fvs. FASEB J. 9**:** 73-80. [DOI] [PubMed] [Google Scholar]
- Record M.T., Courtenay, E.S., Cayley, S., and Guttman, H.J. 1998. Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem. Sci. 23**:** 190-194. [DOI] [PubMed] [Google Scholar]
- Ring B., Yarnell, W., and Roberts, J. 1996. Function of E. coli RNA polymerase σ factor σ70 in promoter-proximal pausing. Cell 86**:** 485-493. [DOI] [PubMed] [Google Scholar]
- Robinson C.R. and Sauer, R.T. 1998. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl. Acad. Sci. 95**:** 5929-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenhagen C., Antelmann, H., Kirstein, J., Delumeau, O., Hecker, M., and Yudkin, M.D. 2003. Binding of σ(A) and σ(B) to core RNA polymerase after environmental stress in Bacillus subtilis. J. Bacteriol. 185**:** 35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker R., Tsodikov, O., McQuade, K., Schlax Jr., P., Capp, M., and Record Jr., M. 2002. Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λPR promoter: Large scale conformational changes in forming kinetically significant intermediates. J. Mol. Biol. 319**:** 649-671. [DOI] [PubMed] [Google Scholar]
- Severinov K., Mooney, R., Darst, S., and Landick, R. 1997. Tethering of the large subunits of Escherichia coli RNA polymerase. J. Biol. Chem. 272**:** 24137-24140. [DOI] [PubMed] [Google Scholar]
- Severinova E., Severinov, K., Fenyo, D., Marr, M., Brody, E.N., Roberts, J.W., Chait, B.T., and Darst, S.A. 1996. Domain organization of the Escherichia coli RNA polymerase σ 70 subunit. J. Mol. Biol. 263**:** 637-647. [DOI] [PubMed] [Google Scholar]
- Sharp M.M., Chan, C.L., Lu, C.Z., Marr, M.T., Nechaev, S., Merritt, E.W., Severinov, K., Roberts, J.W., and Gross, C.A. 1999. The interface of σ with core RNA polymerase is extensive, conserved, and functionally specialized. Genes & Dev. 13**:** 3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto N., Kamigochi, T., and Utiyama, H. 1986. Release of the σ subunit of Escherichia coli DNA-dependent RNA polymerase depends mainly on time elapsed after the start of initiation, not on length of product RNA. J. Biol. Chem. 261**:** 11859-11865. [PubMed] [Google Scholar]
- Siegele D.A., Hu, J.C., Walter, W.A., and Gross, C.A. 1989. Altered promoter recognition by mutant forms of the σ 70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 206**:** 591-603.2661828 [Google Scholar]
- Straus D.B., Walter, W.A., and Gross, C.A. 1987. The heat shock response of E. coli is regulated by changes in the concentration of σ 32. Nature 329**:** 348-351. [DOI] [PubMed] [Google Scholar]
- Susa M., Sen, R., and Shimamoto, N. 2002. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 277**:** 15407-15412. [DOI] [PubMed] [Google Scholar]
- Taylor W.E., Straus, D.B., Grossman, A.D., Burton, Z.F., Gross, C.A., and Burgess, R.R. 1984. Transcription from a heat-inducible promoter causes heat shock regulation of the σ subunit of E. coli RNA polymerase. Cell 38**:** 371-381. [DOI] [PubMed] [Google Scholar]
- Timpe L.C. and Peller, L. 1995. A random flight chain model for the tether of the Shaker K+ channel inactivation domain. Biophys. J. 69**:** 2415-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D.G., Sekine, S., Laptenko, O., Lee, J., Vassylyeva, M.N., Borukhov, S., and Yokoyama, S. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417**:** 712-719. [DOI] [PubMed] [Google Scholar]
- Wassarman K.M. and Storz, G. 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101**:** 613-623. [DOI] [PubMed] [Google Scholar]
- Wu F.Y., Yarbrough, L.R., and Wu, C.W. 1976. Conformational transition of Escherichia coli RNA polymerase induced by the interaction of σ subunit with core enzyme. Biochemistry 15**:** 3254-3258. [DOI] [PubMed] [Google Scholar]
- Zhou Y.N. and Gross, C.A. 1992. How a mutation in the gene encoding σ 70 suppresses the defective heat shock response caused by a mutation in the gene encoding σ 32. J. Bacteriol. 174**:** 7128-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.N., Kusukawa, N., Erickson, J.W., Gross, C.A., and Yura, T. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor σ 32. J. Bacteriol. 170**:** 3640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.N., Walter, W.A., and Gross, C.A. 1992. A mutant σ 32 with a small deletion in conserved region 3 of σ has reduced affinity for core RNA polymerase. J. Bacteriol. 174**:** 5005-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]